Abstract
The olfactory receptor (OR) genes represent the largest multigene family in the genome of terrestrial vertebrates. Here, the high-throughput next-generation sequencing (NGS) approach was applied to characterization of OR gene repertoires in the green anole lizard Anolis carolinensis and the Japanese four-lined ratsnake Elaphe quadrivirgata. Tagged polymerase chain reaction (PCR) products amplified from either genomic DNA or cDNA of the two species were used for parallel pyrosequencing, assembling, and screening for errors in PCR and pyrosequencing. Starting from the lizard genomic DNA, we accurately identified 56 of 136 OR genes that were identified from its draft genome sequence. These recovered genes were broadly distributed in the phylogenetic tree of vertebrate OR genes without severe biases toward particular OR families. Ninety-six OR genes were identified from the ratsnake genomic DNA, implying that the snake has more OR gene loci than the anole lizard in response to an increased need for the acuity of olfaction. This view is supported by the estimated number of OR genes in the Burmese python's draft genome (∼280), although squamates may generally have fewer OR genes than terrestrial mammals and amphibians. The OR gene repertoire of the python seems unique in that many class I OR genes are retained. The NGS approach also allowed us to identify candidates of highly expressed and silent OR gene copies in the lizard's olfactory epithelium. The approach will facilitate efficient and parallel characterization of considerable unbiased proportions of multigene family members and their transcripts from nonmodel organisms.
Keywords: next-generation sequencing, olfactory receptor, Squamata, molecular evolution, pseudogene
Introduction
Natural environments are filled with various odors. These odors are rich in information, and thus, most animals have evolved an acute sense of smell to detect and interpret them. In vertebrates, odor chemicals are mainly detected by olfactory receptors (ORs) that are expressed in the olfactory sensory neurons (reviewed in Mombaerts 2004). To discriminate vast numbers of odor chemicals, the number of vertebrate ORs is highly increased hundreds or thousands of intact OR genes being found in one species (reviewed in Nei et al. 2008). OR genes thus represent the largest multigene family in the genome of terrestrial vertebrates. Discrimination of odor chemicals is based on the “combinatorial coding” manner, in which most odorants are identified not by the activation of a single OR but by the activation pattern of multiple ORs (Su et al. 2009).
Previous studies have suggested that acuity of olfaction in vertebrates is reflected by the copy number of functional OR genes and/or the percentage of pseudogenes within a species (Gilad et al. 2004; Kishida et al. 2007; Steiger et al. 2008, 2009; Hayden et al. 2010). Therefore, comparative study of OR diversity among ecologically divergent species may provide significant insights into adaptive evolution of odor perception. However, identifying individual members of the OR gene repertoire in a species by subcloning and Sanger sequencing strategy is very difficult because of the large number and sequence diversity of the OR genes. At present, the best and the only way to obtain nearly complete OR gene repertoire is in silico screening of the whole-genome sequence, but genomic databases are available only for a mere handful of model organisms.
In vertebrate groups in which genomic data have been published for multiple species (i.e., mammals, birds, and teleost fishes), copy numbers of the OR genes are highly variable between species (Alioto and Ngai 2005; Niimura and Nei 2007; Steiger et al. 2008). In April 2009 when we started the present study, reptilian draft genome sequences were available only for the green anole lizard Anolis carolinensis, and thus, variation of the OR copy number among reptilian taxa was not known. The number of OR genes estimated from the anole lizard draft genome was smaller than those identified for other vertebrate groups (Niimura 2009; Steiger et al. 2009; Kishida and Hikida 2010). However, the lower number of OR genes in the anole lizard may not be representative of reptiles because many reptilian species possess highly developed sense of smell (Pianka and Vitt 2003; Vitt et al. 2003). To understand the evolution of olfactory ability in reptiles, it seems crucial to investigate the OR gene repertoire for organisms other than A. carolinensis, although studies of reptilian OR genes are very limited (e.g., Kishida et al. 2007; Kishida and Hikida 2010).
High-throughput next-generation sequencing (NGS) is rapidly changing methodologies of molecular genetics studies (Mardis 2007). Recent development of Roche GS FLX Titanium DNA sequencing technology enables one to sequence numerous DNA fragments of more than 400 bp in average size (∼1 kbp with the latest specification in February 2012) without the vector-based cloning that tends to introduce a bias in cloned sequences (http://454.com/products-solutions/454-sequencing-system-portfolio.asp). Furthermore, this method can potentially discriminate polymerase chain reaction (PCR) errors from true sequences by sequencing the same DNA regions multiple times. These advantages can make the FLX-based NGS approach suitable for characterizing large multigene families, such as the vertebrate OR gene family. Indeed, this approach has been shown to be effective in characterizing the polymorphic multilocus MHC system (Babik et al. 2009). The OR genes, however, form a more complicated multigene family than the MHC genes, and the accuracy and efficiency of the NGS approach for investigating the vertebrate OR genes need to be evaluated thoroughly.
In the present study, we first attempted to assess the usefulness of the NGS approach for experimental identification of OR genes in the anole lizard, which can be evaluated based on the in silico identified OR gene repertoire from its draft genome sequence. We show that this approach can provide a reliable view of the lizard's OR gene repertoire by recovering a considerable proportion of OR genes encoded in its genome. We then applied this approach to characterization of the OR gene repertoire in the Japanese four-lined ratsnake Elaphe quadrivirgata. The ratsnake and the Burmese python being the second reptilian taxa with a new draft genome sequence (Castoe et al. 2011) are known to have developed a life style that is highly dependent on the olfaction, whereas Anolis lizards are believed to rely on the visual sense for the prey capture and the escape from predators (Pianka and Vitt 2003). Thus, comparison of the OR gene repertoire between the snakes and the anole lizard may provide insights into molecular evolution of the olfactory genes in squamate reptiles.
Materials and Methods
Identification of OR Genes from the Anole Lizard and the Python Genome Assembly
We examined the draft genomic sequences of the green anole lizard (AnoCar2.0, May 2010; http://www.ensembl.org/Anolis_carolinensis/Info/Index/; Alföldi et al. 2011) and the Burmese python (GenBank ID: AEQU000000000; Castoe et al. 2011) to identify the nearly complete OR gene repertoire in each species. OR sequences were identified by a method that was used to find fish vomeronasal-type ORs (Hashiguchi and Nishida 2006) with slight modifications. First, a TBlastN search was conducted with the cutoff E value of 10−10 against the genomic data using several representative vertebrate OR amino acid sequences as queries. Obtained sequences were verified as ORs by BlastP searches against NCBI nonredundant (nr) database. Next, each region of Blast similarity was extended to 1 kb in 5′ and 3′ directions to predict OR-coding sequences. For each of these genomic regions, intronless OR-coding sequences were estimated by the profile hidden Markov model (profile HMM)–based gene prediction with the program WISE2 (Birney et al. 2004). A profile HMM was constructed from the alignment of known OR sequences from human, frog, and fish using the HMMER software package (http://hmmer.janelia.org). Positions of initiation and stop codons of the obtained OR-coding sequences were identified manually.
The anole lizard putative OR sequences were classified into two groups, apparently functional genes and nonfunctional pseudogenes. If a sequence contained any disruptive frameshift and/or stop codon, it was considered as a pseudogene. In this study, partial sequences (less than 600 bp) were also classified as pseudogenes, although some partial genes may have resulted from incomplete genome assembly. The python putative OR sequences were classified into three groups, functional genes, pseudogenes, and truncated (partial) genes, because the python genome contained many truncated OR copies found in very short (∼2 kb) contigs. In the python, all nondisrupted OR sequences of less than 800 bp in length were classified as truncated genes. Each OR sequence identified was searched against the HORDE (the Human Olfactory Data Explorer) #42 database (Olender et al. 2004, http://genome.weizmann.ac.il/horde/) using the FASTA search and classified into the same family as the best-hit human OR sequence. Our family classification followed that by Glusman et al. (2000).
Sample Collection and DNA/RNA Extraction
Genomic DNA of the green anole lizard was extracted using a DNeasy Tissue Kit (QIAGEN) from muscle tissues of a young dead individual obtained from a pet shop in 2002. Genomic DNA of the Japanese four-lined ratsnake was similarly extracted from blood samples of a female individual caught in Shiga Prefecture, Japan through the courtesy of Dr Michihisa Toriba. Total RNA of the anole lizard was extracted from a male individual that was captured at Chichi-jima Island, Ogasawara, Japan in 2008 with permission from the Ministry of the Environment. An upper jaw portion containing both nasal and vomeronasal parts of olfactory organs were excised immediately after killing the animal and cut into 5 mm pieces (see supplementary fig. 1, Supplementary Material online that illustrates the excised portion and provides evidence that it covers these organs). In this study, we were unable to excise nasal and vomeronasal parts separately from each other.
Cells were disrupted in Lysing Matrix D tubes for 30 s at the 6.5 m/s power with Fastprep-24 instrument (MP Biomedicals), from which total RNA was extracted using a mirVana miRNA Isolation Kit (Ambion) according to the manufacture's instruction. After the treatment of resultant RNA samples with TURBO DNase free (Ambion) for 1 h at 37 °C to degrade possibly remaining DNA fractions completely (see supplementary fig. 2, Supplementary Material online), reverse transcription reaction was carried out using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and the random hexamers primer, following the manufacturer's protocol. Incubation time was 10 min at 25 °C, 2 h at 37 °C, and finally 5 s at 85 °C for inactivating the reverse transcriptase.
PCR Amplification and the High-Throughput Sequencing
To amplify OR sequences of the anole lizard and the ratsnake, degenerate primers were designed within conserved regions among tetrapod OR genes. Known OR genes from the anole lizard, human, and frog (Xenopus tropicalis) were mainly used for this purpose. Forward and reverse primers were designed in the third transmembrane region and the fourth intracellular region of OR genes, respectively. Expected length of PCR products with these primers was ∼331 bp long. For amplification from the anole lizard genomic DNA, AcORg_F1 was used as a forward primer and AcORg_R1 as a reverse primer (table 1). Each primer started with GGGC followed by a 6-bp tag for identifying species and PCR templates (genomic DNA or cDNA). GGGC tetranucleotide at the 5′-end of primers was used to eliminate the effect of the 5′-terminal nucleotide on the tag efficiency (Binladen et al. 2007; Valentini et al. 2009). To discriminate three PCR reactions with different templates (the anole lizard genomic DNA, the anole lizard cDNA, and the ratsnake genomic DNA) and forward/reverse strands of each reaction, we used six different tag sequences (table 1).
Table 1.
Primers Used in This Study
| Name | Species | Template | Primer Sequence |
| AcORg_F1 | Anolis carolinensis | Genomic DNA | GGGCTCTGAGATGGCATATGAYCGVTAYKTKGC |
| AcORg_R1 | A. carolinensis | Genomic DNA | GGGCTGTCAGGAACAGGTRGARAARGCYTT |
| AcORm_F1 | A. carolinensis | Nose cDNA | GGGCTCGTAGATGGCATATGAYCGVTAYKTKGC |
| AcORm_R1 | A. carolinensis | Nose cDNA | GGGCTGTACGGAACAGGTRGARAARGCYTT |
| EqORg_F1 | Elaphe quadrivirgata | Genomic DNA | GGGCTCGATGATGGCATATGAYCGVTAYKTKGC |
| EqORg_R1 | E. quadrivirgata | Genomic DNA | GGGCTGCTAGGAACAGGTRGARAARGCYTT |
| F2 | A. carolinensis | Genomic DNA | ATGGCATATGAYCGVTAYNTDGC |
| R2 | A. carolinensis | Genomic DNA | GAACAGGTDGARARWGYYTT |
| F3 | A. carolinensis | Genomic DNA | CATATGAYCGVTAYKTDGCYATHTG |
| R3 | A. carolinensis | Genomic DNA | CAGTAARRTGGGARSHRCADGTDGA |
NOTE.—The first six primers were used for the NGS experiments, whereas the other four primers were used for manual sequencing of clones for amplified products. Tag sequences are indicated by underlines. Note that three forward primers ending in F1 share identical sequences after the tag sequences (the F1 primer sequence) and that three reverse primers ending in R1 do so (the R1 primer sequence). F1–F3 primers are forward primers, and R1–R3 primers are reverse ones. F2 and R2 primers are designed in the same location as F1 and R1 primers, respectively, but have slightly different bases in some positions. F3 and R3 primers are designed in different locations, but the amplified F3–R3 region largely overlaps with the F1–R1 (= F2–R2) region.
PCR was performed in a 10 μl reaction mixture using a PrimeSTAR HS DNA polymerase (Takara Bio), 0.5 μM each primer, and template DNA either from diluted genomic DNA or randomly reverse-transcribed cDNA. PCR reaction cycle scheme was 98 °C for 30 s, followed by 28 cycles of 98 °C for 10 s, 50 °C for 15 s, and 72 °C for 30 s. PCR products were electrophoresed in a 1% agarose gel and purified using a MinElute Gel Extraction Kit (QIAGEN). Concentrations of PCR products were measured using NanoVue Plus Spectrophotometer (GE Healthcare). Equal amounts (150 ng) of individual amplicons were then pooled from the three sources (i.e., the lizard genomic DNA, the lizard cDNA, and the ratsnake genomic DNA) and sequenced as a part of single GS FLX Titanium Genome Analyzer (Roche) sequencing run. This was conducted as an outsourcing service by Takara Bio, Inc. Raw reads data obtained by the FLX sequencing have been deposited to the Read Archive at DDBJ with accession numbers DRA000409–DRA000411.
Assembling the NGS Data
First, reads obtained from the FLX sequencing were divided into the three groups on the basis of the sequence tags (see table 1). Second, these reads were assembled into contigs by Sequencher version 4.8 (Gene Codes) with the default setting. Contigs that consist of less than five FLX reads (hereby designated <5× coverage), and all singletons were excluded from the data set because they are more likely affected by PCR errors and chimeras than contigs with denser coverages (see below). Note that the read number in a contig is equivalent to the read coverage per site because most FLX reads span the amplified region. Contigs with less than 1% nucleotide differences were considered as the same sequence, which originated from alleles of the same locus or PCR errors. Under this criterion, almost all OR gene sequences identified in the anole lizard genome can be recognized as separate genes (data not shown). Consensus sequences of each resultant contig were queried by a BlastX search against NCBI nr database in order to verify that they are really OR gene members. If the BlastX best hit was a previously known OR, it was considered a putative OR-coding sequence. Each OR-coding sequence thus identified was queried against the HORDE #42 Database using the FASTA search and classified into the same family as the best-hit human OR sequence. These OR-coding sequences obtained in this study have been deposited to the DDBJ/EMBL/NCBI Nucleotide Sequence Database with accession numbers shown in table 2.
Table 2.
Summary of Obtained OR Sequences by the Next-Generation Sequencing
| Species | Template | FLX Reads (contigs)a | OR-Related Reads (contigs)b | OR-Coding Reads (contigs)c | DDBJ Accession Number | Specimen Voucher Numberd |
| Anolis carolinensis | Genomic DNA | 6196 (195) | 5182 (71) | 5043 (56) | AB646799–AB646854 | SDNCU-A0007 |
| A. carolinensis | Nose cDNA | 6854 (194) | 6329 (70) | 5966 (40) | FX180060–FX180099 | SDNCU-A0008 |
| Elaphe quadrivirgata | Genomic DNA | 6202 (253) | 5831 (140) | 5505 (96) | AB646855–AB646950 | — |
The number of reads (initially assembled contigs in parentheses) that had the corresponding tag sequences.
The number of reads that constituted the OR-related contigs after excluding non-OR sequences and the OR contigs with <5× coverages as well as unifying sequences with <1% sequence divergences (see text).
The number of reads that constituted putatively legitimate OR-coding sequences after excluding chimeras for the lizard sequences and after excluding contigs with <11× coverages for the ratsnake sequences (see text). Database accession numbers for the resultant contig sequences are also given in the next column.
Whole body frozen specimens are deposited to the Specimen Depository of the Graduate School of Natural Sciences, Nagoya City University.
Assessment of OR Sequences Obtained by the NGS Approach
It is generally known that high-throughput NGS methods are affected by higher error rates than the traditional Sanger sequencing method, depending on different sequence contexts (Moore et al. 2006). Under- or overcalls of homopolymer runs are typical errors in pyrosequencing with Roche GS FLX Titanium Genome Sequencer (Margulies et al. 2005; Moore et al. 2006). In addition, generation of sequence chimeras by PCR amplification also cannot be ignored for PCR-based cloning and sequencing of multilocus genes, such as the OR gene family. To assess the validity of putative OR gene sequences obtained by the GS FLX sequencing (designated FLX-based OR sequences), we corresponded each FLX-based OR sequence from the anole lizard to the OR gene sequences identified in its draft genome (designated DB-based OR sequences) using the FASTA search. Possible PCR-mediated recombination errors as well as homopolymer run-associated sequencing errors were picked up manually by checking the pairwise alignments resulting from the FASTA search.
Phylogenetic Analyses
Phylogenetic trees containing the ratsnake OR sequences were constructed using two different data sets. One data set consists of human (Niimura and Nei 2005), chicken (Niimura 2009), the anole lizard, and the ratsnake ORs. The other data set consists of OR sequences from the ratsnake, the python, and 14 reptilian taxa (Kishida and Hikida 2010). Only the FLX-based OR sequences that consist of more than ten reads (i.e., >10 coverage) were used for the ratsnake (see Results). In each data set, deduced amino acid sequences were aligned using MAFFT program (Katoh et al. 2002), and the alignment was finally inspected and corrected by eye. Phylogenetic trees were constructed by the neighbor joining method with matrices of the Poisson-corrected amino acid distances, using MEGA 4 software package (Tamura et al. 2007). The reliability of each nodal relationship was assessed by 1000 bootstrap replications.
Results
Repertoire of OR Genes Identified from the Anole Lizard and Python Genome Assembly
By a comprehensive data mining approach, we identified 108 putatively functional and 28 disrupted and/or truncated (<600 bp) OR gene sequences from the anole lizard genome assembly. All these DB-based OR sequences were intronless and most of them were tightly clustered within several chromosomal or scaffold regions (see supplementary table 1, Supplementary Material online). The composition of HORDE families in the lizard OR gene repertoire is shown in figure 1A. All the anole lizard OR genes except one belonged to class II. One class I OR gene was classified into family 51. Class II OR genes assigned to the same HORDE families were generally clustered together in a phylogenetic tree (see supplementary fig. 3, Supplementary Material online).
FIG. 1.—
Relative gene composition of OR families (HORDE classification) identified from the anole lizard and the ratsnake. (A) ORs obtained from the anole lizard genome assembly. (B) FLX-based OR sequences from the anole lizard genomic DNA. (C) FLX-based OR sequences from the anole lizard nose cDNA. (D) FLX-based OR sequences from the ratsnake genomic DNA. (E) DB-based OR sequences from the python genome assembly.
From the python genome assembly, we identified 153 functional, 13 disrupted, and 114 truncated OR gene sequences (see supplementary table 2, Supplementary Material online), although this gene repertoire may be somewhat incomplete owing to the lower coverage of the python genome (ca. 17× coverage from Illumina paired-end sequences; Castoe et al. 2011). All these OR sequences were intronless. Chromosomal positions of these OR sequences are unknown because the contigs of python genome are very short (typically ∼2 kbp) and unconnected. The composition of HORDE families in the python OR gene repertoire is shown in figure 1E. Unlike the anole lizard, 17 class I OR genes were identified in the python genome. In these class I OR genes, four genes were classified into family 51, 12 were family 52, and 1 was family 55.
The Anole Lizard OR Sequences Obtained by the NGS Approach
Using the NGS approach, we obtained 6,196 reads from the anole lizard genomic DNA and 6,854 reads from its nose cDNA (table 2). The initial assembling gave rise to 195 and 194 contigs from genomic DNA and cDNA, respectively. After excluding non-OR sequences and OR sequences with <5× coverages as well as unifying possibly identical sequence contigs (i.e., <1% pairwise nucleotide differences: see Materials and Methods), 71 (genomic DNA) and 70 (cDNA) distinct contigs remained. Fifteen (genomic DNA) and 30 (cDNA) artificial chimeric sequences were additionally detected by FASTA searches against the database sequences. Excluding the chimeras from the data set, 56 and 40 distinct OR sequences were finally identified from the lizard genomic DNA and cDNA, respectively (table 2). These sequences were found to contain ten (genomic DNA) and three (cDNA) homopolymer run–associated errors (see supplementary tables 3 and 4, Supplementary Material online).
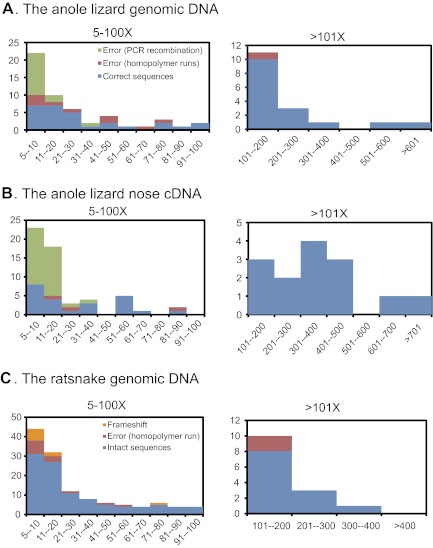
Figure 2 shows histograms of the read coverage for the FLX-based OR sequences. For the sequences obtained from the lizard genomic DNA, most erroneous sequences had <11× coverages (fig. 2A), whereas 15 of 30 chimeras had more than 10× coverages for the sequences obtained from the cDNA (fig. 2B). The higher rate of chimeras in cDNA-originated OR sequences could possibly be attributable to errors during the reverse transcription reaction. Thus, when we applied this approach to the characterization of the ratsnake OR genes amplified from its genomic DNA, contigs of <11× coverages were considered to be possibly incorrect and thus discarded.
FIG. 2.—
Histograms of the coverages or read numbers for each FLX-based contig sequence: (A) the anole lizard genomic DNA, (B) the anole lizard nose cDNA, and (C) the ratsnake genomic DNA.
The composition of HORDE families identified in the lizard's FLX-based OR sequences was very similar to that identified in the DB-based OR sequences (fig. 1A and B). We found that approximately one-third of the FLX-based OR sequences, when queried with their full-length OR gene sequences, was assigned to a different but neighboring HORDE family. However, the HORDE family distribution based on 56 full-length OR genes had no noticeable difference from that from 56 partial OR gene sequences (data not shown). This indicates that the FLX-based sequencing method used in this study can cover almost all families of OR sequences, at least for the anole lizard but possibly for other species as well, even though all gene members of these families were not picked up.
On the other hand, HORDE family composition of OR sequences obtained from the genomic DNA was slightly different from that obtained from the cDNA (fig. 1B and C). For instance, OR sequences of families 2 and 8 were found only from the genomic DNA. Proportion of family 5 OR sequences in the latter (15/40 × 100 = 38%) was slightly larger than that in the former (15/56 × 100 = 27%), although this difference was not significant (P = 0.528, Fisher's exact test). These differences likely reflect mRNA expression levels of different OR genes in the lizard's olfactory epithelium.
Expression of the Anole Lizard OR Genes
Coverage numbers for each FLX-based OR sequence obtained from nose cDNA seem to reflect primarily its expression level (i.e., abundance of the corresponding mRNA). However, they are also highly dependent on other factors, such as the efficiency of PCR amplification in relation to, for example, the matching of primers to individual gene copy sequences. On the other hand, coverage numbers for each FLX-based OR sequence obtained from genomic DNA are considered to reflect only the latter part because the copy number for each gene is equal in the genomic DNA. Thus, relative expression level of each OR-coding sequence may be roughly estimated by comparing its coverage number in cDNA-originated OR sequences with that in genomic DNA–originated sequences, provided that similar numbers of OR-coding reads are obtained from the two sources (table 2). If the former coverage number is significantly higher than the latter number, the corresponding OR gene copy may be considered highly expressed. In a reverse situation, its expression level may be considered relatively low.
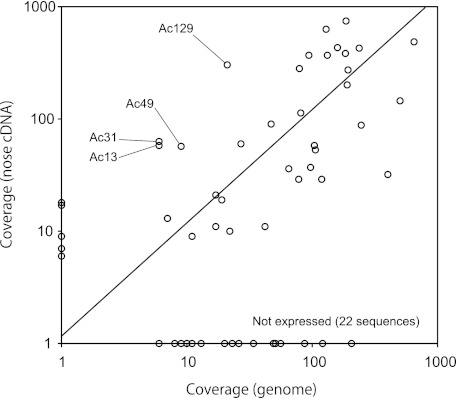
Figure 3 shows comparison of the coverage numbers between the lizard genomic DNA–originated and cDNA-originated OR sequences. Because total numbers of OR-coding reads were similar between the two sources (5,043 of genomic DNA origin and 5,966 of cDNA origin; see table 2), the direct comparison of coverage numbers may provide us with information of the expression level for each gene. For four OR sequences, Ac13 (HORDE family 4), Ac31 (family 1), Ac49 (family 14), and Ac129 (family 9), coverages of their cDNA-originated sequences were more than five times as large as those of the corresponding genomic DNA–originated sequences, suggesting that these OR sequences were highly expressed in the olfactory epithelium of the individual used in this study. Conversely, 22 OR sequences were found only in genomic DNA–originated sequences (fig. 3 and supplementary table 6, Supplementary Material online), suggesting that these ORs were not transcribed. Three of the 22 OR sequences were considered pseudogenes (Ac119, Ac128, and Ac131), but remaining 19 sequences appeared functional. It seems noteworthy that six OR sequences (Ac6, Ac41, Ac73, Ac97, Ac103, and Ac118) were found only in cDNA-originated sequences but that expression levels of these sequences were not clear owing to their low coverage numbers.
FIG. 3.—
Comparison of the coverages or read numbers in FLX-based OR-coding sequences originated from the anole lizard genomic DNA and nose cDNA. Logarithmic scales are used for both axes, and a line stands for an equal level of coverages between the two sources after normalization of total OR-coding read numbers between the two sources (the slope: 5966/5043 = 1.18). To include OR sequences that were not found (i.e., 0× coverage) in the scatter plot, we added one to the read coverage numbers of all sequences.
The Ratsnake OR Genes
We obtained 6,202 reads from the ratsnake genomic DNA by the FLX-based sequencing approach. The initial assembling generated 253 contigs, from which 140 contigs remained after excluding non-OR sequences and OR sequences with <5× coverages as well as unifying sequences with <1% sequence divergences (table 2). Due to the lack of reference genome sequence for the ratsnake, we were unable to specify chimeras in the resultant ratsnake OR sequences. We thus automatically removed contigs with <11× coverages to minimize chimeric sequences (see the reasons outlined earlier). Finally, we identified 96 distinct OR sequences from the ratsnake genomic DNA (see table 2 and supplementary table 5, Supplementary Material online) and used them for subsequent phylogenetic analyses. By translating them to amino acid sequences, we identified eight sequences that contain frameshifts in homopolymer runs. We also found three sequences in which coding frames were disrupted by nucleotide insertions or deletions outside the homopolymer region, although we could not judge whether they represent pseudogenes or sequencing artifacts. If contigs with 5×–10× coverages were included, these numbers considerably elevate (15 sequences with homopolymer run–associated errors and 9 disrupted sequences outside the homopolymer region). Thus, ratsnake OR sequences with such low (<11×) coverages may include a number of indels and/or disrupted stop codons possibly originated from PCR/sequencing artifacts (fig. 2C).
By conducting the FASTA search against the human OR amino acid sequences, each of the ratsnake OR sequences was assigned to the HORDE family. The resultant family occurrence of the ratsnake ORs was similar to that of the anole lizard ORs (fig. 1B and D), although proportions in numbers of family member genes were considerably different between the two species (table 3). Gene proportions of families 2 and 12 in the ratsnake ORs were significantly larger than those in the lizard ORs. Difference in family 12 was significant in the 5% level even after Bonferroni correction (P < 0.0038). Conversely, gene proportions of families 11 and 14 in the ratsnake ORs were significantly smaller than those in the lizard ORs. The family occurrence of the ratsnake ORs was also similar to that of the python ORs (fig. 1D and E), but proportions in gene numbers of a few families were different between these species (table 3). Gene proportions of families 5 and 12 in ratsnake ORs were significantly larger than those in the python ORs. Difference in family 12 was significant in the 5% level even after Bonferroni correction (P < 0.0036). Gene proportions of family 4 and class I ORs in ratsnake were significantly smaller than those in the python ORs.
Table 3.
Numbers of OR Genes Assigned to Each HORDE Family for the Anole Lizard and the Python DB-Based Sequences and the Ratsnake FLX-Based Sequences
| Family | Number of Genes (the anole lizard) | Number of Genes (the python) | Number of Genes (the ratsnake) | P Value (lizard vs. ratsnake) | P Value (python vs. ratsnake) |
| 1 | 6 | 14 | 5 | 1.000 | 1.000 |
| 2 | 2 | 27 | 9 | 0.011a | 1.000 |
| 4 | 10 | 15 | 1 | 0.054 | 0.085 |
| 5 | 37 | 64 | 44 | 0.052 | 0.004a |
| 6 | 12 | 38 | 4 | 0.292 | 0.021a |
| 7 | 0 | 1 | 0 | — | 1.000 |
| 8 | 2 | 6 | 1 | 1.000 | 0.684 |
| 9 | 5 | 4 | 6 | 0.534 | 0.024 |
| 10 | 21 | 45 | 8 | 0.166 | 0.123 |
| 11 | 13 | 18 | 2 | 0.032a | 0.124 |
| 12 | 1 | 5 | 10 | 0.001b | 0.001b |
| 13 | 3 | 6 | 2 | 1.000 | 1.000 |
| 14 | 23 | 19 | 4 | 0.007a | 0.466 |
| Class Ic | 1 | 17 | 0 (1) | 1.000 | 0.009a |
| Not identified | 0 | 1 | 0 | — | — |
| Total | 136 | 280 | 96 | — | — |
Significant in the 5% level by Fisher's exact test.
Significant in the 5% level after Bonferroni correction.
The class I genes of the anole lizard and the ratsnake correspond to families 51 and 52, respectively. Seventeen class I genes of the python include 4 for family 51, 12 for family 52, and 1 for family 55. Coverage of the ratsnake class I OR (8×) is lower than the cutoff (11×).
Evolution of the Ratsnake OR Genes
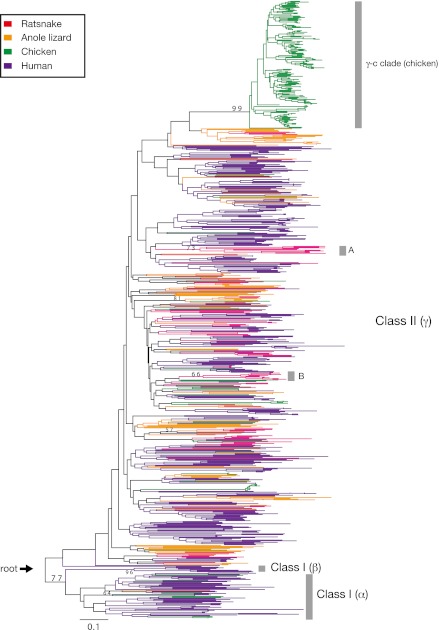
Figure 4 shows a neighbor joining tree constructed using the OR partial sequences from the ratsnake and three vertebrate species (the anole lizard, human, and chicken). All the 96 ratsnake ORs were widely scattered within the class II clade (group γ, Niimura and Nei 2005) without forming large lineage-specific phylogenetic clusters like the γ-c clade for chicken (Niimura and Nei 2005; Steiger et al. 2008). Many of these ratsnake ORs were clustered with the anole lizard ORs, implying their origination before the lineage divergence between these taxa. However, at least two small ratsnake-specific clades were found (i.e., clades A and B in fig. 4).
FIG. 4.—
A neighbor joining tree of 799 OR amino acid partial sequences from four vertebrate species (the ratsnake: 92, the anole lizard: 108, human: 387, and chicken: 212). The number of amino acid sites used for the analysis is 77. Bootstrap values of more than 50% are shown on major internal nodes only. The tree is rooted at an arbitrary position on a lineage between class I and class II ORs as indicated by an arrow.
As was the case with the anole lizard, only one class I OR sequence was potentially present in the ratsnake (table 3). However, this OR sequence had 8× coverage reads, which was lower than the tentative cutoff value (<11×). Thus, this sequence was not included in the phylogenetic tree of figure 4. When included, the ratsnake class I OR sequence did not cluster with the anole lizard counterpart (data not shown). Blast searches against the HORDE database indicated that the ratsnake class I OR was assigned to family 52, whereas the anole lizard class I OR belonged to family 51 (table 3). This implies that the ratsnake class I OR has a different origin from the lizard counterpart.
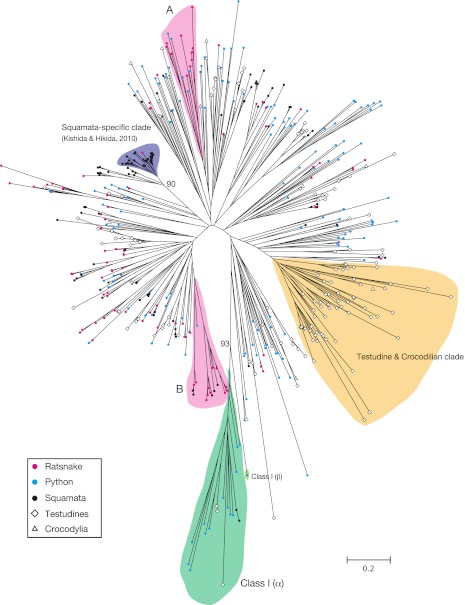
The phylogenetic tree of OR sequences from the ratsnake, the python, and 14 reptilian taxa (fig. 5) showed that a majority of the ratsnake ORs were evolutionarily close to the other squamate ORs. However, some ratsnake ORs were more closely related to turtle ORs than to squamate ORs, and the others were snake specific (e.g., ten genes in clade A). It seems noteworthy that the ratsnake had only one OR gene that belonged to the “Squamata-specific ORs” clade (Kishida and Hikida 2010). One part of the phylogenetic tree was occupied by turtle and crocodilian OR genes without squamate ones, designated the Testudine and Crocodilian clade. Another striking feature is the representation of class I OR group by a number of the python OR genes.
FIG. 5.—
An unrooted neighbor joining tree of 358 OR amino acid partial sequences from the ratsnake, python, and 14 reptilian species (7 squamates, 6 testudines, and 1 crocodilian). The number of amino acid sites used for the analysis is 69. GenBank protein IDs and the sources of the OR genes of the 14 reptilian taxa were shown in supplementary table 7 (Supplementary Material online). Bootstrap values are shown on representative nodes only.
Discussion
Efficiency of the High-Throughput Approach for Characterization of OR Gene Repertoires
Using a tiny part (∼1/30) of the sequencing capacity by a single run of GS FLX Titanium Genome Sequencer, we identified many OR partial sequences from the two squamate species simultaneously. The broad phylogenetic distribution (supplementary fig. 3, Supplementary Material online) and the HORDE family composition (fig. 1B) of FLX-based OR sequences for the anole lizard showed that this approach can quickly characterize considerable unbiased proportions of OR gene members from multiple sources in parallel. Tagging the PCR products from different sources of template DNA is a key procedure in this method. However, this approach could not recover all 136 members of OR genes identified in the anole lizard genome assembly. The apparent recovery rate of OR gene sequences was 41% (56/136 × 100). Two A. carolinensis individuals, one used for the draft genome sequencing (Castoe et al. 2011) and the other used for the FLX-based sequencing (this study), may have the polymorphism in OR gene loci. Thus, the total number of OR genes owned by the latter individual is not necessarily 136. However, this low recovery rate still implies that many OR genes were not identified by the NGS method. Then, how can one pursue higher recovery rates? Isn't it realistic to recover nearly complete OR gene members by the method?
In order to gain perspectives into these questions, we conducted further analyses of the FLX-based sequences together with some manual experiments. First, coverage read number for each FLX-based OR sequence was found to be rather heterogeneous (fig. 2), suggesting that PCR amplification efficiency of OR sequence varies from gene to gene. The partial recovery rate (41%) is most likely due to differences of primer matching to OR gene copies. At an early phase of this work, we designed several primers, with which A. carolinensis OR gene fragments were amplified, cloned into E. coli, and manually sequenced. This preliminary experiment showed that the F1 and R1 primers (see table 1 for their sequences without tag regions) provided the broadest coverage of OR genes, all of which are included in the 56 genomic DNA–originated OR sequences shown in table 2 (data not shown). After the NGS experiments, we examined the matching of the F1 and R1 primers to 43 NGS-collected (either from genomic DNA or cDNA) and 65 NGS-uncollected functional OR genes (see supplementary fig. 4, Supplementary Material online). Whereas the F1 primer appears to have a similar matching to both NGS-collected and NGS-uncollected genes, the R1 primer shows a somewhat reduced matching to NGS-uncollected genes, especially at the fourth to seventh positions from the 3′-end of the primer.
We then designed additional primers (F2, F3, R2, and R3; for details, see table 1) by referring to sequences of the NGS-uncollected OR genes with an expectation that the new primer pairs (i.e., F2–R2 and F3–R3) can cover many of the NGS-uncollected genes as well as a certain proportion of the NGS-collected ones. We manually sequenced 100 clones having an insert of the amplified products (49 clones for F2–R2 and 51 clones for F3–R3). The 49 clones for F2–R2 provided 20 distinct OR genes covering 19 of 136 OR genes identifiable from the lizard draft genome sequence, plus one new OR gene not identified in the draft genome. Seven of the 20 genes were not collected by the NGS approach using the F1–R1 primers. Similarly, the 51 F3–R3 clones provided 14 distinct OR genes covering 10 of the 136 OR genes, plus 4 new OR genes not identified in the draft genome. Six of the 14 genes were not collected by the NGS approach using the F1–R1 primers. As shown in supplementary figure 3 (Supplementary Material online), these OR genes collected using the F2–R2 and F3–R3 pairs appear to be randomly distributed in a phylogenetic tree, as is the case with those collected using the F1–R1 pair.
We also considered a possibility that the ≥5× coverage criterion for identifying OR sequences in contigs restricted the recovery rate and that more intensive sequencing by the NGS approach could pick up genes with a low amplification efficiency. We conducted the BlastN search (%ID >0.99 and >50 bp; see Materials and Methods for the reasoning) of all 6,196 genomic DNA–originated FLX reads (table 2) against the 136 OR gene sequences. Sixty-two potential OR sequences including 52 functional ones (see supplementary fig. 3, Supplementary Material online) were identified by this search. Together with six OR genes not represented in the draft genome (see supplementary table 3, Supplementary Material online), 68 OR sequences may have been collected if we had obtained much more FLX reads so that all these sequences satisfied the ≥5× coverage criterion. Taken together, by expanding the read depth and combining three different primer pairs for the NGS approach, we estimate that as many as 81 A. carolinensis OR sequences could be collected. The cDNA-originated NGS characterization showed that six additional OR genes could be amplified and identified by the F1–R1 primer pair (Ac6, Ac41, Ac73, Ac97, Ac103, and Ac118; supplementary table 6, Supplementary Material online). We consider that these OR genes can be basically identifiable by the NGS approach, though we do not know why they were not represented in the genomic DNA–based FLX reads. Thus, the identifiable A. carolinensis OR genes by the NGS approach can collectively reach 87 (87/136 × 100 = 64%). This is still not the level of exhaustive characterization of all OR gene members, but we do not deny a possibility that the combinatory use of more primer pairs may be able to reach this level in future.
Accuracy of the High-Throughput Approach for Characterization of OR Gene Repertoires
Accuracy of the NGS approach in identifying OR genes was assessed by comparing FLX-based OR sequences obtained from the anole lizard genomic DNA with the corresponding DB-based OR sequences using the FASTA search. In principle, two types of errors could be included in the FLX-based OR sequences: errors caused by PCR amplification, such as nucleotide substitutions and chimera formation, and errors caused by the FLX pyrosequencing, such as nucleotide substitutions and indels usually associated with homopolymer runs (Margulies et al. 2005; Moore et al. 2006). In this study, most nucleotide substitutions originated from PCR errors seem to be excluded from the resultant OR sequences in contigs because each OR sequence is determined at least five times and substitution errors were excluded by generating consensus sequences. Indeed, 35 of 56 FLX-based OR sequences originated from the lizard genomic DNA were completely identical with the corresponding DB-based OR sequences (see supplementary table 3, Supplementary Material online). This indicates the low error rate of nucleotide substitutions in the FLX-based OR sequences.
On the other hand, chimeras were frequently found in the FLX-based OR sequences (fig. 2). As described in Materials and Methods, we employed a PCR condition that minimizes PCR errors and chimera formation during PCR: the use of PrimeSTAR HS DNA polymerase (Takara Bio) that have both high fidelity and high efficiency in amplification, and restriction of amplification cycles to 28 (Lenz and Becker 2008). In spite of this endeavor, formation of some chimeric sequences in amplifying multicopy genes seems unavoidable (Saitoh and Chen 2008). Twelve of 15 chimeras (80%) were found to have <11× coverage in FLX-based OR sequences originated from the lizard genomic DNA (fig. 2A), whereas 28 of 30 chimeras (93%) were found to have 20× or fewer coverage in cDNA-originated OR sequences (fig. 2B). Higher frequency of chimeras in the cDNA-originated OR sequences is possibly caused by the reverse transcription reaction. One way to eliminate artificial chimeras as much as possible is therefore not to use contigs with 20× or fewer coverages. However, this would lead to elimination of considerable numbers of true OR sequences with 11×–20× coverage. Thus, we decided to set the <11× cutoff coverage value for OR sequences originated from the ratsnake genomic DNA.
In previous studies, OR genes in nonmodel vertebrates have been PCR amplified, cloned, and sequenced by the traditional Sanger sequencing method (Buck and Axel 1991; Ngai et al. 1993; Freitag et al. 1995; Kishida et al. 2007; Steiger et al. 2008; Hashiguchi and Nishida 2009; Kishida and Hikida 2010). However, this traditional approach has limitations in extensive identification of OR gene family. Moreover, it seems very difficult to avoid errors associated with PCR (nucleotide substitutions and chimera formation) by sequencing limited numbers of clones manually. The NGS approach can potentially overcome some of these problems by determining much larger numbers of OR sequences than the traditional approach, although some chimeric sequences may still exist in low-coverage contigs. In addition, the NGS method can handle multiple samples (different species and individuals) simultaneously by adding tags to PCR primers. Finally, the NGS approach does not use a step for molecular cloning into bacteria and thus seems freer from the cloning bias than the traditional approach. This is especially advantageous in comparing numbers of cDNA-originated FLX reads to gain insights into gene expression. Taken together, the current approach using the NGS power seems promising toward the efficient and accurate characterization of multigene family genes and their transcripts in nonmodel organisms in future.
Expression of the Anole Lizard OR Genes
In the present study, we identified 40 different OR sequences from the nose cDNA of the anole lizard. It is likely that these OR sequences represent a highly expressed set of OR gene copies in the lizard's olfactory epithelium. In addition, four OR sequences (Ac13, Ac31, Ac49, and Ac129) were suggested to have a notably high level of expression (fig. 3). Among them, Ac129 OR sequence identified from the genome database appeared to be a pseudogene (see supplementary tables 1, 3, and 4, Supplementary Material online). However, Ac129 may be a functional gene in an individual used for this study because Ac129 cDNA-originated sequence, at least within the sequenced 331 bp region, did not contain any disrupted stop codon or frameshift that existed in the Ac129 database sequence. If the Ac129 database sequence does not include a sequencing error, a possible explanation is that Ac129 represents a segregating pseudogene, where both functional and nonfunctional alleles coexist in a locus. Several segregating pseudogenes for ORs have been reported in, for example, human (Menashe et al. 2003, 2007), and one of such OR loci was suggested to relate to differences of odor sensitivity among individuals (Menashe et al. 2007). Investigating the Ac129 polymorphism in future may be interesting to understand the genetic basis of odor sensitivity in the anole lizard.
Comparison of the coverage read numbers of OR sequences originated from genomic DNA versus cDNA indicated that 22 lizard OR genes (19 intact and 3 disrupted sequences) were not detected for their expression in the olfactory epithelium (fig. 3). If this really reflect their lack of expression rather than any biases in our experiments (e.g., low efficiency in the reverse transcription reaction), approximately 39% (22/56 × 100) of the lizard OR genes are not expressed in the adult olfactory epithelium. DNA microarray studies suggested that proportion of silent OR genes that are not expressed in the olfactory epithelium is less than 30% in human and mouse (Zhang et al. 2004, 2007). Iguanian lizards including the green anole lizard have highly developed the visual sense, and most iguanians may not be dependent on a well-developed olfactory system (Zug et al. 2001). The relatively low proportion of expressed lizard OR genes may reflect the reduced role of olfaction in this species. Verification of this speculation should await more rigorous comparison of expressed OR gene repertoire in diverse groups of squamates.
Diversity and Evolution of the Squamate OR Genes
Using the NGS approach, we identified 96 distinct OR gene sequences from the ratsnake genomic DNA. By applying the same criterion (e.g., exclusion of contigs with <11× coverages without the chimeric test), 47 OR gene sequences were identifiable from the anole lizard genomic DNA (see supplementary table 3, Supplementary Material online). Under a simple equal recovery rate assumption (47/136 × 100 = 35%) for the ratsnake, total number of OR gene loci in this species was roughly estimated to be 278 (96/47 × 136). When the coverage cutoff criterion is changed from <11× to <5×, a similar number of the OR gene loci was estimated for the ratsnake (140/71 × 136 = 268).
These estimates are close to the estimated number of OR genes in the Burmese python (280; table 3) for which the draft genome sequence is newly available (Castoe et al. 2011). Thus, the OR gene numbers in these snakes seem larger than those in the anole lizard (136, table 3) and zebra fish (176, Niimura 2009) but much smaller than those in frog (1,638, Niimura 2009) and human (802, Niimura and Nei 2005), even smaller than those in chicken (433) and zebra finch (553) (Niimura 2009; Steiger et al. 2009). Thus, the snakes may have less diverse OR gene repertoire than most nonsquamate tetrapods. This may sound somewhat unexpected because most colubrid and pythonid snakes are known to have an acute sense of smell (Zug et al. 2001; Pianka and Vitt 2003; Vitt et al. 2003). However, many squamate reptiles have highly developed the vomeronasal olfactory system, and their sharpness in olfaction is dependent on both nasal and vomeronasal receptors (Zug et al. 2001; Pianka and Vitt 2003; Vitt et al. 2003). One intriguing possibility is that these snakes took a strategy to diversify vomeronasal receptor genes (i.e., V1Rs and V2Rs) rather than nasally expressed OR genes.
Phylogenetic analyses showed that OR sequences of the anole lizard and the two snakes are broadly distributed in the phylogenetic tree (figs. 4 and 5). This implies that these squamate lineages have kept a number of OR subfamilies that originated before the divergence of mammalian and reptilian/avian lineages. Although large lineage-specific phylogenetic clusters as seen in avian species (e.g., chicken γ-c clade) were not found for the squamates, there was a small snake-specific OR clade (clade A: see figs. 4 and 5). Clade B was also specific to the ratsnake and other squamates (figs. 4 and 5). OR gene members in these specific clades may have recently increased by gene duplications for adaptation to squamate-specific odor environments.
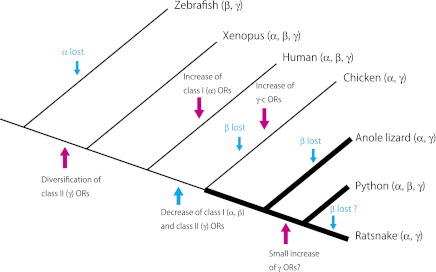
Figure 6 outlines the evolution of squamate OR genes. As reviewed in the literature (Niimura and Nei 2005; Niimura 2009), the repertoire of class I (α) and class II (γ) OR genes was expanded when amphibian ancestors emerged to land. However, without the radiation of the γ-c clade members, birds have relatively small numbers of OR genes (e.g., 94 and 16 for chicken and zebra finch OR genes outside the γ-c clade, respectively; Steiger et al. 2009). Except for the γ-c clade, both squamate and chicken ORs are broadly distributed in the phylogenetic tree (fig. 4), suggesting that most OR subfamilies in birds and squamates originated before their divergence. We thus consider that the OR gene repertoire may have been shrunk for both class I (α and β) and class II (γ) genes in the genome of an ancestral sauropsid lineage. Resultantly, avian and squamate OR gene repertoire may consist of basically small numbers of OR genes. However, we cannot strictly exclude the possibility that ORs in avian and squamate lineages decreased independently. In response to the ecological needs to detect more or less smells, individual squamate members probably fluctuated the number of OR genes in their genome. Most snakes have highly developed sense of smell (Zug et al. 2001), and they may have retained somewhat larger numbers of OR genes than the anole lizard.
FIG. 6.—
Schematic illustration of the evolution of vertebrate class I (α and β) and class II (γ) OR genes. In the phylogenetic tree, branches shown in thick lines indicate the squamate lineage.
An unexpected finding in the python OR gene repertoire was the presence of 17 class I genes that include two group β member (table 3; supplementary table 2, Supplementary Material online). The group β OR gene was not found in the anole lizard OR gene repertoire, and the NGS approach did not identify any group β gene in the ratsnake (table 3; supplementary table 5, Supplementary Material online). Without assuming a horizontal transfer of the group β gene to the python genome, a reasonable explanation could be deletions of the group β gene in multiple lineages leading to the anole lizard, the ratsnake, and even birds (fig. 6). It was shown that group β OR genes in tetrapods are actually orthologous to some teleost fish OR genes (Niimura 2009), implying a possibility that the group β ORs are used to recognize odor chemicals common to aquatic and terrestrial vertebrates. Niimura (2009) thus deduced that the group β ORs may detect both volatile and water-soluble chemicals, such as alcohol. Repeated loss of the group β OR genes in squamates may be related to the ecological differences among species, such as habitat preferences. The Burmese python is known to show water-dependent life style occasionally (The Invasive Species Specialist Group 2010), and this might be related to the retention of the group β OR gene in this species. Characterization of the OR gene repertoire in more squamate taxa will clarify the evolutionary mechanisms of the group β OR genes more fully. Also, we currently do not know why the python has a larger number of class I (α and β) OR genes than the anole lizard and the ratsnake (table 3). Any reasonable explanation to connect the class I gene variation to ecological features may be expected by the further characterization of squamate OR gene repertoires.
In the present study, we investigated the anole lizard OR genes to evaluate the usefulness of the NGS approach in characterizing OR genes in nonmodel vertebrate species. The NGS approach should be broadly applicable to efficient characterization of vertebrate OR genes and their transcripts and therefore promises to expand the scale of future studies on vertebrate olfactory systems. For example, comparative analyses of OR gene transcripts between male and female lizards by the NGS approach may provide a clue to understanding molecular basis of olfactory recognition of conspecific individuals in mating.
Supplementary Material
Supplementary figures 1–4, tables 1–7, and data 1 and 2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank the late Dr Michihisa Toriba (The Japan Snake Institute) and Dr Hideaki Mori (Japan Wildlife Research Center) and Mrs Yoshiaki Ariyama (Ogasawara National Park Ranger Office) and Kosho Yagi (Remix Peponi) for their kind help in collecting samples. We also thank Dr Tomohide Yoshimura (Yoshimura Animal Clinic) for his special instruction in the anatomy of reptiles and Drs Akira Kanamori (Nagoya University) and Junji Sano (Nagoya City University) for experimental advices. Our gratitude is extended to Ms Chiemi Yamada (Nagoya City University) for her kind assistance in processing the NGS data and Drs Shigeru Itoh and Shin Sugiyama (Nagoya University) for their warm encouragement. Dr Hideaki Tagami (Nagoya City University) kindly allowed us to use his laboratory equipment. Dr Yoshihito Niimura and three anonymous reviewers provided valuable comments and suggestions. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant No. 20370033 to Y.K. and No. 22770236 to Y.H.). The A. carolinensis genomic sequence data were produced by the Broad Institute of MIT and Harvard University.
References
- Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babik W, Taberlet P, Ejsmond MJ, Radwan J. New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Resour. 2009;9:713–719. doi: 10.1111/j.1755-0998.2009.02622.x. [DOI] [PubMed] [Google Scholar]
- Binladen J, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. Genewise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12:406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag J, Krienger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:0120–0125. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, et al. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome. 2000;11:1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol Biol. 2006;6:76. doi: 10.1186/1471-2148-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Screening the V2R-type putative odorant receptor gene repertoire in bitterling Tanakia lanceolata. Gene. 2009;441:74–79. doi: 10.1016/j.gene.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Hayden S, et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Invasive Species Specialist Group. The Global Invasive Species Database. 2010. [cited 2012 April 3]. Available from: http://www.issg.org/database. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T, Hikida T. Degeneration patterns of the olfactory receptor genes in sea snakes. J Evol Biol. 2010;23:302–310. doi: 10.1111/j.1420-9101.2009.01899.x. [DOI] [PubMed] [Google Scholar]
- Kishida T, Kubota S, Shirayama Y, Fukami H. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol Lett. 2007;3:428–430. doi: 10.1098/rsbl.2007.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz TL, Becker S. Simple approach to reduce PCR artefact formation leads to reliable genotyping of MHC and other highly polymorphic loci—implications for evolutionary analysis. Gene. 2008;427:117–123. doi: 10.1016/j.gene.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2007;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- Menashe I, et al. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Moore MJ, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A. The family of genes encoding odorant receptors in the channel catfish. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- Niimura Y. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol Evol. 2009;1:34–44. doi: 10.1093/gbe/evp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One. 2007;8:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T, Feldmesser E, Atarot T, Eisenstein M, Lancet D. The olfactory receptor universe—from whole genome analysis to structure and evolution. Genet Mol Res. 2004;3:545–553. [PubMed] [Google Scholar]
- Pianka ER, Vitt LJ. Lizards—windows to the evolution of diversity. London: University of California Press; 2003. [Google Scholar]
- Saitoh K, Chen W-J. Reducing cloning artifacts for recovery of allelic sequences by T7 endonuclease I cleavage and single re-extension of PCR products—a benchmark. Gene. 2008;423:92–95. doi: 10.1016/j.gene.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Steiger SS, Fidler AE, Valcu M, Kempenaers B. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc R Soc B Biol Sci. 2008;275:2309–2317. doi: 10.1098/rspb.2008.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger SS, Kuryshev VY, Stensmyr MC, Kempenaers B, Mueller JC. A comparison of reptilian and avian olfactory receptor gene repertoires: species-specific expansion of group γ genes in birds. BMC Genomics. 2009;10:446. doi: 10.1186/1471-2164-10-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C-Y, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139:45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Valentini A, et al. New perspectives in diet analysis based on DNA barcording and parallel pyrosequencing: the trnL approach. Mol Ecol Resour. 2009;9:51–60. doi: 10.1111/j.1755-0998.2008.02352.x. [DOI] [PubMed] [Google Scholar]
- Vitt LJ, Pianca ER, Cooper WE, Jr, Schwenk K. History and global ecology of squamate reptiles. Am Nat. 2003;162:44–60. doi: 10.1086/375172. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug GR, Vitt LJ, Caldwell JP. Herpetology. An introductory biology of amphibians and reptiles. 2nd ed. San Diego (CA): Academic Press; 2001. [Google Scholar]