Abstract
Recombinant type 3 ryanodine receptor (RyR3) has been purified in quantities sufficient for structural characterization by cryoelectron microscopy and three-dimensional (3D) reconstruction. Two cDNAs were prepared and expressed in HEK293 cells, one encoding the wild-type RyR3 and the other encoding RyR3 containing glutathione S-transferase (GST) fused to its amino terminus (GST-RyR3). RyR3 was purified from detergent-solubilized transfected cells by affinity chromatography using 12.6-kDa FK506-binding protein in the form of a GST fusion as the affinity ligand. Purification of GST-RyR3 was achieved by affinity chromatography by using glutathione-Sepharose. Purified recombinant RyR3 and GST-RyR3 proteins exhibited high-affinity [3H]ryanodine binding that was sensitive to activation by Ca2+ and caffeine and to inhibition by Mg2+. 3D reconstructions of both recombinant RyR3 and GST-RyR3 appeared very similar to that of the native RyR3 purified from bovine diaphragm. Comparison of the 3D reconstructions of RyR3 and GST-RyR3 revealed that the GST domains and, hence, the amino termini of the RyR3 subunits are located in the “clamp” structures that form the corners of the square-shaped cytoplasmic region of homotetrameric RyR3. This study describes the 3D reconstruction of a recombinant ryanodine receptor and it demonstrates the potential of this technology for characterizing functional and structural perturbations introduced by site-directed mutagenesis.
Keywords: recombinant membrane protein, cryoelectron microscopy, single-particle image processing
Calcium release from intracellular stores is essential for many fundamental biological functions. Ryanodine receptors (RyRs) are members of a family of intracellular calcium-release channels, which also includes the inositol 1,4,5-trisphosphate receptors (1–3). Three isoforms of RyR have been identified. Type 1 RyR (RyR1) and RyR2 are the major calcium-release channels in skeletal and cardiac muscle, respectively, whereas RyR3 was found mainly in diaphragm, smooth muscle, and brain. However, recent studies show that all three isoforms are widely expressed in many tissues with different expression levels (4–6). Unlike RyR1 and RyR2, RyR3 is present at low levels and often is coexpressed with other RyR isoforms (6, 7). The RyR isoforms, encoded by three different genes, share a high degree of sequence homology (8–10).
RyRs are the largest known ion channels, assembled as homotetramers with a molecular mass of more than 2 MDa (8–10). Three-dimensional (3D) structures of the native RyR1, RyR2, and RyR3 have been determined to ≈30Å resolution by electron microscopy (EM) of frozen-hydrated, detergent-solubilized receptors in conjunction with single-particle image processing (11–14). Additional structural studies have been performed, mainly on RyR1, to determine the location of binding sites for ligands, including calmodulin, FK506-binding protein (FKBP; ref. 15), and Imperatoxin A (16), and also to characterize conformational changes between the “open” and “closed” states of the channel (17, 18). Similar structural analyses of RyR2 (13) and RyR3 (14) have been reported, but progress on these isoforms has been hindered by difficulties in purifying sufficient quantities of structurally intact receptors (19–21).
Further progress in elucidating structure/function relationships for RyRs would greatly accelerate if expression systems were available for isolating genetically modified receptors that are suitable for biochemical and biophysical characterization. Several laboratories now are expressing cloned RyRs in cultured cell lines, but only in a few cases has purification of the receptors been attempted, and there are no reports of structural characterization of the receptors (22–26). In the present study, we have expressed RyR3 and glutathione S-transferase (GST)-RyR3 cDNAs in HEK293 cells and purified the proteins in a single step by affinity chromatography. Using cryo-EM and single-particle image processing, we have demonstrated that the purified recombinant RyR3 exhibits intact structure nearly identical to that of the native RyR3. To demonstrate the utility of this approach for obtaining detailed structural information, we have compared the 3D reconstructions of RyR3 and GST-RyR3, which reveals that the N terminus of RyR3 is located in the clamp regions at the corners of the cytoplasmic assembly. This approach should be readily applicable to the other RyR isoforms and to mapping the locations of other surface-exposed amino acids.
Materials and Methods
Construction of RyR3 and GST-RyR3 cDNA.
The cloning and construction of cDNAs encoding the rabbit smooth muscle RyR3 have been described previously (24). The cDNA encoding the GST was amplified from the pGEX-3X vector by PCR. A unique NheI site was introduced into the forward primer, and a unique AscI site was added into the reverse primer. A unique AscI site also was introduced into the RyR3 cDNA just before the start codon. The NheI-AscI GST fragment was linked with the AscI-EcoRV(555) RyR3 fragment. The resulting NheI-EcoRV GST-RyR3 fusion fragment then was used to replace the NheI-EcoRV fragment in the full-length RyR3 cDNA to form the GST-RyR3 expression vector. The GST-RyR3 construct also contained the c-myc tag, EQKLISEEDL, inserted after Glu-4318. The c-myc tag first was inserted into the SpeI(12795)-EcoRI(13353) cDNA fragment in pBluescript by the overlap-extension method. The SpeI(12795)-EcoRI(13353) (c-myc) was linked with the EcoRI(11833)-SpeI(12795) fragment to form EcoRI(11833)-EcoRI(13353) (c-myc), which subsequently was used to replace the corresponding fragment in GST-RyR3. The sequences of all PCR fragments were verified by DNA sequencing.
Expression and Purification of RyR3 and GST-RyR3.
HEK293 cells were transfected with RyR3 or GST-RyR3 cDNA as described previously (24, 25). The transfected cells were lysed by the addition of a lysis buffer [20 mM Pipes (pH 7.2 with NaOH)/130 mM NaCl/5 mM Na2AMP/0.5 mM EGTA/0.7 mM CaCl2/2.5 mM DTT/1% CHAPS/0.5% egg lecithin/protease inhibitor mix (1 mM benzamidine/2 μg/ml leupeptin/2 μg/ml pepstatin A/2 μg/ml aprotinin/0.5 mM PMSF)] and incubation on ice for 1 h. Insoluble materials were removed, and sucrose (300 mM, final) and NaCl (400 mM, final) were added to the supernatant (cell lysate). Glutathione-Sepharose beads (80 μl) bound with 225 μg of GST–12.6-kDa FKBP (FKBP12.6) were added to the cell lysate. For the purification of GST-RyR3, only glutathione-Sepharose beads were added. The mixture was incubated at 5°C with rotation for 2 days. The beads were collected by centrifugation and washed four times with the lysis buffer containing 0.5% CHAPS and 0.25% egg lecithin. RyR3 or GST-RyR3 was eluted from the beads by incubating with an eluting buffer (20 mM Pipes, pH 7.2/400 mM NaCl/5 mM Na2AMP/300 mM sucrose/0.5 mM EGTA/0.7 mM CaCl2/2.5 mM DTT/0.4% CHAPS/0.16% egg lecithin/protease inhibitor mix/50 mM glutathione) for 10 min. For one preparation of RyR3, the elution buffer contained 60 μM FK506 instead of glutathione. Eluates were aliquoted, frozen in liquid nitrogen, and stored at −90°C.
[3H]Ryanodine Binding.
Equilibrium [3H]ryanodine binding to purified recombinant RyRs was carried out as described previously (27), with some modifications. [3H]Ryanodine binding was carried out in a total volume of 300 μl of binding solution containing 24–153 ng of purified recombinant proteins, 500 mM KCl, 25 mM Tris, 50 mM Hepes (pH 7.4), 0.8 mM CaCl2, 2.5 mM caffeine, 0.4–33 nM [3H]ryanodine, and the protease inhibitor mix at 37°C for 2 h. The binding mix was diluted with 5 ml of ice-cold washing buffer containing 25 mM Tris⋅HCl (pH 8.0) and 250 mM KCl and was filtered immediately through Whatman GF/B filters presoaked with 1% polyethylenimine. The filters were washed, and the radioactivity associated with the filters was determined by liquid scintillation counting. Nonspecific binding was determined by measuring [3H]ryanodine binding in the presence of 30 μM unlabeled ryanodine. All binding assays were done in duplicate. For determining the effects of Ca2+, caffeine, and Mg2+, [3H]ryanodine binding was carried out in a binding buffer containing 500 mM KCl, 25 mM Tris, 50 mM Hepes (pH 7.4), 0.5 mM EGTA, 0.35 mM CaCl2, 15 nM [3H]ryanodine, and the protease inhibitor mix, plus either 0.8 mM Ca2+, 2.5 mM caffeine, or 5 mM Mg2+ as indicated.
Cryo-EM and Image Processing.
The purified RyR3 or GST-RyR3 was diluted 2- to 3-fold with EM diluting buffer (20 mM Pipes, pH 7.2/400 mM KCl/5 mM Na2AMP/0.7 mM CaCl2/0.5 mM EGTA/0.5% CHAPS/2 mM DTT/2 μg/ml leupeptin). A 5-μl aliquot was applied to a carbon-coated holey EM grid (15). Specimen grids were transferred to a Gatan cryoholder and examined in a Philips EM 420 transmission electron microscope (Eindhoven, the Netherlands). The operating voltage was 100 kV, the electron dose was estimated at 8–10 electrons/Å2, and the temperature of the specimen grid was maintained at −178 ± 2°C. Electron micrographs were recorded at a magnification of ×38,600 and a defocus value of 1.5–2.0 μm. Selected micrographs were digitized by using a Eurocore Hi-Scan microdensitometer (Saint-Denis, France). Image processing was accomplished by using the spider/web software package (28).
3D reconstructions were obtained by following the projection-matching procedure (29). An initial set of 48 projection images that covered evenly the entire angular space was generated from a previous 3D reconstruction of native RyR1. For the recombinant RyR3 (glutathione-eluted), RyR3 (FK506-eluted), and GST-RyR3, a total of 7,690, 5,680, and 11,100 particle images were selected, respectively. After “matching” particles to the projection images by cross-correlation, 5,603, 5,272, and 7,101 high-quality particles were used to generate the first 3D reconstructions. For subsequent iterations, a set of 1,126 projection images was generated from the current 3D reconstruction to more finely sample the angular space, the particles were rematched to these projection images, and a new reconstruction was computed. The cycle was performed three to four times until the reconstruction stabilized. The final 3D reconstructions of RyR3 (glutathione-eluted), RyR3 (FK506-eluted), and GST-RyR3 were computed from 3,598, 4,722, and 5,893 particles, respectively. The final resolutions were estimated to be 34Å by Fourier shell correlation, using a cut-off of 0.5 for both RyR3s and GST-RyR3 (30).
Ca2+-Release Measurements and Immunoblotting.
Cytoplasmic Ca2+ in transfected HEK293 cells was measured with the fluorescent Ca2+ indicator dye fluo-3, and PAGE and immunoblotting were carried out as described previously (24).
Results
Purification and Functional Properties of the Recombinant RyR3 and GST-RyR3.
Fig. 1A shows SDS/PAGE of the affinity-purified RyRs. A high-molecular-weight band was detected clearly in both the RyR3 and GST-RyR3 samples (unless specified otherwise, RyR3 will refer to recombinant type 3 RyR that was eluted from glutathione-Sepharose beads by using glutathione rather than FK506; see Materials and Methods for details). These bands corresponded to RyR3 and GST-RyR3, as confirmed by immunoblotting (Fig. 1B). GST-RyR3 exhibits a lower level of expression and slower migration in the gel compared with RyR3. The lower-molecular-weight bands present in the RyR3 sample are the intact and degraded GST-FKBP12.6. No degradation of either RyR3 or GST-RyR3 was detected. These data indicate that the recombinant RyR3 and GST-RyR3 can be purified in one step by affinity chromatography.
Figure 1.
Purification and functional characterization of recombinant RyR3 and GST-RyR3. (A) Twenty-five microliters of purified recombinant RyR3 (glutathione-eluted) and GST-RyR3 were subjected to a 12% SDS/PAGE gel and stained with Coomassie brilliant blue. (B) A Western blot of 2 μl of purified RyR3 and GST-RyR3 by using the anti-RyR antibody 34C (Affinity BioRegents). (C) HEK293 cells were transfected with 10 μg of RyR3 or GST-RyR3 cDNA. Fluorescence intensity of fluo 3-loaded cells was measured before and after additions of 2.5 mM caffeine (●) and 50 μM ryanodine (○).
To examine whether GST-RyR3 is functional, we determined the response to caffeine and ryanodine of Ca2+ release in HEK293 cells transfected with the GST-RyR3 cDNA. As shown in Fig. 1C, addition of 2.5 mM caffeine to GST-RyR3-transfected cells induced a transient increase in the fluorescence signal. A further increase in fluorescence was observed on the subsequent addition of 50 μM ryanodine. These results are similar to those observed with RyR3 transfected cells (Fig. 1C). Neither caffeine- nor ryanodine-induced Ca2+ release was detected in HEK293 cells transfected with control (pCDNA3) vector DNA or no DNA (data not shown). Thus, these results demonstrate that, like RyR3, GST-RyR3 forms a functional Ca2+ release channel in HEK293 cells.
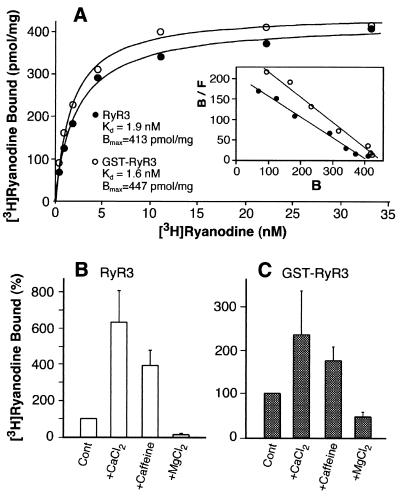
To determine further whether the purified recombinant RyR3 and GST-RyR3 retain functional activity and regulatory properties, we carried out [3H]ryanodine binding to the purified proteins. [3H]Ryanodine binding has been widely used as a functional assay for RyR channel activities, because ryanodine binds to only the open state of the channel (27, 31). Fig. 2A shows that both purified RyR3 and GST-RyR3 proteins are capable of binding [3H]ryanodine. The Kd and Bmax values are 1.9 nM and 413 pmol/mg protein for RyR3 and 1.6 nM and 447 pmol/mg protein for GST-RyR3, which are similar to those obtained with the purified native RyR3 proteins from diaphragm (20, 21). In addition, [3H]ryanodine binding to purified RyR3 and GST-RyR3 was activated by Ca2+ and caffeine and was inhibited by Mg2+ (Fig. 2 B and C). These results suggest that the purified recombinant RyR3 and GST-RyR3 are as functional and regulatable as native purified RyR3.
Figure 2.
Purified recombinant RyR3 and GST-RyR3 proteins exhibit regulated, high-affinity [3H]ryanodine binding. (A) Equilibrium [3H]ryanodine binding to purified RyR proteins was carried out as described in Materials and Methods. The purified GST-RyR3, which was from the same preparation that was used for cryo-EM and 3D reconstructions, exhibits a Kd of 1.6 nM and Bmax of 447 pmol/mg protein. Different preparations of purified RyR3 (glutathione-eluted) were used for [3H]ryanodine binding and for 3D reconstructions. The Kd and Bmax values for the RyR3 were 1.9 ± 0.17 nM and 413 ± 6.3 pmol/mg protein (n = 2). [3H]Ryanodine binding to purified RyR3 (B) and GST-RyR3 (C) was carried out in the presence of 150 nM free Ca2+ (control) or with 0.8 mM Ca2+, 2.5 mM caffeine, or 5 mM Mg2+. Data shown are mean ± average error from three separate experiments. All binding determinations were done in duplicate.
Cryo-EM of RyR3 and GST-RyR3.
Fig. 3 shows electron micrographs of frozen-hydrated, purified, recombinant RyR3 and GST-RyR3. Both micrographs show individual receptors with apparently intact structure. For GST-RyR3, aggregated particles were found often (white arrows in Fig. 3B). This could result from cross-linking among GST-RyR3 proteins through the N-terminal GST domain, which has the potential to dimerize (32, 33). In some of the aggregates, individual receptors were resolved (arrowheads in Fig. 3B), and in these instances the receptors frequently appeared to interact at or near their corners with adjoining receptors, indicating that the N-terminally fused GST moieties are localized at or near the corners of the RyR3's cytoplasmic region. Aggregated particles were seldom found for RyR3. All 3D reconstructions were computed by using images of isolated RyR3s that were not interacting with other receptors.
Figure 3.
Cryo-EM of RyR3 and GST-RyR3. Parts of micrographs of glutathione-eluted RyR3 (A) and GST-RyR3 (B) are shown embedded in a thin layer of vitreous ice. Some individual particles are marked with black arrows, and in B, the white arrows point to aggregated particles. The two arrowheads indicate a receptor in the 4-fold symmetric orientation that apparently is part of an oligomer. It appears to be connected to adjacent receptors by means of interactions near its corners. (Bar = 300Å.)
Compared with micrographs of RyR1 (11, 15), both expressed (Fig. 3) and native RyR3s (14) showed lower contrast and multiple orientations. The lower contrast may have been caused by the sucrose and lipid in the final EM sample buffer. The recombinant RyR3s in the thin layer of vitreous ice showed a higher frequency of non-four-fold symmetric views (Fig. 3) than we have observed for RyR1 in previous studies (11). This is an advantageous property for 3D reconstruction, but it was still necessary to collect some data with the specimen grid tilted by 30° to obtain an adequate sampling of orientations.
3D Reconstruction of RyR3 and GST-RyR3: Localization of the Amino Terminus of RyR3.
3D reconstructions of glutathione-eluted RyR3 (Fig. 4A), FK506-eluted RyR3 (Fig. 4B), and GST-RyR3 (Fig. 4C) are very similar to those determined previously for the three RyR isoforms purified from native tissues (11–14). The reconstructions consist of two major components: a larger cytoplasmic assembly (290 × 290 × 130Å) that is composed of at least 10 distinct domains (labeled by numerals) and a smaller transmembrane assembly (120 × 120 × 70Å, labeled “TA”).
Figure 4.
3D surface representation of RyR3s and GST-RyR3. (A) Glutathione-eluted RyR3 is shown in blue. (B) FK506-eluted RyR3 is shown in green. (C) GST-RyR3 is shown in yellow. The 3D reconstructions are shown in three different views. (Left) “Top” views of the cytoplasmic surface, which interacts with the transverse-tubule in muscle. (Center) “Bottom” views of the surface that would face the sarcoplasmic reticulum lumen. (Right) “Side” views. TA, transmembrane assembly. The numerals on the cytoplasmic assembly indicate the distinguishable domains as numbered previously (11).
Depending on the method of elution, RyR3 could be purified with or without GST-FKBP12.6. Because glutathione dissociates the GST-FKBP12.6/RyR3 complex from glutathione-Sepharose, glutathione-eluted RyR3 contains both RyR3 and GST-FKBP12.6. On the other hand, because FK506 dissociates RyR3 from GST-FKBP12.6 bound to glutathione-Sepharose, FK506-eluted RyR3 should be devoid of GST-FKBP12.6. One might expect then that the glutathione-eluted RyR3 would have GST-FKBP12.6 bound to it, which would contribute additional density to its 3D map. This apparently is not the case. Comparison of the 3D reconstructions of glutathione-eluted RyR3 (Fig. 4A) and of FK506-eluted RyR3 (Fig. 4B) does not reveal any significant differences in either the known FKBP-binding site (between domains 3 and 9, as located on RyR1; ref. 15) or anywhere else. This result is not entirely unexpected because there is evidence that RyR3 does not bind FKBP with high affinity (21, 34). Thus, the interaction of GST-FKBP12.6 with RyR3 apparently is not strong enough to be preserved after elution and cryo-EM under our experimental conditions.
Comparison of the reconstructions of the recombinant RyR3 and GST-RyR3 revealed some structural differences. In Fig. 5A, the two reconstructions (Fig. 4 A and C) are aligned and superimposed. The 3D volume of RyR3 is shown in solid blue and that of GST-RyR3 is shown in transparent yellow. The main difference is located in each of the corners of the cytoplasmic assembly (the so-called “clamp” regions), in the middle of the region defined by domains 7–10 (indicated with a red star). There are some other, smaller differences of less certain significance (e.g., in or near domains 1, 3, and 10).
Figure 5.
Comparison of RyR3 and GST-RyR3. (A) 3D reconstructions (RyR3 in solid blue and GST-RyR3 in transparent yellow) that were mutually aligned and superimposed. A red star in one of the clamp regions indicates the major difference between the two reconstructions. (B) Difference map. The differences (shown in red) were obtained by subtracting the 3D volume of the RyR3 from the GST-RyR3 reconstruction.
Comparison of the two surface representations shown in Fig. 5A may give a misleading impression of the significance of differences between them because the magnitudes of the differences are not indicated. Therefore, we examined the differences in more detail by subtracting the 3D volume of RyR3 from that of GST-RyR3. The difference map shown in Fig. 5B (red regions) confirms that the excess density present in the clamps of GST-RyR3 is indeed the most significant difference from RyR3. The absolute values of the density differences in this region are substantially higher than the threshold density used to create the surface representations (Fig. 4), and the volume of the region at this density threshold matches approximately that expected for GST [based on Mr ≈ 26 (32) and assuming a density of 1.37 g/cm3 (11)]. We attribute the density present in the clamp regions of the GST-RyR3 to the GST, and, therefore, the amino terminus of RyR3 must lie in the clamps.
A minor difference, located in the vicinity of domain 3 (Fig. 5B Left), appears to represent only about one-fourth of the volume corresponding to the main difference in the clamp regions. It seems unlikely that this region corresponds to the GST portion of GST-RyR3. Its small size indicates that it is near the limit of detection, and its significance therefore is uncertain. Furthermore, a region of negative difference was present nearby (not shown), indicating that if this difference is significant, it more likely represents a conformational change rather than the net addition of mass contributed by the GST (in this context, it is important to point out that only a positive difference was detected in the clamp region). If this is so, then the minor difference possibly was caused by the 10-residue c-myc insertion beginning at Glu-4318 and/or the N terminus-fused GST. It is highly unlikely that the main difference in the clamp region is due to the c-myc insertion. An insertion of this size is not expected to be detectable at the resolution achieved in this study. Furthermore, we recently have purified wild-type RyR2 and RyR2 containing the same c-myc insertion as present in GST-RyR3. The 3D reconstructions essentially were identical, and structural alterations resulting from the c-myc insertion were not detectable. More importantly, no differences were found in the clamp regions, which confirms our interpretation that the excess density present in the clamps of GST-RyR3 is due to the GST rather than to the c-myc insert (Z.L., J.Z., S.R.W.C., and T.W., unpublished results).
Discussion
This study describes the purification of functional recombinant RyRs that are suitable for structural characterization by cryo-EM. All previous 3D reconstructions have been carried out on RyRs that were purified from native tissues. It now should be feasible to genetically modify the amino acid sequence of the RyR and to determine the effects of such modifications on RyR structure by cryo-EM. As an example of this type of application, we have localized the amino termini of the RyR3 on the receptor's 3D structure by determining a reconstruction of a modified RyR3 in which a GST domain is fused to its N terminus.
Previously, cryo-EM has been used to determine the binding locations of RyR modulators such as calmodulin and FKBP and to identify RyR domains that undergo movements associated with channel opening and closing (14–18). The amino acid residues involved in these processes, however, have yet to be identified. One way to obtain this information is to determine reconstructions of RyRs in complex with antibodies whose epitopes have been characterized (35). However, RyR is an extremely large membrane protein containing ≈5,000 aa residues per protomer; to comprehensively map the sequence for surface-exposed regions using sequence-specific antibodies would be an arduous undertaking. Alternatively, as illustrated in this paper, it is possible to add sufficient protein mass at a defined location in the sequence to allow its detection in 3D reconstructions. This approach recently has been used successfully in the studies of some large, multisubunit complexes (36, 37).
The GST-RyR3 construct used in this study was engineered to contain, after residue Glu-4318, a 10-residue insertion corresponding a well characterized epitope derived from the protein, c-myc. Little, if any, perturbation of the receptor's structure by the insertion was apparent in the 3D reconstruction at the resolution achieved (≈35Å), and the pharmacology and calcium-release activity were retained (Figs. 1C and 2C). We are encouraged by these results because they suggest a strategy for systematically mapping the locations of surface-exposed segments from RyR's sequence. It should be feasible to isolate a set of recombinant RyRs containing c-myc insertions at predicted surface-exposed sites and to then form immunocomplexes with anti-c-myc antibodies (or Fab fragments). These immunocomplexes then could be analyzed by cryo-EM to locate the antibodies and, thus, the location of the insertion. This approach is perhaps more suitable for localizing surface-exposed regions formed by sequences that are not at the amino or carboxyl termini, where insertion of a large domain such as GST may disrupt the native structure.
Although RyR3 is capable of interacting with GST-FKBP12.6 biochemically, we were unable to detect bound GST-FKBP12.6 in the 3D reconstruction of RyR3. The 3D reconstructions of RyR3 in the presence and absence of GST-FKBP12.6 are essentially identical (Fig. 4 A and B). Apparently, under our experimental conditions, GST-FKBP12.6/RyR3 complexes did not remain associated after their elution with glutathione or when they are applied to a grid and frozen for cryo-EM. Dissociation of FKBPs from RyR1 and RyR3 also has been reported when the RyRs were purified by antibody affinity chromatography (21). Thus, the differences observed in the 3D reconstructions between RyR3 and GST-RyR3 are most unlikely to result from the addition of GST-FKBP12.6 to the RyR3 structure. Furthermore, had this been the case, the addition of GST-FKBP12.6 to the RyR3 structure would have produced a negative volume change rather than a positive one in the difference map (Fig. 5B), which was formed by subtracting the volume of RyR3 from that of GST-RyR3 (VGST-RyR3 − VRyR3).
The localization of the RyR3 N terminus reported here is of particular interest because a number of mutations that cause the skeletal muscle diseases, malignant hyperthermia, and central core disease have been found to involve amino acid residues spanning residues 38–614 of the RyR1 sequence (38). It seems likely that these residues lie within the domains comprising the clamp regions, based on our finding that N-terminally fused GST lies approximately in the center of the cluster of domains that comprise the clamps. Intriguingly, the clamp structures have been shown previously to possess different conformations in the open and closed states of the channel (17). Note that RyR1 and RyR3 are sufficiently similar to make such correlations among the isoforms. Thus, mutations in this region may affect conformational changes associated with channel gating. By the same token, insertion of the GST domain into this critical region would be likely to have some impact on channel gating, which may explain some observed differences in the extent of caffeine-induced Ca2+ release (Fig. 1C) and in the degree of Ca2+ and caffeine activation and Mg2+ inhibition between RyR3 and GST-RyR3 (Fig. 2 B and C).
In conclusion, RyR3, which is the most difficult RyR isoform to purify from natural sources, can be expressed and purified in sufficient quantities for biochemical and structural characterization. 3D reconstructions at (34 Å)−1 resolution of the recombinant RyR3s showed structures essentially identical to those of native RyR3. Comparison of the 3D reconstruction of GST-RyR3 with that of RyR3 reveals that the amino terminus of RyR3 lies in the clamp regions of the cytoplasmic assembly. The methodology developed in this study should be useful for mapping the receptor's linear sequence and functional domains onto the 3D structure of RyR, thereby providing insights into the structural basis of RyR function.
Acknowledgments
This work was supported by the Muscular Dystrophy Association and a National Institutes of Health grant AR40615 to T.W. and by research grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Alberta to S.R.W.C. The authors gratefully acknowledge use of the Wadsworth Center's electron microscopy core facility and the computing facilities of the Center's Resource for Visualization of Biological Complexity (National Institutes of Health National Biotechnological Resource Grant RR01219 from the National Center for Research Resources).
Abbreviations
- RyR3
type 3 ryanodine receptor
- 3D
three-dimensional
- GST
glutathione S-transferase
- EM
electron microscopy
- FKBP
FK506-binding protein
- FKBP12.6
12.6-kDa FKBP
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Clapham D E. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 2.Berridge M J. J Physiol. 1997;499:291–306. doi: 10.1113/jphysiol.1997.sp021927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzini-Armstrong C, Protasi F. Physiol Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 4.Sutko J L, Airey J A. Physiol Rev. 1996;76:1027–1071. doi: 10.1152/physrev.1996.76.4.1027. [DOI] [PubMed] [Google Scholar]
- 5.Giannini G, Conti A, Mammarella S, Scrobogna M, Sorrentino V. J Cell Biol. 1995;128:893–904. doi: 10.1083/jcb.128.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorrentino V, Volpe P. Trends Pharmacol Sci. 1993;14:98–103. doi: 10.1016/0165-6147(93)90072-r. [DOI] [PubMed] [Google Scholar]
- 7.Sorrentino V, Reggiani C. Trends Cardiovasc Med. 1999;9:54–61. doi: 10.1016/s1050-1738(99)00003-1. [DOI] [PubMed] [Google Scholar]
- 8.Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, et al. Nature (London) 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- 9.Otsu K, Willard H F, Khanna V K, Zorzato F, Green N M, MacLennan D H. J Biol Chem. 1990;265:13472–13483. [PubMed] [Google Scholar]
- 10.Hakamata Y, Nakai J, Takeshima H, Imoto K. FEBS Lett. 1992;312:229–235. doi: 10.1016/0014-5793(92)80941-9. [DOI] [PubMed] [Google Scholar]
- 11.Radermacher M, Rao V, Grassucci R, Frank J, Timerman A P, Fleischer S, Wagenknecht T. J Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serysheva I I, Orlova E V, Chiu W, Sherman M B, Hamilton S L, van Heel M. Nat Struct Biol. 1995;2:18–24. doi: 10.1038/nsb0195-18. [DOI] [PubMed] [Google Scholar]
- 13.Sharma M R, Penczek P, Grassucci R, Xin H-B, Fleischer S, Wagenknecht T. J Biol Chem. 1998;273:18429–18434. doi: 10.1074/jbc.273.29.18429. [DOI] [PubMed] [Google Scholar]
- 14.Sharma M R, Jeyakumar L H, Fleischer S, Wagenknecht T. J Biol Chem. 2000;275:9485–9491. doi: 10.1074/jbc.275.13.9485. [DOI] [PubMed] [Google Scholar]
- 15.Wagenknecht T, Radermacher M, Grassucci R, Berkowitz J, Xin H-B, Fleischer S. J Biol Chem. 1997;272:32463–32471. doi: 10.1074/jbc.272.51.32463. [DOI] [PubMed] [Google Scholar]
- 16.Samso M, Trujillo R, Gurrola G B, Valdivia H H, Wagenknecht T. J Cell Biol. 1999;146:493–499. doi: 10.1083/jcb.146.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlova E V, Serysheva I I, van Heel M, Hamilton S L, Chiu W. Nat Struct Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 18.Serysheva I I, Schatz M, van Heel M, Chiu W, Hamilton S L. Biophys J. 1999;77:1936–1944. doi: 10.1016/S0006-3495(99)77035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murayama T, Ogawa Y. J Biol Chem. 1997;272:24030–24037. doi: 10.1074/jbc.272.38.24030. [DOI] [PubMed] [Google Scholar]
- 20.Jeyakumar L H, Copello J A, O'Malley A M, Wu G-M, Grassucci R, Wagenknecht T, Fleischer S. J Biol Chem. 1998;273:16011–16020. doi: 10.1074/jbc.273.26.16011. [DOI] [PubMed] [Google Scholar]
- 21.Murayama T, Oba T, Katayama E, Oyamada H, Oguchi K, Kobayashi M, Otsuka K, Ogawa Y. J Biol Chem. 1999;274:17297–17308. doi: 10.1074/jbc.274.24.17297. [DOI] [PubMed] [Google Scholar]
- 22.Moore R A, Nguyen H, Galceran J, Pessah I N, Allen P D. J Cell Biol. 1998;140:843–851. doi: 10.1083/jcb.140.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhat M B, Zhao J, Zang W-J, Balke C W, Takeshima H, Wier W G, Ma J J. J Gen Physiol. 1997;110:749–762. doi: 10.1085/jgp.110.6.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S R W, Li X, Ebisawa K, Zhang L. J Biol Chem. 1997;272:24234–24246. doi: 10.1074/jbc.272.39.24234. [DOI] [PubMed] [Google Scholar]
- 25.Chen S R W, Ebisawa K, Li X, Zhang L. J Biol Chem. 1998;273:14675–14678. doi: 10.1074/jbc.273.24.14675. [DOI] [PubMed] [Google Scholar]
- 26.Nakai J, Sekiguchi N, Rando T A, Allen P D, Beam K G. J Biol Chem. 1998;273:13403–13406. doi: 10.1074/jbc.273.22.13403. [DOI] [PubMed] [Google Scholar]
- 27.Du G G, Imredy J P, MacLennan D H. J Biol Chem. 1998;273:33259–33266. doi: 10.1074/jbc.273.50.33259. [DOI] [PubMed] [Google Scholar]
- 28.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 29.Penczek P A, Grassucci R A, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra A, Penczek P, Agrawal R K, Gabashvilli I S, Grassucci R A, Jünemann R, Burkhardt N, Nierhaus K H, Frank J. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 31.Inui M, Saito A, Fleischer S. J Biol Chem. 1987;262:1740–1747. [PubMed] [Google Scholar]
- 32.Walker J, Crowley P, Moreman A D, Barrett J. Mol Biochem Parasitol. 1993;61:255–264. doi: 10.1016/0166-6851(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 33.Henderson R M, Schneider S, Li Q, Hornby D, White S J, Oberleithner H. Proc Natl Acad Sci USA. 1996;93:8756–8760. doi: 10.1073/pnas.93.16.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fessenden J D, Wang Y M, Moore R A, Chen S R W, Allen P D, Pessah I N. Biophys J. 2000;79:2509–2525. doi: 10.1016/S0006-3495(00)76492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benacquista B L, Sharma M R, Samso M, Zorzato F, Treves S, Wagenknecht T. Biophys J. 2000;78:1349–1358. doi: 10.1016/S0006-3495(00)76689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Steven A C. Proc Natl Acad Sci USA. 1998;95:14622–14627. doi: 10.1073/pnas.95.25.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spahn C M T, Grassucci R, Penczek P, Frank J. Structure. 1999;7:1567–1573. doi: 10.1016/s0969-2126(00)88347-1. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann-Horn F, Jurkat-Rott K. Physiol Rev. 1999;79:1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]