Abstract
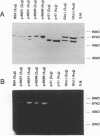
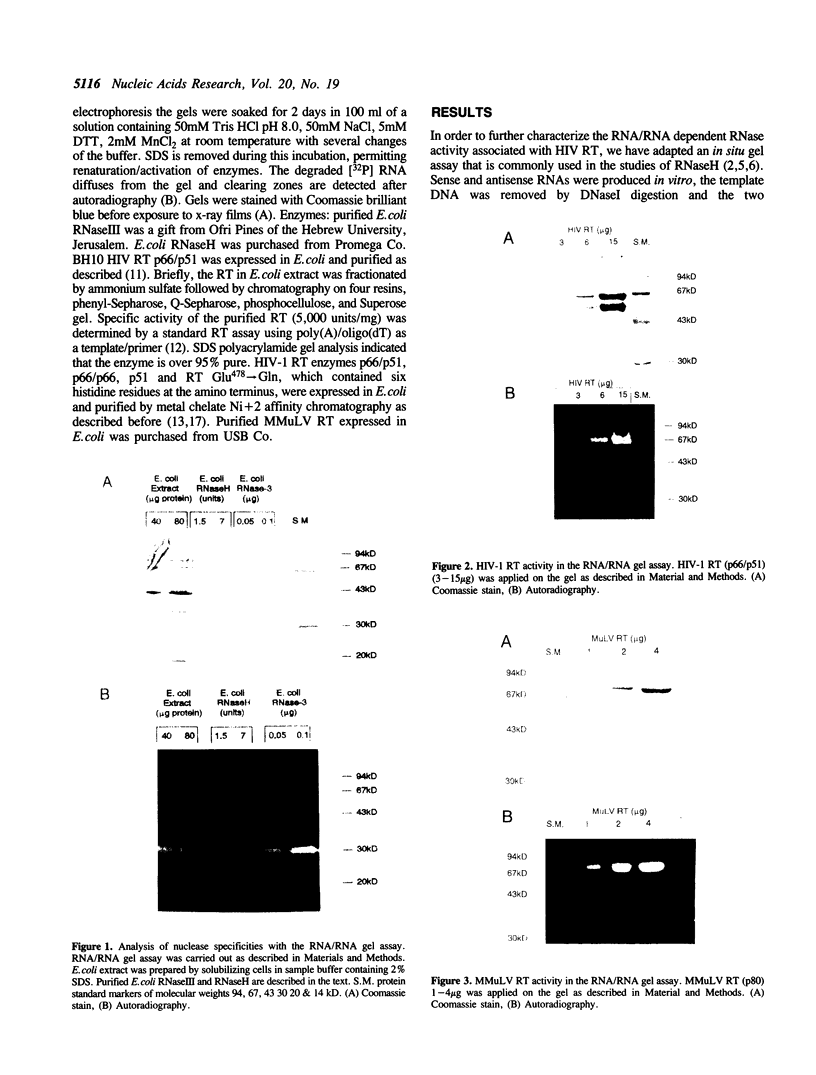
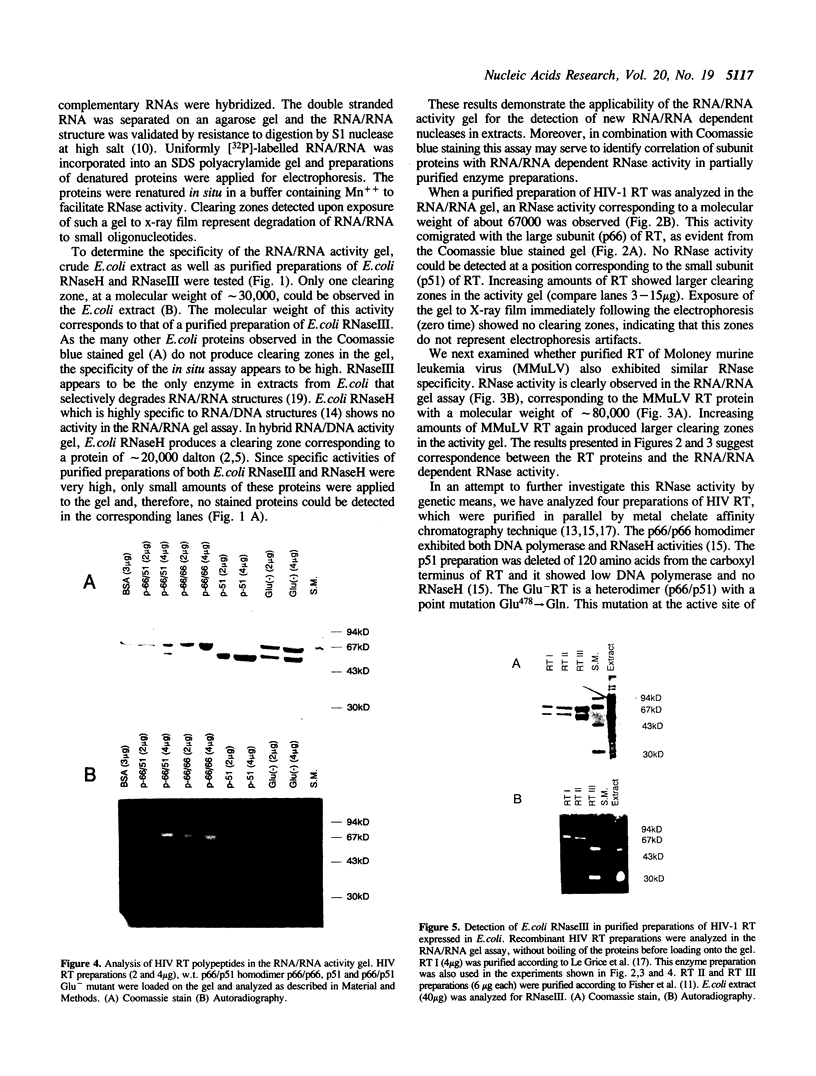
An in situ gel assay was applied to the study of double stranded RNA dependent RNase activity associated with reverse transcriptase (RT) of HIV-1 and murine leukemia virus. Polyacrylamide gels containing [32P] RNA/RNA substrate were used for electrophoresis of proteins under denaturing conditions. The proteins were renatured and in situ enzymatic degradation of 32P-RNA/RNA was followed. E. coli RNaseIII, but not E. coli RNaseH, was active in this in situ gel assay, indicating specificity of the assay to RNA/RNA dependent nucleases. Analysis of purified preparations of HIV-1 RT p66/p51 expressed in E. coli demonstrated an RNA/RNA dependent RNase activity comigrating with the large subunit (p66) of the enzyme. In addition, this activity of the RT was often accompanied by a contaminating RNA/RNA dependent RNase, with a molecular weight approximately 30,000 dalton identical to that of E. coli RNaseIII. As the p51 small subunit of HIV-1 RT and a mutant of RT p66/p51, at Glutamic acid #478, did not exhibit RNA/RNA dependent RNase activity, at least part of the active site of the RNA/RNA dependent RNase activity appeared to reside at the carboxy end of the molecule. As these RT proteins are also deficient of RNaseH, our results suggest overlapping or identical catalytic sites for degradation of the substrates RNA/DNA and RNA/RNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Artzi H., Zeelon E., Gorecki M., Panet A. Double-stranded RNA-dependent RNase activity associated with human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):927–931. doi: 10.1073/pnas.89.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P. L., Ferris A. L., Hughes S. H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992 Feb;66(2):1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991 Apr 5;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Hizi A., Barber A., Hughes S. H. Effects of small insertions on the RNA-dependent DNA polymerase activity of HIV-1 reverse transcriptase. Virology. 1989 May;170(1):326–329. doi: 10.1016/0042-6822(89)90389-9. [DOI] [PubMed] [Google Scholar]
- Hizi A., Hughes S. H., Shaharabany M. Mutational analysis of the ribonuclease H activity of human immunodeficiency virus 1 reverse transcriptase. Virology. 1990 Apr;175(2):575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Hizi A., McGill C., Hughes S. H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman A. D., Banapour B., Levy J. A. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology. 1985 Dec;147(2):326–335. doi: 10.1016/0042-6822(85)90135-7. [DOI] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Hudson G. O., Moomaw E. W., Nodes B. R. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Iwai S., Ohtsuka E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987 May 11;215(2):327–330. doi: 10.1016/0014-5793(87)80171-0. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990 Jan 26;187(2):307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Gonzalez M. A. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res. 1990 Jun 11;18(11):3293–3298. doi: 10.1093/nar/18.11.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]