Abstract
Fulvestrant is a selective estrogen receptor downregulator (SERD) and highly effective antagonist to hormone-sensitive breast cancers following failure of previous tamoxifen or aromatase inhibitor therapies. However, after prolonged fulvestrant therapy, acquired resistance eventually occurs in the majority of breast cancer patients, due to poorly understood mechanisms. To examine a possible role(s) of aberrantly expressed microRNAs (miRNAs) in acquired fulvestrant resistance, we compared antiestrogen-resistant and -sensitive breast cancer cells, revealing the over-expression of miR-221/222 in the SERD-resistant cell lines. Fulvestrant treatment of estradiol (E2)- and fulvestrant-sensitive MCF7 cells resulted in increased expression of endogenous miR-221/222. Ectopic upregulation of miR-221/222 in estrogen receptor-α (ERα)-positive cell lines counteracted the effects of E2 depletion or fulvestrant-induced cell death, thus also conferring hormone-independent growth and fulvestrant resistance. In cells with acquired resistance to fulvestrant, miR-221/222 expression was essential for cell growth and cell cycle progression. To identify possible miR-221/222 targets, miR-221- or miR-222- induced alterations in global gene expression profiles and target gene expression at distinct time points were determined, revealing that miR-221/222 overexpression resulted in deregulation of multiple oncogenic signaling pathways previously associated with drug resistance. Activation of β-catenin by miR-221/222 contributed to estrogen-independent growth and fulvestrant resistance, whereas TGF-β-mediated growth inhibition was repressed by the two miRNAs. This first in-depth investigation into the role of miR-221/222 in acquired fulvestrant resistance, a clinically important problem, demonstrates that these two ‘oncomirs’ may represent promising therapeutic targets for treating hormone-independent, SERD-resistant breast cancer.
Keywords: microRNAs, breast cancer, antiestrogen, drug resistance, fulvestrant
Introduction
Estrogen receptor-α (ERα), the primary mediator of estrogen action, plays a pivotal role in the development and progression of breast cancer (Ariazi et al., 2006). As over 70% of breast cancers overexpress ERα, targeting the receptor by endocrine therapy is considered the most important treatment for patients with ER-positive breast tumors (Ariazi et al., 2006). Tamoxifen, a selective estrogen receptor modulator (Jordan, 2003), and aromatase inhibitors (AIs), which block estrogen synthesis (Smith and Dowsett, 2003), are effective first-line endocrine therapies that significantly improve relapse-free and overall survival of all stages of ER-positive breast cancer (Nicholson and Johnston, 2005). However, acquired resistance to both tamoxifen and AI typically develops after prolonged treatment in a majority of initially responsive breast cancers (Normanno et al., 2005), and nearly 50% of the advanced ER-positive breast cancer patients do not respond to tamoxifen or AI in the first-line setting (Normanno et al., 2005).
Resistance to AIs and tamoxifen is associated with activation of several growth factor signaling pathways, including human epidermal growth factor receptor 2 (HER2) and insulin-like growth factor I receptor, which ‘cross-talk’ with the ERα signaling pathway, resulting in an activation of various mitogen-activated protein kinases (MAPKs) and PI3K/AKT involved in cell survival and proliferation (Ali and Coombes, 2002; Nicholson and Johnston, 2005; Normanno et al., 2005). As the transcriptional program of ERα continues to play a critical role in these acquired resistance mechanisms, second-line endocrine therapies, using alternative ERα-inhibitory mechanisms, have been developed (McDonnell, 2006). One of these, the pure steroidal antiestrogen fulvestrant (Faslodex, ICI 182,780, Astra-Zeneca Corp., London, UK), completely suppresses ERα activity, inactivating both ERα-mediated genomic and non-genomic signaling, and is now approved for the treatment of postmenopausal women following failure of previous antiestrogen therapy (tamoxifen and AIs) (Howell, 2006a). However, despite its potent antitumor effects, fulvestrant does not circumvent the development of antiestrogen resistance (Howell, 2006b). We (Fan et al., 2006) and others (Nicholson et al., 2005; Normanno et al., 2005) have shown that acquired resistance to fulves-trant is an ERα-independent phenomenon, involving constitutive activation of autocrine-regulated growth-stimulatory pathways, ultimately dissociating breast cancer cells from ERα-mediated growth, although the mechanism(s) of upregulation of these mitogenic pathways remains unknown.
MicroRNAs (miRNAs) are an abundant class of endogenous small non-coding RNAs (20–22 nucleotides in length) that regulate crucial biological processes, including development, differentiation, apoptosis and proliferation (Bushati and Cohen, 2007). Imperfect pairing of miRNAs with the 3′ untranslated region of target mRNAs generally results in their destabilization or ribosomal blockade (Bushati and Cohen, 2007). Deregulation of miRNAs is now considered a hallmark of cancer (Calin and Croce, 2006), in which they can function as oncogenes or tumor suppressors (Garzon et al., 2009). In breast cancer, miR-125b, -145, -21 and -155 have been shown the most significantly deregulated miRNAs (Iorio et al., 2005) and miR-7, -128a, -210, and -516-3p have been strongly associated with cancer progression (Foekens et al., 2008). Furthermore, miR-10b may play a role in breast cancer invasion and metastasis (Ma et al., 2007), and miR-206 is inversely correlated with ERα expression in human breast tumors and appears to target ERα (Adams et al., 2007; Kondo et al., 2008). Estradiol (E2)-regulated miRNA signatures, and miRNA regulation of ERα and its downstream transcriptional response to E2 have also recently been reported (Bhat-Nakshatri et al., 2009; Castellano et al., 2009), and an important role for miRNAs in ERα biology is beginning to emerge (Klinge, 2009).
Although altered expression of miRNAs in breast cancer has been widely reported (Iorio et al., 2005, 2008; Heneghan et al., 2009), the functional consequences of aberrant expression of specific miRNAs in antiestrogen-resistant disease remain unclear. Overexpression of miR-221 and miR-222 has been observed in a number of advanced malignancies (Galardi et al., 2007; Gillies and Lorimer, 2007; Visone et al., 2007; Felicetti et al., 2008; Fornari et al., 2008; Garofalo et al., 2009; Pineau et al., 2010), including ERα-negative primary tumors (Zhao et al., 2008), and has also been associated with tamoxifen resistance (Miller et al., 2008). Our previous miRNA microarray data also indicated miR-221/222 upregulation in acquired fulvestrant resistance (Xin et al., 2009). In the current study, we used antiestrogen-sensitive and -resistant breast cancer cell line models to comprehensively investigate the role of miR-221/222 in hormone-independent growth and acquired resistance to fulvestrant. miR-221/222-transfected cells demonstrated that global gene expression changes associated with multiple (ER-independent) growth-promoting, oncogenic pathways are critical for acquired selective estrogen receptor downregulator (SERD) resistance. We further validated that miR-221/222 activated β-catenin signaling and repressed TGF-β-signaling-induced growth inhibition. These results indicate vital, multi-functional roles of miR-221/222 in the acquisition of fulvestrant-resistant breast cancer.
Results
miR-221/222 expression is increased in antiestrogen-resistant breast cancer cell lines and is upregulated by fulvestrant in antiestrogen-sensitive MCF7 cells
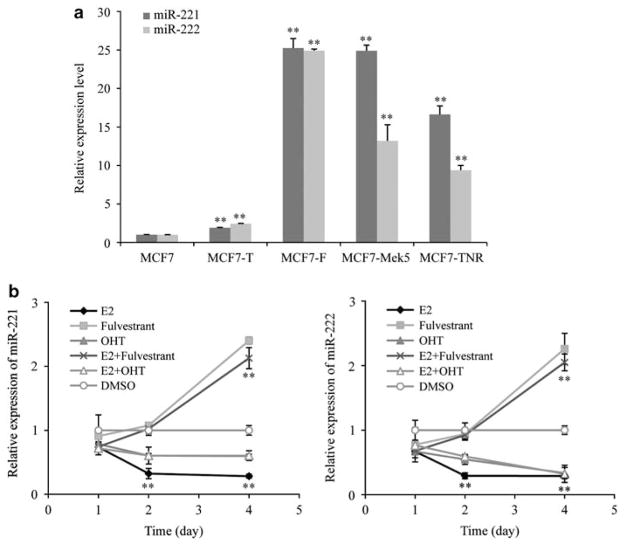
To investigate the association between miR-221/222 expression and antiestrogen resistance, four different antiestrogen-resistant human breast cancer cell lines were compared, each derived by different mechanisms. Tamoxifen-resistant (MCF7-T) and fulvestrant-resistant (MCF7-F) MCF7 sublines were established by prolonged growth of MCF7 cells in tamoxifen- or fulvestrant-containing media, respectively (Fan et al., 2006). MCF7-T cells remained responsive to fulvestrant but with reduced drug sensitivity compared with the parental MCF7 cells, whereas MCF7-F cells were fulvestrant-resistant and also cross-resistant to tamoxifen. The expression profile analysis indicated that subsets of clinically relevant genes (prognostic markers, clinical outcome and ERα signature genes) are altered in these two cell lines (Fan et al., 2006), making them valuable models for investigating the molecular events underlying the development of antiestrogen resistance. Tumor necrosis factor-resistant (MCF7-TNR) cells were obtained by prolonged exposure of MCF7 cells to increasing concentrations of tumor necrosis factor-α (Weldon et al., 2004), whereas MCF7-Mek5 cells were established by stably transfecting MCF7 cells with constitutively active mitogen-activated protein kinase kinase 5 (MEK5) construct (Zhou et al., 2008). Both the MCF7-TNR and -Mek5 cell lines were resistant to tumor necrosis factor-α-(Zhou et al., 2008) and fulvestrant-induced apoptosis (unpublished data). Real-time qRT–PCR results revealed upregulation of miR-221/222 in MCF7-T cells (Figure 1a), consistent with a previous study (Miller et al., 2008). However, in the fulvestrant-resistant cells (MCF7-F, MCF7-Mek5 and MCF7-TNR), miR-221/222 were markedly overexpressed (P<0.01, versus MCF7-T) (Figure 1a), suggesting a stronger association of miR-221/222 with pure anti-estrogen resistance.
Figure 1.
Upregulation of miR-221/222 in breast cancer cell lines. (a) miR-221/222 expression in four antiestrogen-resistant MCF7 cell lines as measured by qRT–PCR analysis and relative to their expression level in parental MCF7 cells (mean±s.e., n = 3). **P<0.01. The following t-test results were obtained: MCF7-F, MCF7-Mek5 or MCF7-TNR versus MCF7-T, P<0.002. (b) Effect of E2, tamoxifen and fulvestrant on endogenous miR-221 (left panel) and miR-222 (right panel) expression. MCF7 cells were serum/E2 starved for 3 days and then treated with 10 nM 17β-estradiol (E2), 1 μM 4-hydroxytamoxifen (OHT), 100 nM fulvestrant alone or in the specified combinations, and cells were collected at the indicated time points. qRT–PCR was performed to determine miR-221/222 expression, relative to those in dimethyl sulfoxide (DMSO)-treated cells (mean±s.e., n = 3). **P<0.01.
Considering the different mechanisms of tamoxifen and fulvestrant action, we next examined the effects of these two drugs on endogenous miR-221/222 expression. To avoid the effects of medium-derived estrogen, MCF7 cells were cultured in estrogen-free medium for 3 days and then treated with different drug combinations. Although expression of miR-221/222 was not significantly altered by 17β-estradiol (E2), 4-hydroxytamoxifen (OHT) or E2 combined with OHT during the first 24 h, these drug treatments actually repressed (P<0.01) miR-221/222 expression at subsequent time points (48–96 h; Figure 1b). By contrast, fulvestrant, alone or in combination with E2, increased (P<0.01) endogenous miR-221/222 expression after 48-h treatment (Figure 1b). As miR-221/222 were overexpressed in fulvestrant-resistant cell lines (Figure 1a), these results suggest that fulvestrant treatment (or removal of ERα) might initially induce miR-221/222 expression during the development of SERD resistance.
miR-221/222 confer estrogen-independent growth and fulvestrant resistance
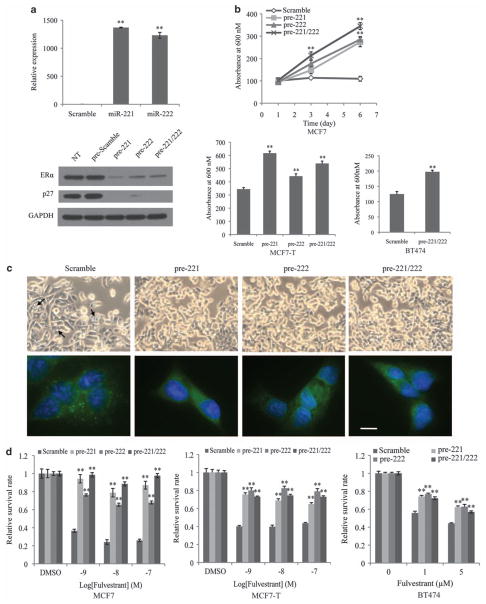
As miR-221/222 expression was induced by fulvestrant and associated with fulvestrant resistance, we next investigated the role of miR-221/222 in the acquisition of fulvestrant resistance. Fulvestrant-sensitive MCF7, MCF7-T and BT474 breast cancer cells were transfected with pre-miRNA to ectopically overexpress miR-221/222, as confirmed by qRT–PCR and immunoblotting analysis for suppressed p27Kip1 and ERα protein levels, two well-known targets of these miRNAs (Galardi et al., 2007; Zhao et al., 2008) (Figure 2a). Although the growth of MCF7 cells was retarded in estrogen-free medium, the proliferation of miR-221 and/or miR-222-overexpressing MCF7 cells was significantly (P<0.01) accelerated (Figure 2b), which was further confirmed using MCF7-T and BT474 cell lines (Figure 2b). After E2 depletion for four days, the scramble control siRNA-treated MCF7 cells exhibited previously described morphological signs of cellular damage (Gozuacik and Kimchi, 2004; Miller et al., 2008), which were not observed in miR-221 and/or miR-222-overexpressing MCF7 cells (Figure 2c, upper panel, arrows indicated). Immunofluorescence staining with anti-LC3B (a marker of autophagy) antibody revealed punctated patterns of LC3 immunofluorescence in scramble control-treated MCF7 cells, whereas LC3 immunofluorescence in miR-221 and/or miR-222-overexpressing cells was diffused (Figure 2c, lower panel). In addition, MCF7 cells overexpressing miR-221 and/or miR-222 displayed a rounded phenotype and were loosely attached to the culture surface (Figure 2c), morphologic characteristics of the aggressive MCF7-F cells (Fan et al., 2006).
Figure 2.
Ectopic expression of miR-221/222 increases ER-independent growth and confers resistance to fulvestrant. (a) Ectopic expression of miR-221/222 in MCF7 cells. MCF7 cells were transiently transfected with 50 nM pre-miRNA precursors. Following a 72-h incubation, cells were subjected to qRT–PCR (upper panel) and immunoblotting with anti-ERα, p27Kip1 and GAPDH antibodies (lower panel). The miR-221/222 levels in scramble control-treated MCF7 cells were normalized as 1 (mean±s.e., n = 3). **P<0.01. (b) ER-independent growth rate of pre-miRNA-transfected cells (MCF7, MCF7-T and BT474). Cells were seeded in E2-free media one day before transfection. MTT assay was performed to determine cell numbers at the indicated time points (absorbance at 600 nM linearly correlated with cell number; mean±s.e., n = 6) **P<0.01. (c) Cell morphology changes in pre-miRNA-transfected MCF7 cells. One day after E2 starvation, MCF7 cells were transfected with pre-221/222 and further cultured in E2-free media for 3 days, followed by phase-contrast microphotography (magnification ×20 objective) and fluorescence microscopy (magnification ×60 oil immersion objective. Bar, 10 μM). (d) Fulvestrant sensitivity of pre-miRNA-transfected cells (MCF7, MCF7-T and BT474). Cells were maintained in E2-free medium containing indicated doses of fulvestrant for 7 days. Relative cell growth rates (drug versus vehicle) were determined by MTT assay (mean±s.e., n = 6). **P<0.01.
To further examine miR-221/222 effects on fulvestrant sensitivity, hormone-sensitive MCF7 cells were treated with different doses of fulvestrant for 7 days, showing strong growth inhibition in the control-transfected cells (Figure 2d, left panel); however, that growth inhibition was markedly blunted in the miR-221 and/or miR-222-overexpressing cells (Figure 2d, Supplementary Figure S1). Ectopically overexpressing miR-221/222 in MCF7-T cells (which have a slightly higher miR-221/222 expression than MCF7 (Figure 1a) but are still sensitive to fulvestrant) also significantly decreased fulvestrant sensitivity (Figure 2d middle panel). Similar results were observed in BT474 cells, which have low miR-221/222 expression (data not shown) but are less fulvestrant sensitive than MCF7 cells (Figure 2d right panel). Immunoblotting analysis (Figure 2a and Supplementary Figure S2) further suggested that targeting p27Kip1 may be one of the mechanisms by which miR-221/222 confers the ERα-independent, fulvestrant-resistant phenotype. Taken together, these results indicate that miR-221/222 over-expression in ERα-positive cell lines counteracts the effects of E2 depletion or fulvestrant-induced cell death, thus also conferring hormone-independent growth and fulvestrant resistance.
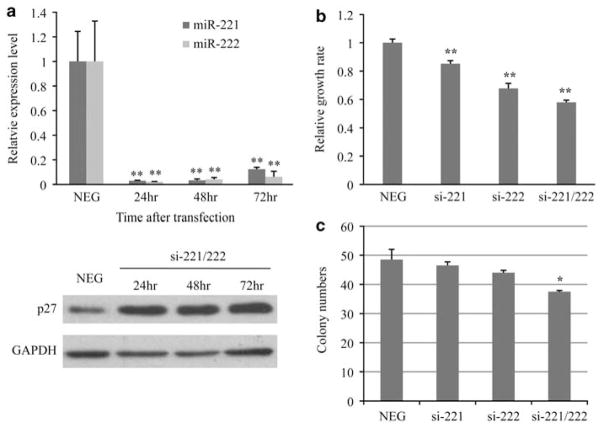
Knockdown of miR-221/222 in MCF7-F cells inhibits cell proliferation
To further investigate the role of miR-221/222 in fulvestrant resistance, MCF7-F cells were transfected with 2′-O-Me-221 (si-221) and 2′-O-Me-222 (si-222), or control 2′-O-Me-eGFP (NEG) antagomirs. After 24 h, miR-221 and miR-222 expression was dramatically reduced in the si-221/222-transfected MCF7-F cells, and p27Kip1 protein levels were restored (Figure 3a; Supplementary Figure S3). BrdU cell proliferation assays indicated that si-221 or si-222 significantly decreased MCF7-F cell growth by decreasing the cell population in S-phase, with si-222 demonstrating stronger inhibitory activity (Figure 3b), perhaps due to cross-inhibition of miR-221 by si-222 (Supplementary Figure S4). Cotransfection of si-221 and si-222 resulted in further inhibition of cell proliferation (Figure 3b). As clonogenic activity of MCF7-F cells was greater than the parental MCF7 cells (Supplementary Figure S5), we then examined the role of miR-221/222 in the increased clonogenicity. MCF7-F cells were cotransfected with antagomirs and a Block-iT Fluorescence Oligo (Invitrogen Corp., Carlsbad, CA, USA) for sorting transfected cells for subsequent clonogenicity assays. As shown in Figure 3c, si-221/222 significantly decreased the clonogenic activity of MCF7-F cells. Collectively, these studies demonstrate that miR-221/222 play a key role in supporting the proliferation of fulvestrant-resistant cells.
Figure 3.
miR-221/222 knockdown inhibits cell proliferation in fulvestrant-resistant MCF7-F cells. (a) Knockdown of miR-221/222 in MCF7-F cells. MCF7-F cells were transiently cotransfected with 100 nM of 2′-O-Me-antagomirs (si-221 and/or si-222). Following a 24, 48 or 72-h incubation, qRT–PCR (upper panel) and immunoblotting (lower panel) with anti-p27Kip1 and GAPDH antibodies were performed. The miR-221/222 levels in negative control-treated MCF7-F cells were normalized as 1 (mean±s.e., n = 3). **P<0.01. (b) Inhibited proliferation of MCF7-F cells after knockdown of miR-221 and/or miR-222. The growth rate was determined using BrdU incorporation assay (mean±s.e., n = 6). **P<0.01. (c) Clonogenic activity of MCF7-F cells. Sorted antagomir-treated MCF7-F cells were cultured in growth medium for 2 weeks and colonies containing >50 cells were scored (mean±s.e., n = 3). *P<0.05.
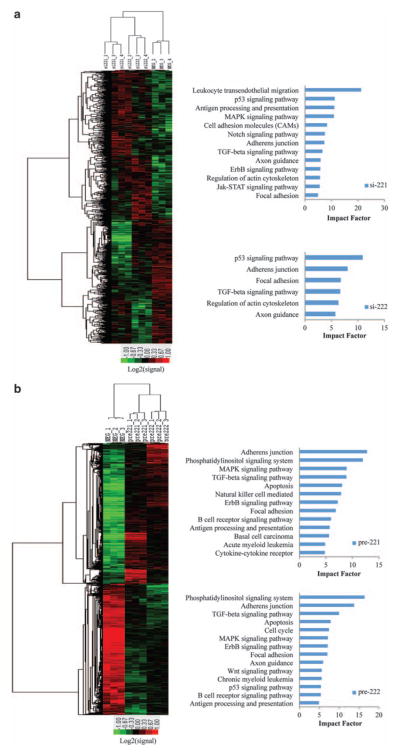
miR-221/222 regulate global gene expression and multiple signaling pathways involved in acquired fulvestrant resistance
The underlying mechanism of miR-221/222-mediated fulvestrant resistance appears to include downregulation of the cell cycle inhibitors p27Kip1 and p57Kip2 (Galardi et al., 2007; Fornari et al., 2008), in addition to ERα (Zhao et al., 2008; Di Leva et al., 2010). However, recent bioinformatic analyses (John et al., 2004) predict that a single miRNA is capable of regulating hundreds of genes, resulting in extensive miRNA-induced mRNA degradation (Filipowicz et al., 2008), and global gene expression changes (Huang et al., 2007; Nam et al., 2009). To gain further insight into the functional role of miR-221/222 in acquired fulvestrant resistance, we performed mRNA expression profiling after knocking down miR-221 or miR-222 in MCF7-F cells. As the mRNA and protein levels of p27Kip1 (used as a control) did not strongly correlate (Supplementary Figure S6), we used a moderate stringency to identify significantly regulated genes (P-value <0.05 and a fold-change >1.2 or <−1.2). Two-dimensional unsupervised hierarchical clustering (average linkage clustering method) was utilized to visualize common expression patterns between samples. Clustering results of all significantly up- or downregulated probes demonstrated a si-221 and si-222 cluster, and a separate control cluster (Figure 4a). After downregulation of miR-221 or miR-222, respectively, we identified 1347 or 1029 upregulated, and 912 or 909 downregulated probes, with 428 probes upregulated and 224 probes downregulated by both (Supplementary Figure S7). For in-depth analysis, we also included our previous gene expression profiles of pre-miRNA-treated MCF7 cells (Di Leva et al., 2010), and the clustering results indicated a similar pattern (Figure 4b).
Figure 4.
miR-221/222-regulated genes and signaling pathways. (a) Left panel: Two-dimensional hierarchical clustering of genes differentially expressed in si-221 or si-222-transfected MCF7-F cells compared with negative control-transfected MCF7-F cells. Each row represents a single gene. Red, genes with higher expression levels; green, genes with lower expression levels. Right panel: representative-signaling pathways significantly regulated by miR-221 or -222-regulated genes. Impact factor strength is shown. (b) The same analysis was performed for pre-221 or pre-222-transfected MCF7 cells.
To investigate the functional role of miR-221/222-regulated genes, pathway-express analysis (Draghici et al., 2007) was used, and genes regulated by si-221 or si-222 in MCF7-F cells were significantly enriched in 23 or 17 KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto, 2000) pathways (Supplementary Table S1), represented by p53, TGF-β, MAPK, Notch, ErbB, and Jak-STAT signaling pathway (Figure 4a). Genes regulated by pre-221 or pre-222 in MCF7 cells were enriched in 21 or 22 KEGG pathways, represented by phosphatidylinositol signaling system, MAPK, TGF-β, ErbB, p53, apoptosis and Wnt signaling pathways (Figure 4b). Representative genes in these pathways are listed in Table 1. Our previous study demonstrated that both EGFR/ErbB2 and Wnt/β-catenin pathways play a role in supporting estrogen-independent cell growth of MCF7-F (Fan et al., 2006) and the Cignal Finder Cancer Pathway Reporter Assay (SABiosciences, Frederick, MD, USA) also showed upregulation of Wnt signaling in MCF7-F cells compared with MCF7, as well as Notch, p53, MAPK, whereas downregulation of TGF-β signaling (Supplementary Figure S8). These pathways are regulated by miR-221/222 (described above), further supporting the multifunctional role of miR-221/222 in the acquisition of fulvestrant resistance.
Table 1.
Cell signaling pathways altered by miR-221/222 in MCF7 and MCF7-F cells
| Gene symbol | Genebank | Fold-change
|
Description | ||
|---|---|---|---|---|---|
| MCF7-F versus MCF7 | pre221/222 in MCF7 | si221/222 in MCF7-F | |||
| TGF-β signaling pathway | |||||
| TGFBR2 | NM_003242 | 4.31 | 1.87 | Transforming growth factor-β receptor II (70/80 kDa) | |
| BMPR2 | NM_033346 | 3.92 | 1.78 | −1.27 | Bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| GDF15 | NM_004864 | 4.86 | −1.47 | Growth differentiation factor 15 | |
| INHBA | NM_002192 | 5.59 | −1.74 | Inhibin-β a (activin a, activin ab-α polypeptide) | |
| INHBC | NM_005538 | −2.79 | 1.38 | Inhibin-β C | |
| SMAD1 | NM_005900 | 1.55 | SMAD family member 1 | ||
| SMAD2 | NM_005901 | −1.87 | 1.28 | −1.33 | SMAD family member 2 |
| SMAD3 | NM_005902 | 11.57 | 1.60 | SMAD family member 3 | |
| SMAD4 | NM_005359 | −1.58 | 1.41 | SMAD family member 4 | |
| ID1 | NM_181353 | 2.52 | 1.52 | Inhibitor of DNA binding 1, dominant-negative helix–loop–helix protein | |
| ID3 | NM_002167 | 1.25 | 1.66 | Inhibitor of DNA binding 3, dominant-negative helix–loop–helix protein | |
| THBS1 | NM_003246 | 11.62 | −2.01 | 1.24 | Thrombospondin 1 |
| ZFYVE16 | NM_014733 | 5.69 | −1.39 | Zinc finger, FYVE domain containing 16 | |
| Wnt signaling pathway | |||||
| WNT5A | NM_003392 | 12.91 | −1.28 | Wingless-type MMTV integration site family, member 5A | |
| FZD5 | NM_003468 | 2.96 | 3.03 | Frizzled homolog 5 (Drosophila) | |
| FZD8 | NM_031866 | 2.08 | 3.73 | Frizzled homolog 8 (Drosophila) | |
| CTNNB1 | NM_001904 | 4.90 | 1.56 | Catenin (cadherin-associated protein)-β 1, 88 kDa | |
| JUN | NM_002228 | −2.02 | 1.52 | Jun oncogene | |
| MAPK8 | NM_139049 | −1.37 | 1.51 | Mitogen-activated protein kinase 8 (JNK1) | |
| MAPK9 | NM_139070 | 1.63 | Mitogen-activated protein kinase 9 (JNK2) | ||
| NFAT5 | NM_173215 | 2.50 | −1.25 | Nuclear factor of activated T-cells 5, tonicity-responsive | |
| ErbB signaling pathway | |||||
| TGFA | NM_003236 | 2.47 | 1.55 | Transforming growth factor-α | |
| AREG | NM_001657 | −1.26 | −1.96 | 1.50 | Amphiregulin |
| CDKN1B | NM_004064 | −2.57 | 1.26 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | |
| HBEGF | NM_001945 | 1.90 | −1.43 | Heparin-binding EGF-like growth factor | |
| PAK2 | NM_002577 | −1.71 | 1.20 | 1.20 | p21 protein (Cdc42/Rac)-activated kinase 2 |
| SOS1 | NM_005633 | 8.20 | 1.62 | Son of sevenless homolog 1 (Drosophila) | |
| SOS2 | NM_006939 | 5.87 | −1.32 | Son of sevenless homolog 2 (Drosophila) | |
| Notch signaling pathway | |||||
| NOTCH1 | NM_017617 | 1.53 | Notch homolog 1, translocation-associated (Drosophila) | ||
| NUMB | NM_003744 | 1.92 | Numb homolog (Drosophila) | ||
| DTX3 L | NM_138287 | 2.99 | 3.61 | −1.34 | Deltex 3-like (Drosophila) |
| DVL2 | NM_004422 | −1.57 | Dishevelled, dsh homolog 2 (Drosophila) | ||
| JAG1 | NM_000214 | 11.35 | 1.63 | 1.28 | Jagged 1 (Alagille syndrome) |
| KAT2A | NM_021078 | 1.73 | −1.31 | K (lysine) acetyltransferase 2A | |
| Jak-STAT signaling pathway | |||||
| JAK2 | NM_004972 | 1.88 | −2.06 | −1.33 | Janus kinase 2 (a protein tyrosine kinase) |
| STAT1 | NM_139266 | 10.88 | 2.16 | −1.34 | Signal transducer and activator of transcription 1, 91 kDa |
| STAT2 | NM_005419 | 4.51 | −1.50 | Signal transducer and activator of transcription 2, 113 kDa | |
| STAT3 | NM_139276 | 4.24 | −1.50 | Signal transducer and activator of transcription 3 (acute phase) | |
| SOS1 | NM_005633 | 8.20 | 1.62 | Son of sevenless homolog 1 (Drosophila) | |
| SOS2 | NM_006939 | 5.87 | −1.32 | Son of sevenless homolog 2 (Drosophila) | |
| SOCS4 | NM_199421 | −1.41 | 1.33 | Suppressor of cytokine signaling 4 | |
| SOCS7 | NM_014598 | −1.76 | 1.34 | Suppressor of cytokine signaling 7 | |
| GHR | NM_000163 | 4.31 | Growth hormone receptor | ||
| IFNB1 | NM_002176 | −2.61 | Interferon-β 1, fibroblast | ||
| IFNGR1 | NM_000416 | 8.70 | 1.68 | Interferon-γ receptor 1 | |
| IL15 | NM_000585 | 3.40 | −1.52 | Interleukin 15 | |
| IL15RA | NM_002189 | 2.06 | −1.32 | Interleukin 15 receptor-α | |
| LIFR | NM_002310 | 1.68 | 1.54 | Leukemia inhibitory factor receptor-α | |
| OSMR | NM_003999 | 9.95 | 1.79 | −1.34 | Oncostatin M receptor |
| MAPK signaling pathway | |||||
| DUSP1 | NM_004417 | −1.25 | Dual specificity phosphatase 1 | ||
| DUSP10 | NM_144729 | 2.25 | 2.65 | −1.33 | Dual specificity phosphatase 10 |
| DUSP2 | NM_004418 | 1.50 | Dual specificity phosphatase 2 (EC:3.1.3.16 3.1.3.48) | ||
| FGFR4 | NM_022963 | −3.81 | 1.52 | Fibroblast growth factor receptor 4 (EC:2.7.10.1) | |
| FLNA | NM_001456 | −1.29 | −1.52 | 1.22 | Filamin A-α (actin-binding protein 280) |
| FLNB | NM_001457 | −2.70 | Filamin B-β (actin-binding protein 278) | ||
| GNA12 | NM_007353 | −3.47 | −3.06 | Guanine nucleotide-binding protein (G protein)-α 12 | |
| HSPA8 | NM_153201 | −1.89 | 1.25 | Heat shock 70 kDa protein 8 | |
| IL1R1 | NM_000877 | −3.44 | −2.31 | Interleukin 1 receptor, type I | |
| MAP2K5 | NM_145162 | 12.39 | −1.31 | Mitogen-activated protein kinase kinase 5 | |
| MAP3K4 | NM_006724 | −3.57 | 1.83 | −1.21 | Mitogen-activated protein kinase kinase kinase 4 |
| MAP3K5 | NM_005923 | 3.60 | 1.50 | Mitogen-activated protein kinase kinase kinase 5 (EC:2.7.11.25) | |
| MAP3K8 | NM_005204 | 8.35 | 1.54 | Mitogen-activated protein kinase kinase kinase 8 (EC:2.7.11.25) | |
| MAPK8 | NM_139049 | −1.37 | 1.51 | Mitogen-activated protein kinase 8 (JNK1) | |
| MAPK9 | NM_139070 | 1.63 | Mitogen-activated protein kinase 9 (EC:2.7.11.24) (JNK2) | ||
| MAPT | NM_173727 | −1.69 | −1.70 | 1.23 | Microtubule-associated protein tau |
| MKNK1 | NM_198973 | 2.26 | −1.86 | −1.28 | MAP kinase-interacting serine/threonine kinase 1 |
| PLA2G10 | NM_003561 | 2.02 | 1.84 | 1.42 | Phospholipase A2, group X |
| PLA2G12A | NM_030821 | −2.64 | −1.54 | Phospholipase A2, group XIIA (EC:3.1.1.4) | |
| PPM1A | NM_177952 | −2.98 | 1.22 | Protein phosphatase 1A (formerly 2C), magnesium-dependent-α isoform | |
| PPP3CA | NM_000944 | −1.98 | 1.23 | Protein phosphatase 3 (formerly 2B), catalytic subunit-α isoform (calcineurin A-a) | |
| PPP3R1 | NM_000945 | −1.82 | 1.37 | Protein phosphatase 3 (formerly 2B), regulatory subunit B-α isoform | |
| PPP5C | NM_006247 | 1.32 | Protein phosphatase 5, catalytic subunit | ||
| RASA2 | NM_006506 | 3.95 | −1.22 | RAS p21 protein activator 2 | |
| STK4 | NM_006282 | 5.37 | −1.37 | Serine/threonine kinase 4 | |
| TNFRSF1A | NM_001065 | 4.67 | −1.27 | Tumor necrosis factor receptor superfamily, member 1A | |
| p53 signaling pathway | |||||
| PERP | NM_022121 | 2.35 | −1.21 | PERP, TP53 apoptosis effector | |
| PMAIP1 | NM_021127 | 2.19 | −1.58 | Phorbol-12-myristate-13-acetate-induced protein 1 | |
| CASP3 | NM_004346 | −2.51 | Caspase 3, apoptosis-related cysteine peptidase | ||
| CDKN1A | NM_078467 | 2.03 | 2.73 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | |
| FAS | NM_000043 | 2.61 | Fas (Tumor necrosis factor receptor superfamily, member 6) | ||
| TNFRSF10B | NM_147187 | 5.05 | 1.79 | Tumor necrosis factor receptor superfamily, member 10b | |
| IGFBP3 | NM_000598 | 265.29 | 2.03 | Insulin-like growth factor-binding protein 3 | |
| GADD45A | NM_001924 | 2.21 | 2.32 | −1.42 | Growth arrest and DNA-damage-inducible-α |
| GADD45G | NM_006705 | −1.32 | Growth arrest and DNA-damage-inducible-γ | ||
| SERPINE1 | NM_000602 | 2.77 | −3.04 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | |
| SESN1 | NM_014454 | 3.03 | 1.51 | Sestrin 1 | |
| SESN2 | NM_031459 | 1.36 | −1.34 | Sestrin 2 | |
| THBS1 | NM_003246 | 11.62 | −2.01 | 1.24 | Thrombospondin 1 |
| SHISA5 | NM_016479 | 3.23 | 1.69 | −1.30 | Shisa homolog 5 (Xenopus laevis) |
| TP73 | NM_005427 | 1.34 | Tumor protein p73 | ||
| MDM2 | NM_006882 | 13.22 | −1.46 | Mdm2 p53-binding protein homolog (mouse) | |
| PTEN | NM_000314 | −2.23 | −1.54 | Phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) | |
| Focal adhesion | |||||
| CAV1 | NM_001753 | −1.59 | −20.66 | 1.30 | Caveolin 1, caveolae protein, 22 kDa |
| CAV2 | NM_198212 | −2.32 | −7.38 | 1.38 | Caveolin 2 |
| CDC42 | NM_044472 | −2.24 | 1.46 | Cell division cycle 42 (GTP-binding protein, 25 kDa) | |
| PAK1 | NM_002576 | −1.51 | 1.39 | p21 protein (Cdc42/Rac)-activated kinase 1 | |
| PAK2 | NM_002577 | −1.71 | 1.29 | p21 protein (Cdc42/Rac)-activated kinase 2 | |
| BCAR1 | NM_014567 | 7.26 | 1.59 | Breast cancer anti-estrogen resistance 1 | |
| BCL2 | NM_000657 | −1.88 | B-cell CLL/lymphoma 2 | ||
| DIAPH1 | NM_005219 | 4.79 | −1.25 | Diaphanous homolog 1 (Drosophila) | |
| FLNA | NM_001456 | −1.29 | −1.52 | 1.22 | Filamin A-α (actin-binding protein 280) |
| FLNB | NM_001457 | −2.70 | Filamin B-β (actin-binding protein 278) | ||
| PPP1CB | NM_002709 | −3.04 | 1.40 | Protein phosphatase 1, catalytic subunit-β isoform (EC:3.1.3.16) | |
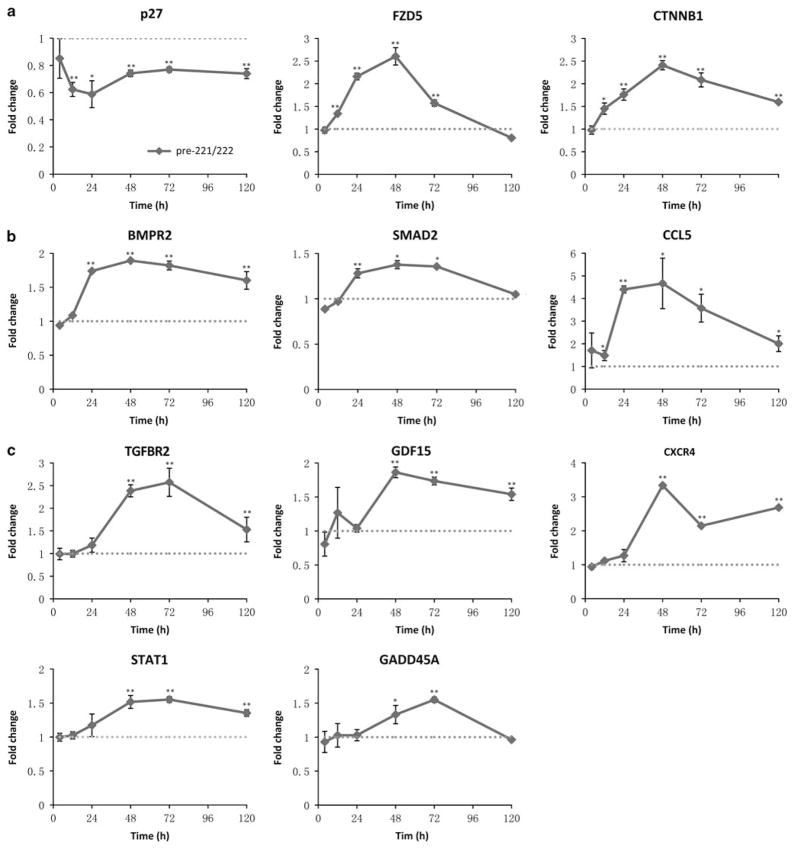
Time-course gene expression after ectopic expression of miR-221/222
As overexpressing miR-221/222 conferred estrogen-independent growth and fulvestrant resistance to MCF7 cells, we examined time-course changes in gene expression associated with this phenomenon. Based on the above gene expression profiles, 10 genes upregulated by miR-221/222 in MCF7 cells (Supplementary Table S2) were chosen for further study performing qRT–PCR at 4, 12, 24, 48, 72 and 120 h after transfection of MCF7 cells with pre-miR-221/222. p27Kip1, a known target of miR-221/222, decreased by 12 h after transfection (Figure 5a). Two key components of the Wnt/β-catenin pathway, FZD5 and CTNNB1 (β-catenin), were upregulated by 12 h (Figure 5a). Increased expression of TGF-β pathway components, BMPR2 and SMAD2, was observed by 24 h after transfection, whereas TGFBR2 was increased by 48 h (Figures 5b and c). The cytokines CCL5 and GDF15, and cytokine receptors CRCR4 were upregulated by 24 and 48 h, respectively (Figures 5b and c). Expression of the STAT1 transcription factor increased by 48 h. Increased expression of GADD45A, which is involved in p53, MAPK, and cell-cycle pathways was seen by 48 h (Figure 5c). Of the genes downregulated by miR-221/222, only one was decreased by 12 h after transfection (Supplementary Figure S9), suggesting that miR-221/222 downregulation of their targets occurs mainly at the protein level in MCF7 cells.
Figure 5.
Time-course gene expression after ectopic expression of miR-221/222 in MCF7 cells. (a) qRT–PCR results showing genes with altered expression within 12 h after transfection of pre-miR-221/222 (solid lines), compared with scramble control (dash lines, normalized to 1). (b) (c) Genes with altered expression within 24 h or 48 h after transfection (mean±s.e., n = 2, with two independent experiments). *P<0.05, **P<0.01.
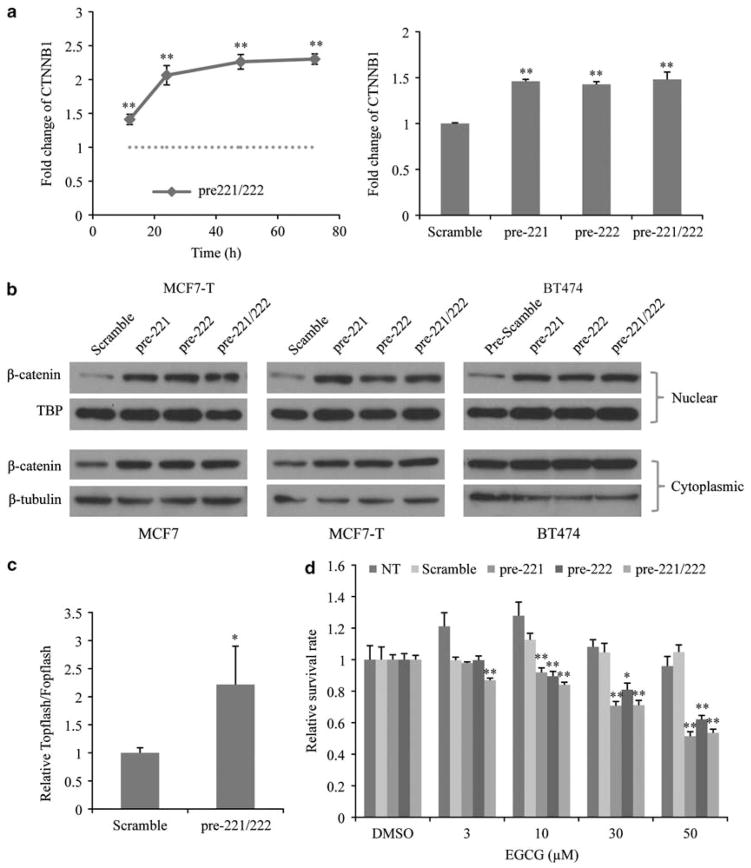
Regulation of β-catenin expression and transcriptional activity by miR-221/222
β-Catenin is a key regulator in the Wnt signal transduction cascade (Takahashi-Yanaga and Kahn, 2010), and we previously demonstrated a vital role of β-catenin for acquired fulvestrant resistance as upregulation and activation of β-catenin contributed to estrogen-independent growth of MCF7-F (Fan et al., 2006). As β-catenin mRNA levels rapidly (12 h) increased in MCF7 cells when overexpressing miR-221/222 (Figure 5a) and knockdown of miR-221 and/or miR-222 decreased β-catenin protein levels in MCF7-F cells (Supplementary Figure S10), we hypothesized that miR-221/222 serves as a positive regulator of β-catenin expression and transcriptional activity. To test this hypothesis, we first confirmed the upregulation of β-catenin mRNA levels by miR-221/222 in MCF7-T and BT474 cells (Figure 6a), which also increased fulvestrant resistance as previously indicated (Figure 2d). As only nuclear β-catenin has been shown to be transcriptionally active (Shitashige et al., 2008), we examined nuclear β-catenin protein levels. After overexpressing miR-221 and/or miR-222, elevated nuclear β-catenin levels were observed in all three cell lines (MCF7, MCF7-T and BT474; Figure 5b), whereas β-catenin levels in the cytoplasm were also increased in MCF7 and MCF7-T cells. Wnt signaling reporter analysis using TOPflash and FOPflash plasmids (Korinek et al., 1997) confirmed that overexpression of miR-221/222 in MCF7 cells upregulated β-catenin-mediated gene transcription (Figure 6c). To further investigate whether the increased β-catenin activity contributed to miR-221/222-mediated estrogen-independent growth, epigallocatechin-3-gallate was used to inhibit β-catenin activity (Kim et al., 2006). Cell proliferation assays revealed that inhibiting β-catenin activity blocked the growth of miR-221 and/or miR-222-overexpressing MCF7 cells, but not parental MCF7 cells or scramble control-treated cells (Figure 6d). Taken together, these results implicate miR-221/222 as a positive regulator of β-catenin expression and transcriptional activity, and the subsequent transactivation of β-catenin target genes appears to contribute to the acquisition of estrogen-independent growth.
Figure 6.
miR-221/222 upregulates and activates β-catenin. (a) Upregulated β-catenin mRNA in MCF7-T and BT474 cells. qRT–CR results indicated the time-course changes of β-catenin mRNA in MCF7-T cells (right panel) and the β-catenin mRNA level 48 h after transfection in BT474 cells (left panel) (mean±s.e., n = 2, with two independent experiments). **P<0.01. (b) Increased β-catenin protein level in the nuclear fraction (MCF7, MCF7-T and BT474). Immunoblotting results showed the protein level of β-catenin in nucleus and cytoplasm, whereas TATA binding protein (TBP) was used as loading control for nuclear proteins (upper panel) and β-tubulin as the control for cytoplasmic proteins (lower panel). (c) Increased β-catenin transcriptional activity. MCF7 cells were transfected with TOPflash or FOPflash reporter plasmids, together with β-gal pasmid for 48 h. Luciferase activities were measured using a luminometer, and β-gal assays were performed to normalize transfection efficiencies. (mean±s.e., n = 3). *P<0.05. (d) Inhibition of β-catenin activity prevents cell growth of pre-miRNA-treated MCF7 cells. Cells were grown in phenol red-free medium with 5% csFBS in the presence of doses of epigallocatechin-3-gallate (EGCG) for 6 days and cell survival rates were determined by MTT assay. (mean±s.e., n = 6). *P<0.05, **P<0.01.
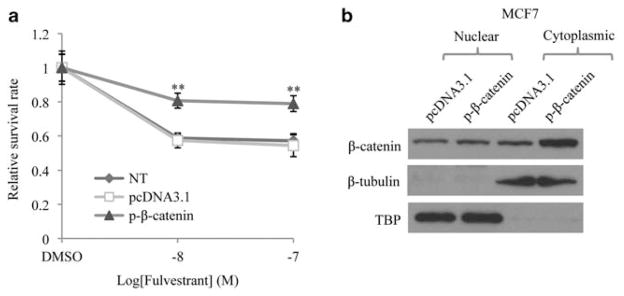
β-Catenin overexpression partially confers fulvestrant resistance
Having demonstrated that miR221/222-activated β-catenin supports estrogen-independent growth, we next examined whether β-catenin alone is enough to confer fulvestrant resistance. MCF7 cells were transiently transfected with β-catenin-overexpressing plasmids and then treated with varying doses of fulvestrant. Cell proliferation assays indicated that although β-catenin alone was not able to increase cell growth in estrogen-free medium (Supplementary Figure S11), cells over-expressing β-catenin were more vulnerable to fulvestrant-induced growth inhibition compared with empty vector-treated cells or parental cells (Figure 7a). Immuno-blotting analysis indicated that overexpression of β-catenin occurred mostly in the cytoplasmic fraction (Figure 7b), suggesting the existence of other factors in MCF7 that repress β-catenin nuclear translocation. These results demonstrated that although β-catenin contributes to acquired fulvestrant resistance, β-catenin overexpression alone was not sufficient for estrogen-independent growth, further suggesting a requirement for additional miR-221/222-regulated genes.
Figure 7.
Overexpression of β-catenin partially confers MCF7 cells resistance to fulvestrant. (a) Decreased fulvestrant sensitivity of β-catenin-overexpressing MCF7 cells. Cells were cultured in phenol red-free medium with 10% csFBS and MTT assay was performed 6 days after adding fulvestrant. (mean±s.e., n = 6). **P<0.01. (b) Immunoblotting results indicating the protein level of β-catenin.
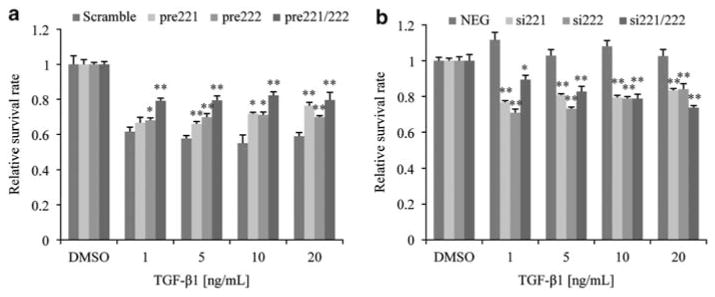
miR-221/222 modulate the effects of TGF-β
An association between TGF-β signaling pathway and antiestrogen resistance has been reported (Buck and Knabbe, 2006). Based on our expression profiling results showing that miR-221/222-regulated genes in MCF7 and MCF7-F cells were enriched for the TGF-β pathway (Figure 4) and cell proliferation assays revealing a growth inhibitory effect of TGF-β1 on MCF7 but not MCF7-F cells (Supplementary Figure S12) (Kalkhoven et al., 1995; Sovak et al., 1999), we hypothesized that the altered sensitivity to TGF-β-induced growth inhibition was associated with miR-221/222 expression. To examine this possibility, pre-miRNA-treated MCF7 cells were treated with TGF-β1. Overexpression of miR-221 and/or miR-222 increased MCF7 cell survival (versus scramble control; Figure 8a), and knockdown of miR-221 and/or miR-222 in MCF7-F cells resulted in growth inhibition by TGF-β1 (Figure 8b). As TGF-β can act as a tumor suppressor or enhancer in breast cancer (Zugmaier et al., 1989; McEarchern et al., 2001), miR-221/222 may serve as an important mediator of the transition of TGF-β from growth suppressor to tumor promoter.
Figure 8.
miR-221/222 alter cell sensitivity to TGF-β1-induced growth inhibition. (a) TGF-β1 sensitivity of MCF7 cells. Cells were treated with doses of TGF-β1 for 4 days and cell growth rates (drug versus vehicle) were determined by MTT assay. (mean±s.e., n = 6). *P<0.05, **P<0.01. (b) TGF-β1 sensitivity of MCF7-F cells determined as in (a).
Discussion
Acquired fulvestrant resistance is associated with loss of ERα expression, E2-independent growth, and activation of multiple growth-stimulatory pathways (Nicholson et al., 2005; Normanno et al., 2005; Fan et al., 2006). Recent studies (Miller et al., 2008; Zhao et al., 2008), including our own (Xin et al., 2009), suggest a prominent role of miR-221 and -222 in the acquisition of antiestrogen resistance. In the current study, we demonstrate that marginal miR-221/222 upregulation (~2-fold) is associated with tamoxifen resistance but marked overexpression (up to 25-fold) of miR-221/222 is characteristic of fulvestrant resistance (Figure 1a). Ectopic expression of miR-221/222 counteracts the well-known effects of E2 starvation and fulvestrant-induced cell damage and death in fulvestrant-sensitive cells, whereas strikingly increasing cellular proliferation and conferring SERD resistance (Figure 2). However, downregulation of miR-221/222 significantly suppresses cell growth and clonogenic activity of fulvestrant-resistant cells (Figure 3). Interestingly, both E2 and tamoxifen repress expression of endogenous miR-221/222 in MCF7 cells, but fulvestrant dramatically induces miR-221/222 (Figure 1b), suggesting that constitutive upregulation of miR-221/222 seen in fulvestrant-resistant cells is likely initiated by prolonged SERD treatment, attenuating the ERα/E2-mediated repression of miR-221/222, in accord with our recent study on a negative miR-221/222-ERα regulatory loop (Di Leva et al., 2010). As it is well established that maintenance of a strong ERα-positive phenotype is characteristic of tamoxifen-resistant breast cancer cells (Wijayaratne et al., 1999; Fan et al., 2006), upregulation of miR-221/222 in tamoxifen-resistant cells (Figure 1a; Miller et al., 2008) must be due to a different mechanism(s). Interestingly, upregulation of these two miRNAs has recently been reported in CD44+CD24−/lowlineage− human breast cancer stem cells (Shimono et al., 2009), which are believed to be largely responsible for propagating the drug-resistant phenotype (O’Brien et al., 2009), further stressing the importance of miR-221/222 in breast cancer progression and acquisition of antiestrogen resistance.
By analyzing miR-221/222-regulated gene expression profiles (knockdown of miR-221/222 in FR and over-expression of miR-221/222 in MCF7), we demonstrate that multiple oncogenic signaling pathways are regulated by miR-221/222 (Figure 4), which we (Fan et al., 2006) and others (Nicholson et al., 2005; Normanno et al., 2005) have reported contribute to the acquisition of fulvestrant resistance. One of the most strikingly regulated is the Wnt pathway, showing upregulation of CTNNB1 (β-catenin) and FZD5 12 h after miR-221/222 overexpression (Figures 5a and 6a). As miRNAs can also upregulate target gene expression (Vasudevan et al., 2007; Chen et al., 2010), these may represent direct miR-221/222 targets. However, using two target prediction programs (TargetScan (Lewis et al., 2003) and PITA (Kertesz et al., 2007)), we were not able to identify seed sequence binding sites for miR-221/222 in β-catenin and FZD. In addition, mRNA stability assays using 5,6-dichloro-1–D-ribofuranosylbenzimidazole demonstrated that β-catenin upregulation occurs at the transcriptional level (data not shown). Thus, the mechanism of miR-221/222-induced activation of β-catenin transcription remains unclear. Interestingly, miR-221/222 also dramatically increases β-catenin protein levels, particularly in the nuclear fraction (Figure 6b), which is not achievable by introducing β-catenin-overexpressing plasmids alone (Figure 7b).
β-Catenin is known to be sequestered in the cytoplasm by the destruction complex APC, AXIN, GSK-3β and CK1α, resulting in proteasomal degradation. Engagement of Wnt receptors inhibits destruction complex formation, allows for β-catenin release, translocation to the nucleus and subsequent activation of β-catenin target gene transcription (Takahashi-Yanaga and Kahn, 2010). In addition to upregulating β-catenin transcription, miR-221/222, by increasing Wnt5A and FZD5 expression (Table 1), may activate Wnt signaling and/or facilitate β-catenin nuclear accumulation by repressing Wnt pathway inhibitors AXIN2, SFRP2, CHD8 and NLK, the predicted targets of miR-221/222 (determined using DIANA microT 4.0 (Maragkakis et al., 2009)). The observation that miR-221/222-activated β-catenin supports estrogen-independent growth (Figures 6c and d), whereas overexpressing β-catenin alone does not (or even decreases cell proliferation under estrogen-free conditions; Supplementary Figure S11), further indicates that miR-221/222 trigger multiple events, which are required for the development of SERD resistance: β-catenin contributes to decreased fulvestrant sensitivity, whereas other factors further promote autocrine-regulated proliferation.
The most commonly regulated pathway by miR-221/222 in MCF7 and MCF7-F cells is the TGF-β signaling pathway (Figure 4), with increased TGFBR2, BMPR2 and SMAD2 after transfection (24–48 h; Figures 5b and c). SMADs (SMAD1/2/3), key regulators of TGF-β signaling activity, are upregulated by miR-221/222; however, miR-221/222 also induced two TGF-β-repressed genes, ID1 and ID3 (Table 1), in accord with downregulated TGF-β signaling activity in MCF7-F cells (with high miR-221/222 expression, Supplementary Figure S8 and Figure 1a). Owing to the great diversity of cell-specific gene responses elicited by TGF-β family members (Massague, 2000), whether the TGF-β pathway is activated or depressed by miR-221/222 is not clear, but the growth inhibitory effect of TGF-β is suppressed by miR-221/222 (Figure 8). As a potent inhibitor of primary human mammary epithelial cells and most breast cancer cell lines (Zugmaier et al., 1989; Basolo et al., 1994), TGF-β is activated by antiestrogens and participates in antiestrogen-induced growth inhibition (Buck et al., 2004). However, by stimulating angiogenesis, inducing extracellular matrix degradation, increasing invasion and metastasis, and inhibiting antitumor immune responses, TGF-β can promote tumor progression during latter stages (Gorsch et al., 1992; McEarchern et al., 2001). Also, defects in autocrine growth-inhibitory loops involving TGF-β signal transduction have been recognized as playing a central role in the development of antiestrogen resistance (Buck and Knabbe, 2006). Thus, miR-221/222 may serve as a switch to turn off the tumor-suppressing effects of TGF-β and enhance its tumor-promoting function during the acquisition of fulvestrant resistance.
p53 signaling is also among the pathways that are activated by miR-221/222 (Figure 4), with upregulation of several p53 target genes, including p21Cip1, PERP, CASP3, FAS, IGFBP3, GADD45A and SESNs (SESTRINs) (Table 1). MCF7-F cells having elevated miR-221/222 also show higher p53 signaling activity compared with MCF7 (Supplementary Figure S8, Figure 1a). Oncogene activation induced by miR-221/222 (Table 1) and altered cell morphology (reduced cell–cell contact, Figure 2c) might drive the activation of p53 (Vousden and Prives, 2009). Although the first identified tumor suppressor, additional roles for p53 have recently been reported, including regulation of metabolism, acting as antioxidants and even the antiapoptotic potential to increase cell survival (Janicke et al., 2008; Vousden and Prives, 2009). miR-221/222 may utilize these newly ascribed-p53 functions to confer fulvestrant resistance. For example, p21Cip1 may also function to inhibit apoptosis, and GADD45A and SESTRINs allow cells to survive until cellular damage has been resolved (Vousden and Prives, 2009). Interestingly, PTEN, a recently identified miR-221/222 target (Garofalo et al., 2009), was not regulated by miR-221/222 in MCF7 cells but slightly induced by miR-221/222 knockdown in MCF7-F cells (Supplementary Figure S13), implying cell-specific regulation by miR-221/222.
In the focal adhesion pathway, two major components of the caveolae plasma membranes, CAV1 and CAV2 (Table1 and Supplementary Figure S9), were downregulated by miR-221/222, consistent with our previous report (Di Leva et al., 2010). Although a predicted target of miR-221/222 (TargetScan (Lewis et al., 2003)), downregulation of CAV2 was not observed until 72 h, but miR-221/222 overexpression rapidly (12 h) downregulated CAV1, despite the fact that CAV1 lacks a perfect seed sequence miR-221/222 binding site. The mechanism(s) underlying this phenomenon requires further investigation. As expression of both CAV1 and CAV2 has been shown to inversely correlate with HER2/neu expression and tumor size in human breast cancer (Sagara et al., 2004), CAV1 and CAV2 downregulation may increase cell proliferation and enhance epidermal growth factor receptor (EGFR) signaling activity.
miR-221/222 have been considered to be oncogenes or tumor suppressors, depending on tumor type (Croce, 2009). Based on our results, ERα status may serve as a marker for the role of miR-221/222 in breast cancer. In ERα-positive cells, which depend on E2/ERα signaling for proliferation, fulvestrant treatment (Figure 1b) or E2 starvation (Di Leva et al., 2010) initially induce miR-221/222, which function as tumor suppressors by downregulating the receptor (Figure 2a). However, prolonged inhibition of ERα activity by fulvestrant treatment or E2 starvation releases miR-221/222 from the negative regulatory loop, resulting in constitutive upregulation of miR-221/222. In this scenario, miR-221/222 act as ‘oncomirs’ by targeting cell cycle inhibitors (p27Kip1, p57Kip2), other tumor suppressors (Garofalo et al., 2009), activating oncogenic signaling pathways (including Wnt, Notch and MAPK), and modulating TGF-β and p53 signaling to support ERα-independent proliferation and promote tumor progression. Targeting these two oncomirs may be a potential therapeutic strategy for preventing the development of fulvestrant resistance or resensitizing breast tumors to this potent and effective estrogen antagonist.
Materials and methods
Materials and cell lines
Pre-miR miRNA precursor for ectopic expression of miR-221, miR-222 and the scramble control were purchased from Applied Sciences/Ambion (Austin, TX, USA). Chemically stabilized antisense miRNA inhibitors (antagomirs) targeting miR-221 and miR-222 (2′-O-Me-221-GAAACCCAGCAGA CAAUGUAGCUL and 2′-O-Me-222-GAGACCCAGUAGC CAGAUGUAGCUL), as well as a negative control antagomir (2′-O-Me-enhanced green fluorescent protein (eGFP)-AAGGCAAGCUGACCCUGAAGUL) (Visone et al., 2007), were synthesized by Eurofins MWG Operon (Huntsville, AL, USA). Other biologicals used in this study have been described previously (Fan et al., 2006). BT474 cells were purchased from American Type Culture Collection (Manassas, VA, USA). See Supplementary materials for antibodies and plasmids.
Cell culture, transfection and clonogenicity assays
MCF7 and MCF7-F cells were cultured as previously described (Fan et al., 2006). BT474 cells were cultured according to American Type Culture Collection- suggested conditions. Pre-miRNAs or antagomirs were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Plasmids were transfected with Fugene HD Transfection Reagent (Roche Diagnostics Co., Indianapolis, IN, USA) according to the manufacturer’s instructions. The clonogenic activity was examined as previously described (Fan et al., 2006). See Supplementary materials for details and other methods used.
Microarray data analysis
To examine global gene expression profiles, MCF7-F cells were treated with miR-221 or miR-222 antagomirs or scramble control for 72 h, followed by RNA preparation/labeling and hybridization to Affymetrix GeneChip HG-U133 plus 2.0 arrays, according to Affymetrix standard protocols (Affymetrix, Santa Clara, CA, USA). Gene expression data were then processed and analyzed using Affymetrix Microarray Analysis Suite version 5.0 (MAS5), as described previously (Fan et al., 2006; Li et al., 2009), and deposited in Gene Expression Omnibus (accession number: GSE19777). See Supplementary materials for details of further analysis.
Statistical analysis
For MTT cell viability, BrdU cell proliferation, clonogenicity and qRT–PCR assays, statistical significance was analyzed by unpaired Student’s t test. P-values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr Man-Wook Hur for providing the TOPflash and FOPflash plasmids, Jim Powers for assistance with fluorescence microscopy, Dr Meiyun Fan for advice on experiments and Dr Curt Balch for critical comments and paper preparation. The IUB Light Microscopy Imaging Center provided microscopy resources. The microarray studies were carried out using the facilities of the Center for Medical Genomics (Dr H Edenberg, Director and Dr J McClintick) at Indiana University School of Medicine. The Center for Medical Genomics is supported in part by the Indiana Genomics Initiative of Indiana University, which is supported in part by the Lilly Endowment, Inc. This work was supported in part by NIH grants CA085289, CA113001 and CA125806, American Cancer Society Research Scholar Grant 09-244-01, and the Walther Cancer Foundation (Indianapolis).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6:181–202. [PubMed] [Google Scholar]
- Basolo F, Fiore L, Ciardiello F, Calvo S, Fontanini G, Conaldi PG, et al. Response of normal and oncogene-transformed human mammary epithelial cells to transforming growth factor beta 1 (TGF-beta 1): lack of growth-inhibitory effect on cells expressing the simian virus 40 large-T antigen. Int J Cancer. 1994;56:736–742. doi: 10.1002/ijc.2910560521. [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MB, Pfizenmaier K, Knabbe C. Antiestrogens induce growth inhibition by sequential activation of p38 mitogen-activated protein kinase and transforming growth factor-beta pathways in human breast cancer cells. Mol Endocrinol. 2004;18:1643–1657. doi: 10.1210/me.2003-0278. [DOI] [PubMed] [Google Scholar]
- Buck MB, Knabbe C. TGF-beta signaling in breast cancer. Ann N Y Acad Sci. 2006;1089:119–126. doi: 10.1196/annals.1386.024. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol. 2010;30:3902–3913. doi: 10.1128/MCB.01237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- Felicetti F, Errico MC, Segnalini P, Mattia G, Care A. MicroRNA-221 and -222 pathway controls melanoma progression. Expert Rev Anticancer Ther. 2008;8:1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- Gorsch SM, Memoli VA, Stukel TA, Gold LI, Arrick BA. Immunohistochemical staining for transforming growth factor beta 1 associates with disease progression in human breast cancer. Cancer Res. 1992;52:6949–6952. [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Kerin MJ. MicroRNAs as novel biomarkers for breast cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. Pure oestrogen antagonists for the treatment of advanced breast cancer. Endocr Relat Cancer. 2006a;13:689–706. doi: 10.1677/erc.1.00846. [DOI] [PubMed] [Google Scholar]
- Howell A. Fulvestrant (‘Faslodex’): current and future role in breast cancer management. Crit Rev Oncol Hematol. 2006b;57:265–273. doi: 10.1016/j.critrevonc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, et al. Using expression profiling data to identify human micro-RNA targets. Nat Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Casalini P, Tagliabue E, Menard S, Croce CM. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44:2753–2759. doi: 10.1016/j.ejca.2008.09.037. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. Tamoxifen: a most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–213. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E, Roelen BA, de Winter JP, Mummery CL, van den Eijndenvan Raaij AJ, van der Saag PT, et al. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6:1151–1161. [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhang X, Rieger-Christ KM, Summerhayes IC, Wazer DE, Paulson KE, et al. Suppression of Wnt signaling by the green tea compound (−)-epigallocatechin 3-gallate (EGCG) in invasive breast cancer cells. Requirement of the transcriptional repressor HBP1. J Biol Chem. 2006;281:10865–10875. doi: 10.1074/jbc.M513378200. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen Regulation of MicroRNA expression. Curr Genomics. 2009;10:169–183. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, et al. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med Genomics. 2009;2:34. doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- McDonnell DP. Mechanism-based discovery as an approach to identify the next generation of estrogen receptor modulators. FASEB J. 2006;20:2432–2434. doi: 10.1096/fj.06-1202ufm. [DOI] [PubMed] [Google Scholar]
- McEarchern JA, Kobie JJ, Mack V, Wu RS, Meade-Tollin L, Arteaga CL, et al. Invasion and metastasis of a mammary tumor involves TGF-beta signaling. Int J Cancer. 2001;91:76–82. doi: 10.1002/1097-0215(20010101)91:1<76::aid-ijc1012>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RI, Hutcheson IR, Hiscox SE, Knowlden JM, Giles M, Barrow D, et al. Growth factor signalling and resistance to selective oestrogen receptor modulators and pure anti-oestrogens: the use of anti-growth factor therapies to treat or delay endocrine resistance in breast cancer. Endocr Relat Cancer. 2005;12(Suppl 1):S29–S36. doi: 10.1677/erc.1.00991. [DOI] [PubMed] [Google Scholar]
- Nicholson RI, Johnston SR. Endocrine therapy—current benefits and limitations. Breast Cancer Res Treat. 2005;93(Suppl 1):S3–10. doi: 10.1007/s10549-005-9036-4. [DOI] [PubMed] [Google Scholar]
- Normanno N, Di Maio M, De Maio E, De Luca A, de Matteis A, Giordano A, et al. Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr Relat Cancer. 2005;12:721–747. doi: 10.1677/erc.1.00857. [DOI] [PubMed] [Google Scholar]
- O’Brien CS, Howell SJ, Farnie G, Clarke RB. Resistance to endocrine therapy: are breast cancer stem cells the culprits? J Mammary Gland Biol Neoplasia. 2009;14:45–54. doi: 10.1007/s10911-009-9115-y. [DOI] [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumor-igenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara Y, Mimori K, Yoshinaga K, Tanaka F, Nishida K, Ohno S, et al. Clinical significance of Caveolin-1, Caveolin-2 and HER2/neu mRNA expression in human breast cancer. Br J Cancer. 2004;91:959–965. doi: 10.1038/sj.bjc.6602029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci. 2008;99:631–637. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348:2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- Sovak MA, Arsura M, Zanieski G, Kavanagh KT, Sonenshein GE. The inhibitory effects of transforming growth factor beta1 on breast cancer cell proliferation are mediated through regulation of aberrant nuclear factor-kappaB/Rel expression. Cell Growth Differ. 1999;10:537–544. [PubMed] [Google Scholar]
- Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, et al. MicroRNAs (miR)-221 and miR-222, both over-expressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Weldon CB, Parker AP, Patten D, Elliott S, Tang Y, Frigo DE, et al. Sensitization of apoptotically-resistant breast carcinoma cells to TNF and TRAIL by inhibition of p38 mitogen-activated protein kinase signaling. Int J Oncol. 2004;24:1473–1480. [PubMed] [Google Scholar]
- Wijayaratne AL, Nagel SC, Paige LA, Christensen DJ, Norris JD, Fowlkes DM, et al. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology. 1999;140:5828–5840. doi: 10.1210/endo.140.12.7164. [DOI] [PubMed] [Google Scholar]
- Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, et al. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430–434. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou C, Nitschke AM, Xiong W, Zhang Q, Tang Y, Bloch M, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugmaier G, Ennis BW, Deschauer B, Katz D, Knabbe C, Wilding G, et al. Transforming growth factors type beta 1 and beta 2 are equipotent growth inhibitors of human breast cancer cell lines. J Cell Physiol. 1989;141:353–361. doi: 10.1002/jcp.1041410217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.