Abstract
Aptamers represent an emerging strategy to deliver cargo molecules, including dyes, drugs, proteins or even genes, into specific target cells. Upon binding to specific cell surface receptors aptamers can be internalized, for example by macropinocytosis or receptor mediated endocytosis. Here we report the in vitro selection and characterization of RNA aptamers with high affinity (Kd = 20 nM) and specificity for the human IL-6 receptor (IL-6R). Importantly, these aptamers trigger uptake without compromising the interaction of IL-6R with its natural ligands the cytokine IL-6 and glycoprotein 130 (gp130). We further optimized the aptamers to obtain a shortened, only 19-nt RNA oligonucleotide retaining all necessary characteristics for high affinity and selective recognition of IL-6R on cell surfaces. Upon incubation with IL-6R presenting cells this aptamer was rapidly internalized. Importantly, we could use our aptamer, to deliver bulky cargos, exemplified by fluorescently labeled streptavidin, into IL-6R presenting cells, thereby setting the stage for an aptamer-mediated escort of drug molecules to diseased cell populations or tissues.
Keywords: Aptamers, cargo delivery, cellular uptake, Interleukin-6 receptor, RNA-protein interaction, SELEX
Introduction
Interleukin-6 (IL-6) is a multifunctional cytokine that is involved in many immune and inflammatory responses. It belongs to the family of the four-helix bundle cytokines.1 IL-6 interacts with its natural receptor, namely IL-6R, and two molecules of the glycoprotein 130 (gp130), the transducer of the IL-6 signal.2 IL-6R appearance is restricted to a few cell types, including hepatocytes, monocytes, macrophages and some lymphocytes.2 In contrast, gp130 can be found on nearly all cell types. Cells that do not produce the membrane bound IL-6R (mIL-6R) can be stimulated by IL-6 via a soluble form of the IL-6R (sIL-6R). The sIL-6R is produced by cells positive for mIL-6R via alternative splicing or proteolytic cleavage of the membrane bound form.3,4 The sIL-6R in complex with IL-6 enables IL-6R lacking cells to respond to IL-6 mediated signaling.2
IL-6 and its receptor are involved in the progression of various inflammatory diseases, such as Crohn's disease and rheumatoid arthritis (RA), and certain cancers, for example multiple myeloma or hepatocellular carcinoma.5 One promising therapeutic strategy to tackle the mentioned diseases could be the specific delivery of drugs into cells by targeting IL-6R.
Aptamers are nucleic acids (DNA or RNA) consisting of about 15–100 nt, which are able to bind target molecules with high affinity and specificity, exhibiting Kd values in the picomolar range. Aptamers can be obtained by an in vitro selection process, termed SELEX (systematic evolution of ligands by exponential enrichment).6,7 Starting with a combinatorial oligonucleotide library containing up to 1015 different nucleic acid species, the SELEX process leads to the enrichment of an oligonucleotide population (polyclonal aptamers) that interacts with the target molecule due to defined three-dimensional structures. In respect of recognition properties aptamers are comparable to antibodies even though they reveal remarkable advantages, such as low immunogenicity and toxicity, longer shelf-life, and lower production costs. Up to now, aptamers were evolved for a wide variety of target molecules, including ions,8 fluorescent dyes,9 antibiotics,10 peptides,11 and proteins.12 Furthermore aptamers targeting viruses13 as well as whole cells14 have previously been selected and characterized for potential therapeutic applications. Next to their appliance in diagnostics, aptamers can serve as stimulating15 or inhibiting16 ligands, respectively. One very promising approach is the application of cell-specific aptamers as delivery vehicles for drug molecules into target cells or tissues. One auspicious example is the successful selection of an RNA aptamer binding to PSMA (prostate-specific membrane antigen) presented on the surface of prostate cancer cells.17 Upon binding, anti-PSMA aptamers undergo receptor-mediated internalization. Coupling those nucleic acids to cargo molecules, such as toxins,18 siRNAs,19 chemotherapeutics, and nanoparticles20 improves delivery to the cells of interest. This can allow to lower the dose required for effective treatment and is therefore considered a promising therapeutic approach which had been validated in animal studies.21
Thus, aptamers are very promising tools for cell-specific targeting. However, there is only a limited number of appropriate representatives to date. Therefore, new aptamers specific for cell surface proteins or whole cells are urgently needed.
We set out to enhance the toolbox of cell-targeting and drug delivering molecules. Thus, we decided to select aptamers specific for the human IL-6R, a cell surface receptor presented on a variety of cells often connected to numerous diseases, like the already mentioned multiple myeloma. Our studies yielded a 19-nt RNA aptamer, namely AIR-3A, which specifically recognizes cells that present IL-6R. This aptamer is able to transport cargo molecules of different sizes, exemplified by fluorescent dyes and streptavidin, into living IL-6R presenting cells. In view that IL-6 actively participates in inflammation associated cancer,22,23 selective targeting of IL-6R presenting tumor cells with toxic substances coupled to aptamers might be a valuable strategy to broaden established IL-6 or IL-6R directed treatment regimens.
Results
In vitro selection of sIL-6R-specific RNA aptamers
Aiming for RNA aptamers binding to cells that present IL-6R on their surface, we used the extracellular soluble part of the receptor (sIL-6R) as target molecule during the in vitro selection experiment. In the first selection cycle the RNA library R1 containing 1013 individual RNA molecules was incubated with sIL-6R, immobilized on magnetic beads. RNA molecules binding to sIL-6R were captured by magnetic separation and amplified by RT-PCR. After in vitro transcription the enriched RNA library was used as starting material for the subsequent selection cycle. The stringency of the selection process was enhanced gradually by increasing the number of washing steps with each additional selection cycle. After 16 selection rounds the enriched RNA library was analyzed for sIL-6R binding by filter retention analysis (FRA), which revealed a significantly increased affinity of the cycle 16 RNA for sIL-6R in comparison to the initial RNA library R1 (Fig. S1).
Subsequently, the corresponding cycle 16 dsDNA library was cloned and the sequences of 20 clones were determined (Fig. 1). We identified six individual sIL-6R-binding RNA aptamers all sharing a G-rich consensus motif.
Figure 1. Sequences of RNA aptamers binding the soluble part of the IL-6 receptor (sIL-6R). Sequences are printed in 5′–3′ direction omitting flanking primer-binding sites. Bold letters indicate conserved positions. F, frequency; consensus sequence is underlined and additionally given below. H: A, C or U; W: A or U.
Affinity and specificity of the RNA aptamer AIR-3 for sIL-6R
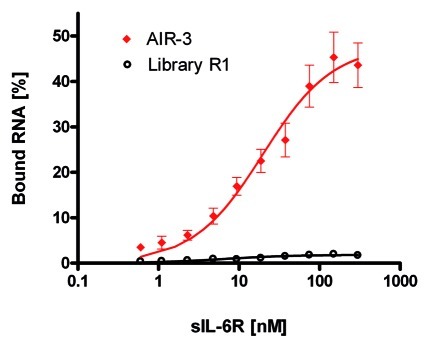
Initial FRAs revealed that among other tested aptamers the RNA aptamer AIR-3 exhibited the highest affinity for sIL-6R (Table S1 and Fig. S2). We further determined the dissociation constant (Kd) for AIR-3 binding to sIL-6R more precisely by repeating the FRA for ten times. Thereby, the initial RNA library R1 served as a control. After quantification resulting curves were fitted assuming a 1:1 binding stoichiometry between both interacting partners. AIR-3 revealed a high affinity for sIL-6R with a calculated Kd-value of 19.7 ± 4.2 nM (Table 1 and Fig. 2). No binding of the RNA library R1 was detectable. Further FRAs revealed that AIR-3 did not bind to the control proteins IL-6 and lysozyme (data not shown).
Table 1. Aptamer—target interactions.
| Aptamer | Target | Method1 | Kd (nM) |
|---|---|---|---|
| AIR-3 |
sIL-6R |
FRA |
19.7 ± 4.2 |
| AIR-3A |
Hyper-IL-6 |
FRA |
60.4 ± 16.0 |
| AIR-3A | Cells | FCM | 8.5 ± 1.0 |
1FRA, filter retention assay; FCM, flow cytometry
Figure 2. Aptamer AIR-3 binds the soluble IL-6 receptor (sIL-6R) with high affinity. Filter retention assays. Constant amounts (<1 nM) of 32P-radioactively labeled aptamer AIR-3 (red diamonds) and RNA starting library R1 (black circles) were incubated with increasing amounts of sIL-6R (0–300 nM). Protein-bound RNA was visualized by autoradiography. Fractions of bound RNA molecules were plotted against the concentration of sIL-6R (logarithmic scale). Data points represent mean values of 10 independent measurements.
Influence of AIR-3 on the interactions between sIL-6R and its ligands IL-6 and gp130
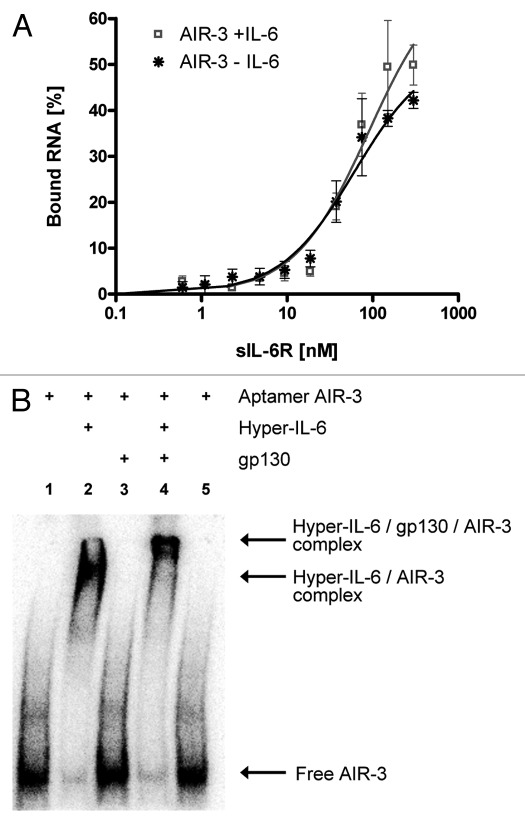
To determine whether aptamer AIR-3 competed with the cytokine IL-6 for binding to sIL-6R, filter retention assays were employed. Increasing amounts of sIL-6R were pre-incubated with or without its ligand IL-6. Constant amounts of radioactively labeled aptamer AIR-3 were added. Figure 3A displays the results of the FRAs revealing no competition between IL-6 and the RNA aptamer AIR-3 for binding to sIL-6R.
Figure 3. Aptamer AIR-3 does not compete with IL-6 or gp130 for binding to IL-6R. (A) Filter retention assays. Constant amounts of IL-6 (1 μM) were pre-incubated with increasing amounts of sIL-6R (0–300 nM) before radiolabeled aptamer AIR-3 (<1 nM) was added to the complex. For data evaluation see legend to Figure 2. (B) Interaction of aptamer AIR-3 (<1 nM) with Hyper-IL-6 (500 nM) or with Hyper-IL-6/gp130 complex (1:2 stoichiometry) analyzed by gel-electrophoretic mobility shift assay. Lanes 1 and 5, free aptamer; remaining lanes contained aptamer + 500 nM Hyper-IL-6 (2), + 1 µM gp130 (3) or both proteins (4).
Knowing that the aptamer AIR-3 specifically interacts with sIL-6R even in the presence of its natural ligand IL-6, we investigated the interaction of aptamer AIR-3 with the designer cytokine Hyper-IL-6, a fusion protein consisting of human IL-6 and the human sIL-6R connected by a flexible polypeptide chain.24 First, we performed FRAs to proof the ability of AIR-3 to interact with Hyper-IL-6 (Fig. S3).
The ability of aptamer AIR-3 to disrupt the interaction between Hyper-IL-6 and the second receptor subunit gp130 was examined by electrophoretic mobility shift assay (EMSA). This EMSA confirmed that AIR-3 did bind to Hyper-IL-6 as illustrated by a retarded migration rate in the native PAGE if compared with sole RNA (Fig. 3B, lanes 1 and 2). Additionally, a supershift confirmed the interaction between AIR-3 and Hyper-IL-6 complexed with gp130 (Fig. 3B, lane 4). The complex formation between Hyper-IL-6 and gp130 was not disrupted by AIR-3. No unspecific binding of AIR-3 to gp130 could be observed (Fig. 3B, lane 3).
Minimization of the RNA aptamer AIR-3
All identified sIL-6R specific aptamers contained a conserved G-rich motif (Fig. 1). According to this observation the aptamer AIR-3 was truncated finally yielding a 19 nucleotides short aptamer, termed AIR-3A (5′-GGGGAGGCUGUGGUGAG17G18G-3′). Three variants of AIR-3A namely G17U, G18U and G17U/G18U were generated to serve as controls. These variants bear G to U nucleotide exchanges at position 17, position 18 or at both positions, respectively. The abilities of these four truncated RNA molecules to bind Hyper-IL-6 were subsequently determined by FRA (Fig. S4, Table 1). The truncated aptamer AIR-3A exhibited a high affinity for Hyper-IL-6, whereas the variants G17U, G18U and G17U/G18U did not show any significant binding.
Structural analyses of AIR-3A and its derived variants
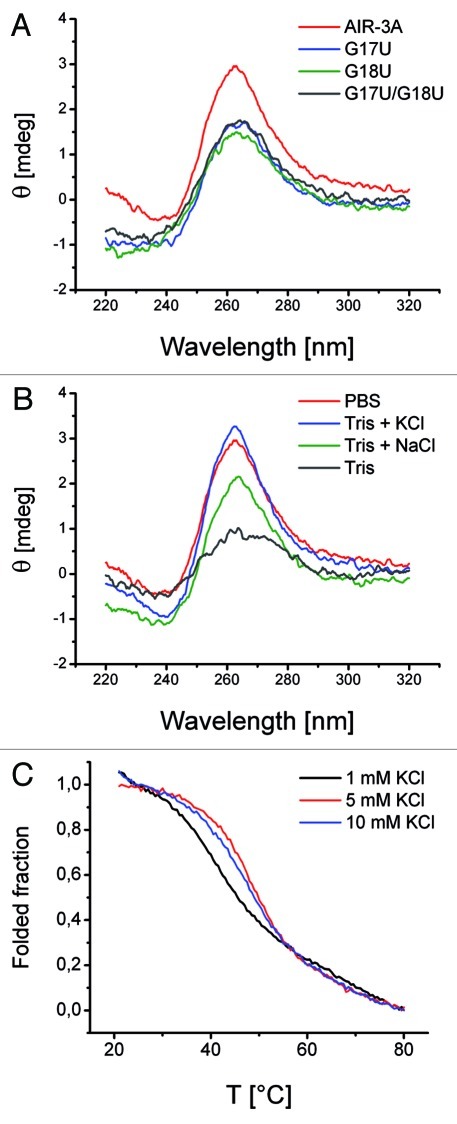
In a first CD spectroscopic experiment the spectrum of the RNA aptamer AIR-3A, solved in 1 × PBS (containing Na+ or K+ ions), revealed two main peaks, a maximum at 265 nm and a minimum at 240 nm (Fig. 4A), suggesting the formation of G-quadruplexes with parallel strand orientation.25,26 The variants G17U, G18U, and G17U/G18U, each solved in PBS as well, did clearly show a decrease in signal intensity at the characteristic wavelength. The G to U nucleotide exchanges seemed to destabilize or destruct the G-quadruplexes in all variants.
Figure 4. Structural analyses of AIR-3A and its variants. (A) CD spectroscopic analyses of AIR-3A, G17U, G18U and G17U/G18U (5 μM in PBS). (B) CD spectrum of AIR-3A (5 μM) in PBS or Tris buffer (50 mM) without or in the presence of potassium or sodium ions (100 mM each), respectively. (C) Melting profile of AIR-3A (2.5 µM; taken at 295 nm) in 10 mM Tris buffer containing 1 mM, 5 mM or 10 mM KCl, respectively.
The formation of stable G-quadruplexes requires the presence of special metal ions like K+ or Na+.27 Therefore we measured the CD spectra of AIR-3A in T-HCl buffer omitting monovalent sodium and potassium cations, respectively (Fig. 4B). The quadruplex formation was obviously reduced as the resulting peak amplitudes of the spectra decreased. After addition of potassium or sodium chloride both peaks increased again indicating that these ions are prerequisites for structural stability.
To confirm the potential G-quadruplex folding of AIR-3A we performed UV melting transitions at 295 nm.28 First, we solved AIR-3A in 10 mM Tris buffer (pH 7.5) including 5 mM KCl. We could show that the UV melting profile (Tm value 48.4°C) was characterized by a hypochromic shift as typically observed for nucleic acids containing G-quadruplex structures (Fig. 4C and Table S2). We evaluated the stability of the AIR-3A G-quadruplex by determining its melting temperatures at different potassium concentrations (0 mM, 1 mM, and 10 mM, respectively). The melting temperature decreased as a result of the reduction of the KCl concentration (Fig. 4C and Table S2). In absence of K+ cations melting curves did not show any hypochromic shift. Therefore it was not possible to determine the corresponding Tm value. Thus folding was significantly dependent on K+ cations.
The melting temperature of AIR-3A did not dependent on RNA concentration over a range from 1 to 10 µM since the Tm corresponding values did not change significantly (Fig. S5A and Table S2).
All three variants G17U, G18U, and G17U/G18U did not exhibit a hypochromic shift within the corresponding UV melting profiles neither in absence nor in presence of 5 mM KCl (Fig. S5B,Table S2). Therefore we could not calculate any Tm values.
Taken together, these results indicated that only AIR-3A exhibited a stable intramolecular G-quadruplex under physiological conditions.
Interaction of AIR-3A with IL-6R presenting cells
Binding of AIR-3A to BAF/gp130/IL6R/TNF cells
We subsequently determined whether AIR-3A was able to bind IL-6R presenting cells. Therefore, we used two murine cell lines, BAF/gp130/IL6R/TNF-cells, a parental pro-B cell line transfected with the cDNA of human gp130, human IL-6R, and TNF, as well as BAF/gp130 cells, that were transfected solely with the cDNA of human gp130, the latter serving as control.
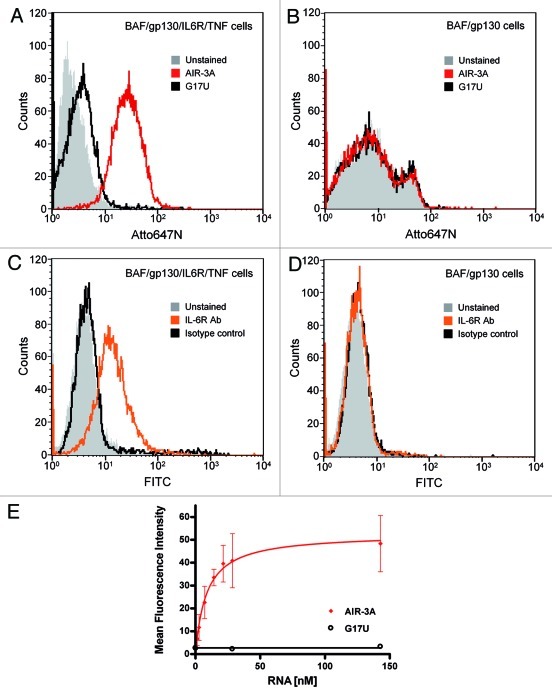
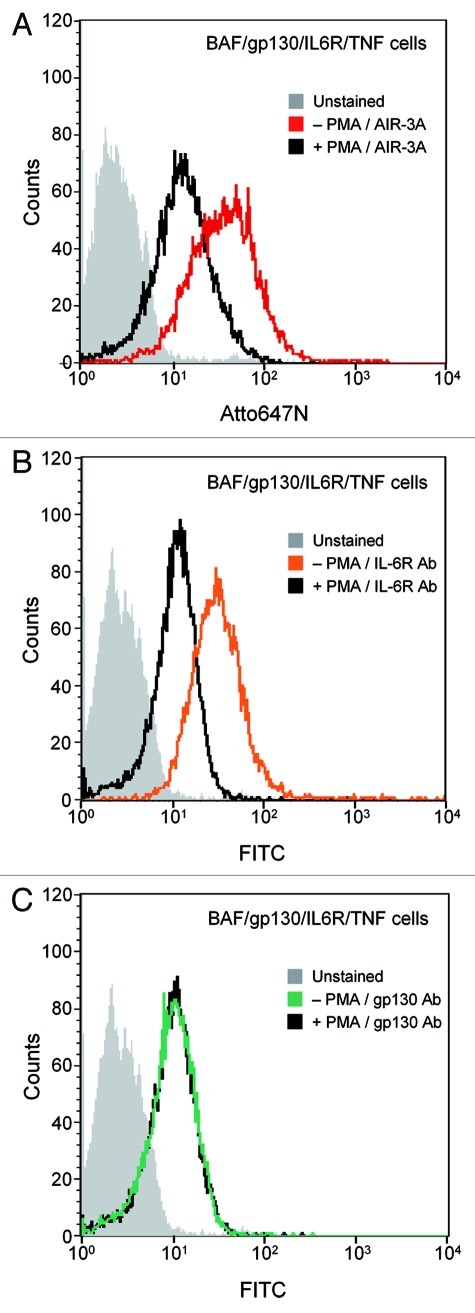
Atto647N-labeled aptamer AIR-3A and the control variant G17U were assayed for their binding capacities on IL-6R presenting BAF/gp130/IL6R/TNF cells by flow cytometry. AIR-3A but not G17U did bind to those cells (Fig. 5A). Both RNAs failed to bind to cells lacking IL-6R (Fig. 5B). IL-6R expression on BAF/gp130/IL6R/TNF cells but not on BAF/gp130 cells was confirmed using a monoclonal human IL-6R specific antibody. This antibody did bind to BAF/gp130/IL6R/TNF cells but not to IL-6R-negative BAF/gp130 cells (Fig. 5C and D, respectively).
Figure 5. Aptamer AIR-3A specifically binds to BAF/gp130/IL6R/TNF cells presenting the human IL-6 receptor. (A)/(B) Interactions of the IL-6R specific RNA aptamer AIR-3A (Atto647N-labeled, red) or the variant G17U (Atto647N-labeled, black) with BAF/gp130/IL6R/TNF cells (A) and BAF/gp130 cells (B) were analyzed by flow cytometry. Binding of AIR-3A was restricted to BAF/gp130/IL6R/TNF cells. BAF/gp130 cells do neither interact with RNA aptamer AIR-3A nor with the variant G17U, respectively (grey, unstained cells). (C)/(D) Cell surface expression of human IL-6R on BAF/gp130/IL6R/TNF cells (C) and BAF/gp130 cells (D) was confirmed by flow cytometry analyses using an antibody specific for human IL-6R. For detection a FITC-labeled goat anti-mouse antibody was used. IL-6R detection was restricted to BAF/gp130/IL6R/TNF cells. For BAF/gp130 cells no IL-6R expression was detectable (grey, unstained cells; orange, mouse anti-hIL6R antibody; black, isotype control). (E) AIR-3A binds to BAF/gp130/IL6R/TNF cells in a concentration dependent manner. Increasing amounts (0–150 nM) of AIR-3A (Atto647N-labeled, red) and variant G17U (Atto647N-labeled, black) were incubated with BAF/gp130/IL6R/TNF cells. Binding was monitored by flow cytometry. Mean fluorescence intensities were plotted against corresponding RNA concentrations. The apparent Kd-value of AIR-3A binding to IL-6R presenting cells was calculated to be 8.5 ± 1 nM.
To analyze whether the binding capacity of the RNA aptamer in serum containing medium might be disturbed by contaminating RNases we performed binding assays in selection buffer, cell culture medium and serum containing medium at 4°C or 37°C, respectively (Fig. S6). In all cases no interference with aptamer binding could be detected within one hour of incubation.
We next determined the apparent Kd-value for the interaction of AIR-3A with IL-6R on corresponding cells. Therefore we incubated 500,000 BAF/gp130/IL6R/TNF cells with increasing amounts of fluorescently labeled aptamer AIR-3A or the non-binding control G17U. Flow cytometry analysis revealed an apparent affinity (Kapp) of 8.5 ± 1.0 nM for AIR-3A (Fig. 5E and Table 1).
Influence of PMA-stimulated IL-6R-shedding on AIR-3A binding
To further analyze if the human IL-6R is indeed the target molecule of AIR-3A on BAF/gp130/IL6R/TNF cells we pretreated the cells with PMA (phorbol-12-myristate-13-acetate). PMA is known to stimulate the shedding of the IL-6R ectodomain from cells expressing the membrane bound IL-6R by activating the protease ADAM17. Subsequently, this leads to the release of sIL-6R from the cell surface.29
In accordance with our interaction data, PMA treatment, led to a significantly reduced binding of aptamer AIR-3A to target cells (Fig. 6A). To confirm the shedding of the IL-6R on BAF/gp130/IL6R/TNF cells treated with PMA we performed flow cytometry analyses using antibodies against human IL-6R and gp130, as a control, respectively. Thus we could show that PMA stimulation did only influence the expression pattern of IL-6R on the cell surface and not of gp130 (Fig. 6B and C).
Figure 6. AIR-3A specifically binds to IL-6R on BAF/gp130/IL6R/TNF cells. (A) BAF/gp130/IL6R/TNF cells treated without (red) or with PMA (black) were analyzed for binding of AIR-3A (Atto647N-labeled). Untreated BAF/gp130/IL6R/TNF cells revealed a higher binding of IL-6R specific aptamer AIR-3A in comparison to PMA-treated cells (grey, unstained cells). (B) BAF/gp130/IL6R/TNF cells without (orange) or with (black) PMA (phorbol-12-myristate-13-acetate) treatment were analyzed for IL-6R presentation using a mouse antibody specific for human IL-6R. For untreated BAF/gp130/IL6R/TNF cells a higher IL-6R presentation on the cell surface was detectable in comparison to PMA-stimulated cells. (C) PMA-treated (black) or untreated (green) BAF/gp130/IL6R/TNF cells were analyzed for gp130 presentation using a mouse antibody specific for human gp130. Flow cytometry analyses revealed the same presentation pattern of gp130 on both cell types. Consequently, the production of gp130 was not influenced by PMA. For detection a FITC-labeled goat anti-mouse antibody was used.
Binding of AIR-3A to further cell lines
We broadened the analysis of AIR-3A binding to further cell lines presenting or lacking IL-6R (Fig. S7). Flow cytometric profiles indicated that AIR-3A as well as the IL-6R-binding mAb recognized their target on IL-6R-presenting U937 cells. In contrast the same probes failed to bind HeLa and HEK293 cells, respectively.
IL-6 receptor mediated internalization of derivatized aptamer AIR-3A
Internalization of fluorescently labeled AIR-3A
In order to test the applicability of aptamer AIR-3A to serve as delivery vehicle, we analyzed whether the AIR-3A is being taken up through an active process by BAF/gp130/IL6R/TNF cells.
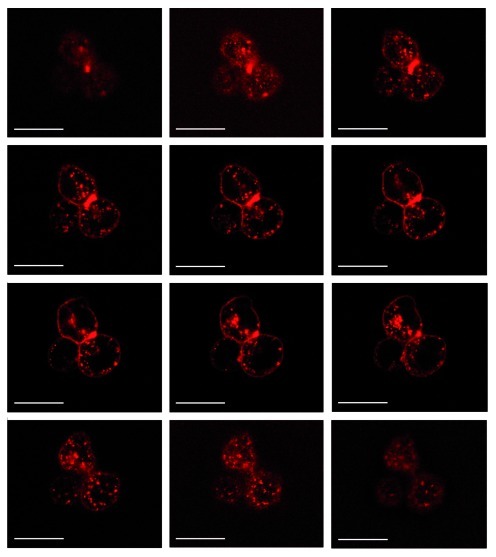
To achieve this we first incubated fluorescently labeled AIR-3A with target cells. Potential internalization was visualized by confocal fluorescence microscopy (Fig. 7). Intracellular fluorescence signals were detected only when incubated with cells presenting IL-6R. Binding of fluorescently labeled G17U to BAF/gp130/IL6R/TNF cells was not detectable (data not shown).
Figure 7. Internalization of AIR-3A bound to BAF/gp130/IL6R/TNF cells. Cells were incubated with Atto645N-labeled AIR-3A for 30 min at 37°C and visualized via confocal laser scanning microscopy. All panels show fluorescence micrographs at various depths within the cells (z-stack starting from upper left to lower right; stack size: 15.6 μm; scanning step depths: 1.3 μm; scale bars: 20 μm).
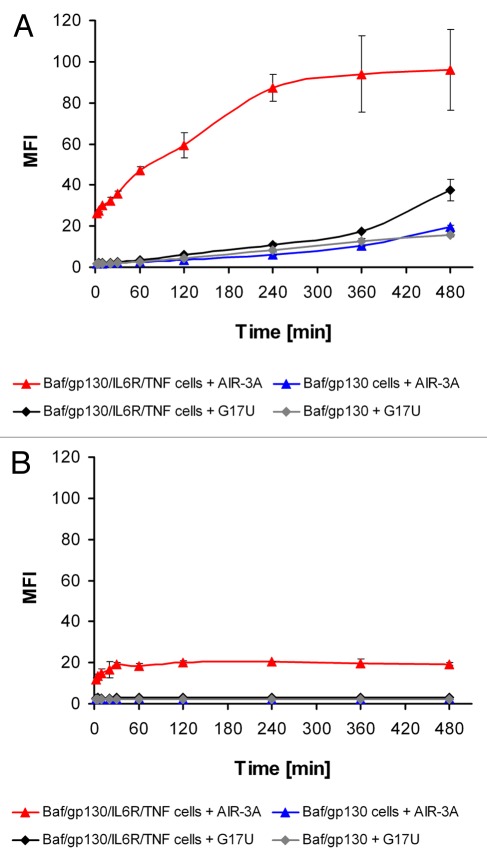
To reveal time depending binding of AIR-3A to IL-6R presenting cells fluorescently labeled RNAs were incubated with BAF/gp130/IL6R/TNF or BAF/gp130 cells for up to 8 h in serum containing medium. If incubated at 37°C AIR-3A binding to receptor presenting cells was detectable within less than 2 min reaching the maximal signal intensity after around 4 h (Fig. 8A). The interaction of G17U with these cells as well as the interaction of both RNAs with BAF/gp130 cells were much lower and showed different kinetics. At 4°C, however, only AIR-3A was able to bind to IL-6R presenting cells (Fig. 8B). The maximal signal intensity peaked earlier (30 min) but remained significantly lower if compared with 37°C.
Figure 8. Time course of AIR-3A binding to IL-6R presenting cells. Cells were incubated with Atto647N-labeled oligonucleotides (25 nM) at 37°C (A) or 4°C (B) for up to 8 h in serum containing medium and subsequently analyzed by flow cytometry. Median fluorescence intensities (MFI) resulting from two independent measurements were plotted against the incubation time.
IL-6R-mediated internalization of aptamer-streptavidin complexes
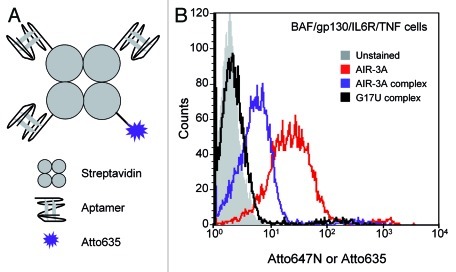
After having shown that the aptamer is able to mediate membrane translocalization of a fluorescence dye, we next investigated whether larger cargos, such as streptavidin, could also be translocated by AIR-3A. We therefore built-up a ternary complex of AIR-3A, a fluorescence dye (Atto635), and streptavidin (Fig. 9A). The former two components, both biotinylated, were mixed in a 3:1 ratio and subsequently combined with one part of streptavidin. This mixture was then incubated with IL-6R presenting cells. Complex formation was monitored by native PAGE (Fig. S8). The corresponding ternary complex in which AIR-3A was replaced by the inactive variant G17U was used as negative control. As expected the G17U-streptavidin-Atto635 control did not interact with the cells whereas the AIR-3A-streptavidin-Atto635 complex bound to BAF/gp130/IL6R/TNF cells (Fig. 9B).
Figure 9. Binding of AIR-3A-streptavidin complexes to BAF/gp130/IL6R/TNF cells. (A) Schematic illustration of aptamer-streptavidin-Atto635 conjugates. Aptamers and Atto635 were covalently modified with a biotin group and non-covalently assembled with streptavidin in a 3:1:1 ratio to gain ternary complexes. (B) Ternary complexes consisting of streptavidin, Atto635 and AIR-3A or G17U, respectively, were analyzed for binding to BAF/gp130/IL6R/TNF cells. Flow cytometric analyses revealed clearly binding of AIR-3A-streptavidin-Atto635 complexes (blue), but not any binding of the G17U-streptavidin-Atto635 complexes (black). Interaction of the IL-6R specific RNA aptamer AIR-3A (Atto647N-labeled, red) with BAF/gp130/IL6R/TNF cells served as positive control (grey, unstained cells).
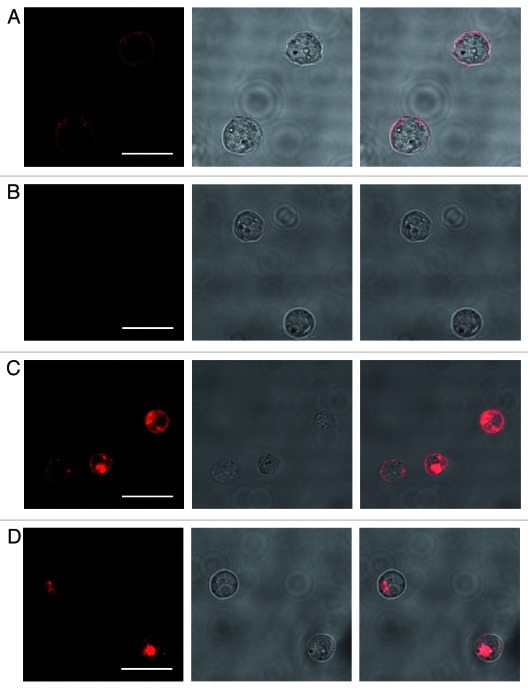
Furthermore, confocal microscopy revealed that even the large complex was internalized upon target cell recognition (Fig. 10). However, internalization was found to be less pronounced compared with the dye-construct and the residual portion still bound to the cell surface could be removed by trypsin treatment (Fig. 10A and B). Incubation on ice did not result in internalization clearly indicating that the process is receptor-mediated rather than related to simple passive diffusion (Fig. 10C and D).
Figure 10. IL-6R-mediated internalization of AIR-3A-streptavidin complexes by BAF/gp130/IL6R/TNF cells. (A) and (B) BAF/gp130/IL6R/TNF cells were incubated with Atto645N-labeled AIR-3A-streptavidin complexes for 30 min at 37°C without (A) or (B) with subsequent trypsin digestion. After incubation at 37°C internalized complexes could be detected. (C) and (D) BAF/gp130/IL6R/TNF cells were incubated with Atto645N-labeled AIR-3A-streptavidin complexes for 30 min at 4°C without (C) and with (D) subsequent trypsin treatment (degradation of surface proteins) before microscopy. After incubation at 4°C no internalized complexes could be detected. For each experiment panels of fluorescence micrographs, transmitted light micrographs and overlays of both (from left to right) are shown. (scale bars: 20 μm)
Discussion
In vitro selections of aptamers have been performed for more than two decades. During this period a large variety of aptamers has been successfully selected and applied for numerous purposes both in vitro and in vivo. Aptamers have been used in vitro for protein purification30 and quantification31 or for enzyme-linked oligonucleotide assays.32 The in vivo usage included inhibition of enzymes,33 interference with ligand-receptor interactions16 or imaging purposes.34 More recently, aptamers, that bind to cell surface proteins, became attractive tools to regulate the function of their protein targets in an agonistic15 or antagonistic manner.16,35
Since most cell-surface proteins undergo recycling processes, such as ligand-induced internalization, aptamers that recognize and bind to those cell-surface proteins might be infiltrated into corresponding cells and thus be used for a cell specific delivery of cargo molecules. This would open new strategies for aptamer-based drugs.
Currently, delivery approaches of cargo molecules into target cells comprise both covalently and non-covalently connected constructs. Non-covalent assemblies include the encapsulation of siRNAs into lipid particles.36 Covalent conjugation was also successfully performed for siRNAs and lipids, like cholesterol.37 One major drawback in most cases, however, is the non-specific cellular uptake of these conjugates. To gain cell-specific uptake and to reduce off-target effects, strategies for selective drug deliveries are desirable.
So far, only few aptamers have been used to specifically escort cargo molecules, like siRNAs,19 protein toxins,18 chemotherapeutics,38 nanoparticles,39 and enzymes,40 to or into specific cells or tissues of interest.
As already mentioned in the introduction, one of the best-studied aptamers for cell-specific delivery is the aptamer for prostate specific membrane antigen (PSMA), a well-known tumor marker of prostate cancer.17 Another promising delivery approach deals with the gp120 aptamer that was coupled to siRNAs specific for HIV-mRNAs and subsequently used to interfere with virus infectivity and reproduction12 (for review see ref. 41).
Thus, aptamers that target cell surface proteins comprise a promising and emerging class of molecules that might be used for the targeted delivery of drug-like agents to special cells or tissues, going along with a high therapeutic potential and a relatively low cytotoxicity. The development of further aptamers to specifically be internalized by cells would be a very attractive strategy for cell-specific escort purposes and therapies.
In this study, we describe the selection and characterization of RNA aptamers targeting the human Interleukin-6 receptor (IL-6R). This receptor is primarily produced by hepatocytes, monocytes, macrophages and some lymphocytes.2 Its native ligand is the cytokine IL-6. This cytokine plays a pivotal role in a variety of medical conditions, for example in multiple myeloma IL-6 was shown to be responsible for the progression of this type of cancer.42 Additionally, in the case of IL-6 it was recently shown that F4/80 tumor infiltrating macrophages secreted high amounts of IL-6, which increased STAT3 phosphorylation and cellular growth in the tumor cells.22,23 Therefore, targeting IL-6R expressing tumor cells with an IL-6R specific aptamer carrying a cellular toxin or other interfering molecules seems to be a novel and promising strategy.
To facilitate in vitro selection of IL-6R-specific aptamers we used the recombinantly produced soluble extracellular portion of the receptor, sIL-6R. Sixteen rounds of in vitro selection yielded six sIL-6R-binding RNA aptamers (Fig. 1). Aptamer AIR-3 revealed best binding properties with high affinity and selectivity. Competition experiments (Fig. 3) revealed that AIR-3 did neither interfere with cytokine IL-6 nor with gp130 recognition of IL-6R. Thus, the aptamer AIR-3 seemed not to interfere with the IL-6 mediated formation of the active signaling IL-6R complex.
For minimization of the IL-6R aptamers we took into account that the initially obtained six individual aptamers shared one common G-rich consensus motif of 19 nucleotides (Fig. 1). Consequently, aptamer AIR-3 was truncated from 106 to 19 nt, the latter corresponding to a molecular weight of only 6 kDa. This shortened version, named AIR-3A, still bound IL-6R with high affinity (Table 1). The variants G17U, G18U and G17U/G18U derived from AIR-3A by replacing a G to U in position 17, 18 or in both positions, lacked any binding activities.
As it is known that G-rich regions in nucleic acids can adopt G-quadruplex conformations we wanted to inquire if structural changes in G17U, G18U and G17U/G18U might have caused the loss of their ability to bind IL-6R and therefore performed CD spectroscopic and UV-melting analyses. AIR-3A as well as G17U in turn fulfilled the criteria for the formation of G-quadruplexes as both possessed a minimum of four interspersed GG dinucleotides. CD analyses and UV-melting studies showed that AIR-3A but not G17U adopted a parallel G-quadruplex structure (Fig. 4, Table S2). Thus a structural distortion of the G-quadruplex was probably the reason why G17U lost its affinity for IL-6R. The variants G18U and G17U/G18U did not fulfill the criteria for G-quadruplex formation. Indeed they did not adopt G-quadruplex structures as revealed by biophysical structural analyses.
Next to these structural investigations we wanted to deepen our knowledge about the functional properties of AIR-3A. Using flow cytometry we revealed that AIR-3A, but not the control G17U, did bind to Baf/gp130/IL-6R/TNF cells that were stably transfected with the cDNA encoding human IL-6R (Fig. 5). AIR-3A as well as G17U did not bind to Baf/gp130 cells lacking IL-6R. However, in case of U937 cells that endogenously present about 2.800 IL-6R molecules on their surface,43 AIR-3A did bind (Fig. S7). Contrarily, cells that do not present IL-6R endogenously, like HEK293 cells, or only a few IL-6R molecules, like HeLa cells,44 were neither bound by AIR-3A nor by an IL-6R-specific monoclonal antibody.
We further analyzed whether the medium components, especially serum, could interfere with the interaction between IL-6R-presenting cells and AIR-3A. We could not detect any difference between selection buffer, medium and serum containing medium (Fig. S6). Advantageously, G-quadruplex structures can reveal enhanced stabilities to serum and cellular nucleases compared with differently structured nucleic acid molecules.45 However, for future in vivo applications, additional modification of the aptamer might be a prerequisite to further improve stability and shelf-life of AIR-3A.
Next to cell-specific binding we could show that fluorescently labeled AIR-3A was additionally internalized by IL-6R-presenting cells at 37°C and not at 4°C (Figs. 7 and 8). Due to the fact that IL-6R endocytosis also only occurs at 37°C,46,47 we conclude a temperature- and receptor dependent endocytotic process for AIR-3A uptake. Alternative forms of endocytosis like macropinocytosis48 presumably can by excluded in case of AIR-3A, at least within the first minutes. However, the exact mechanism of the AIR-3A uptake remains to be verified. There are only few studies that propose a model for aptamer uptake. In cancer cells, Reyes-Reyes et al. for example proved macropinocytosis as basic process for the uptake of AS1411, a nucleolin binding G-rich DNA aptamer serving as anticancer agent.45 For successful cargo delivery by aptamers into target cells sole uptake is not the only hurdle to overcome. Furthermore the endosomal escape—the transfer of cargo molecules via the endosomal membrane into the cytoplasm—is an additional prerequisite. Currently, all conceivable steps involved are poorly understood.49
In the study described here the IL-6R specific RNA aptamer AIR-3A was proven to serve as specific carrier for the internalization of larger cargos, exemplified by the protein streptavidin (MW about 67,2 kDa). In other words AIR-3A—only 19 nt short—was capable of specifically carrying a cargo protein with a molecular weight 10 times higher than its own into cells. Due to the high potential of AIR-3A to additionally serve as delivery vehicle for siRNAs, toxins, or nanoparticles, this aptamer might become interesting for the treatment of various diseases connected to IL-6R presenting cells. AIR-3A may also serve as a tool to study cellular uptake of nucleic acids in greater detail on its own. Further structural investigations of AIR-3A and its derivatives, including crystallization of free aptamer or in complex with sIL-6R as well as the selection of aptamers capable of blocking the active receptor are under way.
Materials and Methods
Chemicals
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich. Buffers were prepared using de-ionized water obtained from a water purification system (Millipore). Selection buffer for SELEX consisted of 137 mM NaCl; 2.7 mM KCl; 6.5 mM Na2HPO4; 1.5 mM KH2PO4 and 3 mM MgCl2 at pH 7.5.
Oligonucleotides
See Table 2. (1) All RNAs were synthesized, optionally modified (5′-Biotin, 5′-Atto647N, or 5′-Cy5), and purified by IBA. (2) DNA library R1, containing 60 randomized nucleotides (N60), and corresponding primers were purchased from Metabion. The T7 promoter region is underlined.
Table 2. Oligonucleotides.
| Oligonucleotide | Sequence (printed in 5′–3′ direction) | Note |
|---|---|---|
| AIR-3A |
GGGGAGGCUGUGGUGAGGG |
(1) |
| G17U |
GGGGAGGCUGUGGUGAUGG |
(1) |
| G18U |
GGGGAGGCUGUGGUGAGUG |
(1) |
| G17U/G18U |
GGGGAGGCUGUGGUGAUUG |
(1) |
| DNA library R1 |
AATGCTAATACGACTCACTATAGGAAGAAAGAGGTCTGAGACATTCT–N60–CTTCTGGAGTTGACGTTGCTT |
(2) |
| T7 primer R1 |
AATGCTAATACGACTCACTATAGGAAGAAAGAGGTCTGAGACATT |
(2) |
| RT primer R1 | AAGCAACGTCAACTCCAGAAG | (2) |
(1) All RNAs were synthesized, optionally modified (5′-Biotin, 5′-Atto647N, or 5′-Cy5), and purified by IBA. (2) DNA library R1, containing 60 randomized nucleotides (N60), and corresponding primers were purchased from Metabion. The T7 promoter region is underlined.
Cell lines
BAF/gp130 cells and BAF/gp130/IL6R/TNF cells were kindly provided by Dr. Athena Chalaris (University Kiel, Germany). Those and all other cell lines used were cultured at 37°C and 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM, PAA, E15–810) supplemented with 10% fetal bovine serum (FBS, PAA, K41–001), penicillin (60 mg/L, PAA, P11–010) and streptomycin (100 mg/L, PAA, P11–010). Culture medium for BAF/gp130 cells was further supplemented with Hyper-IL-6 (10 ng/mL).24 Culture medium for BAF/gp130/IL-6R/TNF cells was further supplemented with Interleukin-6 (10 ng/mL). For stimulation of BAF/gp130/IL6R/TNF cells with phorbol-12-myristate-13-acetate (PMA, AppliChem, A0903), cells were incubated for 2 h at 37°C in presence of 100 nM PMA. The PMA stock solution (10 mM) was prepared immediately before usage.
Protein preparation/cloning and expression of IL-6, sgp130Fc, sIL-6R and Hyper-IL-6
The proteins IL-6,50 sIL-6R,51 sgp130Fc,52 and Hyper-IL-624 were produced as previously described.
Fill in reaction and in vitro transcription
The initial single stranded DNA library R1 contained 60 randomized nucleotides (N60) flanked by two constant primer-binding sites. The forward primer (T7 primer R1) contained the sequence for the T7 RNA polymerase promoter region. The DNA library R1 was converted into dsDNA by a two-step fill in reaction. First the ssDNA library was hybridized with the reverse primer in a hybridization reaction: equal amounts (1 µM) of DNA library R1 and reverse primer (RT primer R1) were mixed in 1 × PCR buffer B (Solis BioDyne), heated to 80°C for 5 min and slowly cooled down to RT. The second strand synthesis was completed using the Klenow fragment (Thermo Fisher Scientific, EP0051) under the following conditions: 1.25 U Klenow fragment per 100 µL; 0.5 µM hybridization product; 500 µM of each dNTP in Klenow buffer (Thermo Fisher Scientific); temperature profile: 1 h 37°C, 10 min 75°C. Double stranded nucleic acids (0.1 µM) were directly used for in vitro transcription by T7 RNA polymerase for 3 h at 37°C in transcription buffer (40 mM TRIS-HCl, pH 7.9) containing T7 RNA polymerase (0.25 U/µL), NTPs (2.5 mM each) and MgCl2 (15 mM). The derived RNA library R1 was purified on 8% denaturing polyacrylamide gels.
Biotinylation of sIL-6R and immobilization on Streptavidin-coated Dynabeads
For immobilization of a biotinylated target protein on Streptavidin-coated magnetic beads (Dynabeads® M-280 Streptavidin, Invitrogen, 112.06D) 100 µg of sIL-6R were mixed with a 3-fold molar excess of Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, 10538723) in a final volume of 100 µL selection buffer followed by incubation on ice for 15 min and further 15 min at RT.53 The excess of non-reacted and hydrolyzed biotin reagent was removed by dialysis against selection buffer using a Slide-A-Lyzer® dialysis cassette (MWCO 10K; Thermo Fisher Scientific). The biotinylated protein was immobilized on 5 mg Dynabeads and suspended in selection buffer (including 1.25 µg BSA/µL).
In vitro selection procedure
In the first round of the in vitro selection process 500 pmol of the RNA library (~1013 molecules) were incubated with 100 pmol sIL-6R immobilized on magnetic beads in selection buffer (containing 1 µg BSA/µL) for 30 min at 37°C. Unbound RNA molecules were removed by magnetic separation. After washing with 200 µL selection buffer, bound RNA molecules were eluted in 50 µL water by heating the sample to 80°C for 3 min and amplified by reverse transcription and polymerase chain reaction (RT-PCR). Therefore the following RT-PCR reaction was prepared: 1 × PCR buffer B; 0.2× First-Strand Buffer (Invitrogen); forward and reverse primer (1 µM each); 1.5 mM MgCl2; 0.3 mM dNTPs; 2 mM DTT. For reverse primer hybridization the mixture was heated to 65°C for 5 min and cooled down on ice. The RT-PCR was started after addition of 15 U SuperScriptTM III Reverse Transcriptase (Invitrogen, 18080–044) and 5 U FIREPol® (Solis BioDyne, 01–01–02000) per 100 µL reaction. Following settings were used: 10 min at 54°C for the reverse transcription; for PCR amplification: 30 sec at 95°C; 30 sec at 60°C and 30 sec at 72°C for an appropriate number of PCR cycles. For the subsequent rounds of selection the derived dsDNAs were transcribed into a RNA library as described above. To increase the stringency during the following rounds of selection the number of washing steps was raised by one each round. After 16 rounds of this in vitro selection process the dsDNA library was cloned via TOPO TA Cloning (pCR2.1, Invitrogen, K456001) and individual clones were sequenced.
Filter retention assay (FRA)
To investigate the binding of RNA molecules to the target protein, filter retention assays were performed. Nucleic acids were radioactively labeled during T7 transcription by incorporation of [α-32P]-ATP (3,000 Ci/mmol, Hartmann Analytic, SCP-207). After gel purification, constant amounts of labeled RNA (<1 nM) were incubated with increasing amounts of the target protein (0–500 nM) in 1× selection buffer. After incubation the samples were filtered through a pre-equilibrated nitrocellulose membrane (0.45 µm, Carl Roth) on a vacuum manifold (Minifold® I Dot-Blot-System; Schleicher & Schuell) and washed four times with selection buffer. The nitrocellulose membrane was dried and exposed to a phosphor-imaging screen (Bio-Rad). The amount of radioactively labeled RNA on the filter was quantified using Quantity One® software (Version 4.6.6, Bio-Rad) and used for the calculation of the bound RNA fraction. For the determination of characteristic Kd-values (dissociation constants) and Bmax-values (maximal binding) data were fitted using a one site-binding model with the aid of the program GraphPad Prism. The following equation was used:
RNAbound = (Bmax × cProtein) / (Kd + cProtein)
Electrophoretic mobility shift assay (EMSA)
RNA-protein-interactions were investigated using a native electrophoretic mobility shift assay. Radioactively labeled RNAs (<1 nM) were incubated with proteins of interest in selection buffer for 30 min at room temperature. 6× DNA Loading Dye (Thermo Fisher Scientific, R0611) was added and samples were loaded on 5% non-denaturing polyacrylamide gels (acrylamide/bisacrylamide 37.5:1) and electrophoresed at 60 V for 2–3 h in 1× TBE buffer. The gel was dried on a vacuum dryer at 70°C for 2 h, exposed to a phosphor imager screen over night, and detected as described above.
Circular dicroism (CD) spectroscopy
For CD spectroscopy RNAs were dissolved in four different buffers (PBS, 50 mM TRIS-HCl (pH 7.5), and 50 mM TRIS-HCl (pH 7.5) optionally containing 100 mM KCl or NaCl; final RNA concentration 5 µM). CD spectra were recorded using a Jasco J-815 CD spectrometer at 25°C. Each spectrum was accumulated for two times and corresponding values were averaged.
UV spectroscopy
For UV-melting experiments RNAs were dissolved in 10 mM TRIS-HCl (pH 7.5) optionally containing 1 mM, 5 mM or 10 mM KCl. RNA concentrations ranged from 1 µM to 10 µM. UV-melting studies of prepared RNAs were conducted on a Varian Cary Bio 300 UV-Visible Spectrophotometer with a temperature controller. Each sample (1200 µL) was filled into a quartz cuvette (1-cm path length), covered with a thin layer of mineral oil, transferred to the spectrophotometer, heated to 80°C and cooled down to 20°C for two times with a heating or cooling rate of 0.5°C min−1. Absorbance was recorded at 295 nm every 30 sec. Each melting curve was analyzed using the method of van’t Hoff to determine the Tm value.54,55
Flow cytometry
The presentation of human IL-6R and human gp130 on the surface of BAF/gp130/IL6R/TNF or BAF/gp130 cells was determined by flow cytometry using antibodies specific for human IL-6R and human gp130, respectively. Five hundred-thousand cells were washed two times in 1× selection buffer and suspended in 350 µL 1 × selection buffer. After addition of a murine primary antibody binding to human IL-6R (B-R6, antibodies-online, ABIN123898) or human gp130 (R&D Systems, MAB228), respectively, or an isotype-specific control antibody (Tetra-His antibody, Qiagen, 34670), in final concentrations of 0.3 ng/µL, cells were incubated for 30 min on ice. Three washing steps with 350 µL 1 × selection buffer followed. Cells were incubated with an APC- or FITC-labeled secondary antibody (1:350 diluted; Th.Geyer, 550826, or Santa Cruz Biotechnology, sc-2078, respectively) in 350 µL 1 × selection buffer for 30 min at 4°C. Cells were washed as previously described and resuspended in 350 µL 1 × selection buffer. Fluorescence intensities were determined by a FACSCalibur flow cytometer (BD Biosciences) counting 10.000 events and evaluated using the BD CellQuest software (Version 3.2.1).
Binding of the IL-6R-specific aptamer AIR-3A or its variant G17U to the surface of defined cells was determined by flow cytometry. Therefore 500,000 cells were washed twice in 1 × selection buffer, serum containing or lacking culture medium. Cy5- or Atto®647N-labeled RNAs (25 nM each) were incubated with 500,000 cells in corresponding solutions between 1 min and 8 h at 37°C or 4°C. Afterwards cells were washed, suspended in 350 µL of the same buffer and analyzed by flow cytometry as described above.
To measure the relative affinities of Atto®647N-labeled aptamer AIR-3A and the variant G17U for BAF/gp130/IL6R/TNF-cells, increasing concentrations (0 nM–150 nM) of corresponding RNAs were mixed with 500,000 cells each. After incubation at 37°C for 20 min cells were treated as previously described and analyzed by flow cytometry. Median fluorescence intensities (MFI) were plotted against RNA concentrations using GraphPad Prism software.
Internalization studies of aptamer AIR-3A using laser scanning microscopy
To visualize the internalization of aptamer AIR-3A of into BAF/gp130/IL6R/TNF-cells 500,000 cells were incubated with Atto®647N-labeled aptamer AIR-3A (25 nM) in 350 µL 1 × selection buffer for 30 min at 37°C. Cells were washed two times with 350 µL 1 × selection buffer and resuspended in 50 µL 1 × selection buffer only.
Suspensions were directly placed on a glass slide and covered by a coverslip. Samples were imaged with LSM 510 ConfoCor2 system (Carl Zeiss). Basic adjustments used: HeNe-Laser (633 nm), 5–15% laser power, 92–896 µm pinhole diameter, beam splitters: HFT 514/633 nm and NFT 545 nm, LP 650 nm filter.
Binding and internalization studies of aptamer-streptavidin-complexes
For preparation of aptamer-streptavidin-complexes, biotinylated RNAs (30 µM) and biotinylated fluorescent dye Atto635 (10 µM; ATTO-TEC, AD 635–71) were mixed in 1 × selection buffer. Streptavidin (New England Biolabs, N7021S) was added to a final tetrameric concentration of 10 µM and the reaction mixture was incubated for 10 min at room temperature. The formation of fluorescently labeled aptamer-streptavidin-complexes was analyzed by native PAGE (10% PAA; acrylamide/bisacrylamide 37.5:1). Resulting bands were visualized by fluorescence detection (filter color: red; BP: 695 nm) using a VersaDoc Imaging System (Bio-Rad).
Binding of Alexa635-labeled complexes to BAF/gp130/IL6R/TNF-cells was tested by flow cytometry. Complexes (25 nM) were mixed with 500,000 cells in selection buffer, incubated for 20 min at 37°C. Cells were analyzed as described above.
To visualize the internalization of aptamer-streptavidin-complexes into BAF/gp130/IL6R/TNF-cells 500,000 cells were incubated with the Atto635-fluorescently labeled complexes (25 nM) in 350 µL 1 × selection buffer for 10 min at 4°C or 37°C. In order to distinguish between cell surface bound complexes and intracellular complexes, cells were optionally treated with trypsin (PAA, L11–004). This protease unspecifically degrades proteins of the cell surface and protein components of the complexes outside the cells. Cells were washed two times with 350 µL 1 × selection buffer and resuspended in 50 µL 1 × selection buffer only.
Suspensions were placed on glass slides, covered by coverslips and imaged using the LSM 510 ConfoCor2 system (Carl Zeiss) as described above.
Supplementary Material
Supplementary file supplied by authors.
Acknowledgments
We are grateful to Elena Wasiljew and Viswatej Avutu for excellent technical assistance and Andrea Rentmeister for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft [SFB 877, projects A1, A6 to J.G. and S.R.-J.; MA 3442/1–1 to G.M.], the cluster of excellence ‘inflammation at interfaces' (to J.G. and S.R.-J.), and the Bundesministerium für Forschung und Bildung (to G.M.).
Glossary
Abbreviations:
- IL-6
Interleukin-6
- IL-6R
Interleukin-6 receptor
- sIL-6R
soluble Interleukin-6 receptor
- gp130
glycoprotein 130
- RA
rheumatoid arthritis
- SELEX
systematic evolution of ligands by exponential enrichment
- PMA
phorbol-12-myristate-13-acetate
- FRA
filter retention assay
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/18062
References
- 1.Kallen KJ, zum Buschenfelde KH, Rose-John S. The therapeutic potential of interleukin-6 hyperagonists and antagonists. Expert Opin Investig Drugs. 1997;6:237–66. doi: 10.1517/13543784.6.3.237. [DOI] [PubMed] [Google Scholar]
- 2.Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006;80:227–36. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 3.Horiuchi S, Koyanagi Y, Zhou Y, Miyamoto H, Tanaka Y, Waki M, et al. Soluble interleukin-6 receptors released from T cell or granulocyte/macrophage cell lines and human peripheral blood mononuclear cells are generated through an alternative splicing mechanism. Eur J Immunol. 1994;24:1945–8. doi: 10.1002/eji.1830240837. [DOI] [PubMed] [Google Scholar]
- 4.Müllberg J, Schooltink H, Stoyan T, Gunther M, Graeve L, Buse G, et al. The soluble interleukin-6 receptor is generated by shedding. Eur J Immunol. 1993;23:473–80. doi: 10.1002/eji.1830230226. [DOI] [PubMed] [Google Scholar]
- 5.Febbraio MA, Rose-John S, Pedersen BK. Is interleukin-6 receptor blockade the Holy Grail for inflammatory diseases? Clin Pharmacol Ther. 2010;87:396–8. doi: 10.1038/clpt.2010.1. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Rajendran M, Ellington AD. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem. 2008;390:1067–75. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 9.Holeman LA, Robinson SL, Szostak JW, Wilson C. Isolation and characterization of fluorophore-binding RNA aptamers. Fold Des. 1998;3:423–31. doi: 10.1016/S1359-0278(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 10.Schürer H, Stembera K, Knoll D, Mayer G, Blind M, Forster HH, et al. Aptamers that bind to the antibiotic moenomycin A. Bioorg Med Chem. 2001;9:2557–63. doi: 10.1016/S0968-0896(01)00030-X. [DOI] [PubMed] [Google Scholar]
- 11.Helmling S, Maasch C, Eulberg D, Buchner K, Schroder W, Lange C, et al. Inhibition of ghrelin action in vitro and in vivo by an RNA-Spiegelmer. Proc Natl Acad Sci USA. 2004;101:13174–9. doi: 10.1073/pnas.0404175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopinath SC, Sakamaki Y, Kawasaki K, Kumar PK. An efficient RNA aptamer against human influenza B virus hemagglutinin. J Biochem. 2006;139:837–46. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 14.Raddatz MS, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew Chem Int Ed Engl. 2008;47:5190–3. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 15.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15:675–82. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann AP, Somasunderam A, Nieves-Alicea R, Li X, Hu A, Sood AK, et al. Identification of thioaptamer ligand against E-selectin: potential application for inflamed vasculature targeting. PLoS One. 2010;5:pii: e13050. doi: 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–33. [PubMed] [Google Scholar]
- 18.Chu TC, Marks JW, 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–92. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 19.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 20.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci USA. 2011;108:1850–5. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–49. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiechl G, Bauer B, Fuss I, Lang SA, Moser C, Ruemmele P, et al. Tumor development in murine ulcerative colitis depends on MyD88 signaling of colonic F4/80+CD11bhighGr1low macrophages. J Clin Invest. 2011;121:1692–708. doi: 10.1172/JCI42540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Fischer M, Goldschmitt J, Peschel C, Brakenhoff JP, Kallen KJ, Wollmer A, et al. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol. 1997;15:142–5. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 25.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J Biol Chem. 1994;269:21858–69. [PubMed] [Google Scholar]
- 26.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–7. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halder K. Hartig JS. RNA Quadruplexes. In: Astrid Sigel, Helmut Sigel, Sigel RKO, eds. Structural and Catalytic Roles of Metal Ions in RNA. Cambridge: The Royal Society of Chemistry, 2011. [PubMed] [Google Scholar]
- 28.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–8. doi: 10.1016/S0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 29.Chalaris A, Gewiese J, Paliga K, Fleig L, Schneede A, Krieger K, Rose-John S, Scheller J. ADAM17-mediated shedding of the IL6R induces cleavage of the membrane stub by gamma-secretase. Biochim Biophys Acta. 2010;1803:234–45. doi: 10.1016/j.bbamcr.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Romig TS, Bell C, Drolet DW. Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J Chromatogr B Biomed Sci Appl. 1999;731:275–84. doi: 10.1016/S0378-4347(99)00243-1. [DOI] [PubMed] [Google Scholar]
- 31.Kirby R, Cho EJ, Gehrke B, Bayer T, Park YS, Neikirk DP, et al. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal Chem. 2004;76:4066–75. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 32.Yan XR, Gao XW, Yao LH, Zhang ZQ. Novel methods to detect cytokines by enzyme-linked oligonucleotide assay. Sheng Wu Gong Cheng Xue Bao. 2004;20:679–82. [ ] [PubMed] [Google Scholar]
- 33.Müller J, Freitag D, Mayer G, Potzsch B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J Thromb Haemost. 2008;6:2105–12. doi: 10.1111/j.1538-7836.2008.03162.x. [DOI] [PubMed] [Google Scholar]
- 34.Charlton J, Sennello J, Smith D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem Biol. 1997;4:809–16. doi: 10.1016/S1074-5521(97)90114-9. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Li DQ, Zhong J, Wu XL, Chen Q, Peng H, et al. IL-17RA aptamer-mediated repression of IL-6 inhibits synovium inflammation in a murine model of osteoarthritis. Osteoarthritis Cartilage. 2011;19:711–8. doi: 10.1016/j.joca.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 37.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 38.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105:17356–61. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, Jeong YY, Jon S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano. 2010;4:3689–96. doi: 10.1021/nn901877h. [DOI] [PubMed] [Google Scholar]
- 40.Chen CH, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci USA. 2008;105:15908–13. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer C, Hahn U, Rentmeister A. Cell-specific aptamers as Emerging Therapeutics. J Nucleic Acids. doi: 10.4061/2011/904750. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lattanzio G, Libert C, Aquilina M, Cappelletti M, Ciliberto G, Musiani P, et al. Defective development of pristane-oil-induced plasmacytomas in interleukin-6-deficient BALB/c mice. Am J Pathol. 1997;151:689–96. [PMC free article] [PubMed] [Google Scholar]
- 43.Taga T, Kawanishi Y, Hardy RR, Hirano T, Kishimoto T. Receptors for B cell stimulatory factor 2. Quantitation, specificity, distribution, and regulation of their expression. J Exp Med. 1987;166:967–81. doi: 10.1084/jem.166.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyers L, Fontaine V, Content J. Modulation of interleukin-6 receptors in human cells. Ann N Y Acad Sci. 1989;557:388–93, discussion 94-5. doi: 10.1111/j.1749-6632.1989.tb24031.x. [DOI] [PubMed] [Google Scholar]
- 45.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–29. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zohlnhöfer D, Graeve L, Rose-John S, Schooltink H, Dittrich E, Heinrich PC. The hepatic interleukin-6 receptor. Down-regulation of the interleukin-6 binding subunit (gp80) by its ligand. FEBS Lett. 1992;306:219–22. doi: 10.1016/0014-5793(92)81004-6. [DOI] [PubMed] [Google Scholar]
- 47.Korolenko TA, Heinrich PK, Hemmann U, Weiergraber O, Dittrich E, Graeve L. Cell endocytosis of the complex interleukin-6-soluble interleukin-6 receptor and its intralysosomal degradation. Biull Eksp Biol Med. 1997;124:527–9. doi: 10.1007/BF02445668. [ ] [DOI] [PubMed] [Google Scholar]
- 48.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 49.Thiel KW, Giangrande PH. Intracellular delivery of RNA-based therapeutics using aptamers. Ther Deliv. 2010;1:849–61. doi: 10.4155/tde.10.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dam M, Mullberg J, Schooltink H, Stoyan T, Brakenhoff JP, Graeve L, et al. Structure-function analysis of interleukin-6 utilizing human/murine chimeric molecules. Involvement of two separate domains in receptor binding. J Biol Chem. 1993;268:15285–90. [PubMed] [Google Scholar]
- 51.Mackiewicz A, Schooltink H, Heinrich PC, Rose-John S. Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J Immunol. 1992;149:2021–7. [PubMed] [Google Scholar]
- 52.Jostock T, Mullberg J, Ozbek S, Atreya R, Blinn G, Voltz N, et al. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur J Biochem. 2001;268:160–7. doi: 10.1046/j.1432-1327.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- 53.Mayer G, Wulffen B, Huber C, Brockmann J, Flicke B, Neumann L, et al. An RNA molecule that specifically inhibits G-protein-coupled receptor kinase 2 in vitro. RNA. 2008;14:524–34. doi: 10.1261/rna.821908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–97. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–20. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file supplied by authors.