Abstract
We have analyzed the off-target activity of two siRNAs (F7-1, F7-2) that knock-down human blood coagulation factor 7 mRNA. F7-1 modulates a significant number of non-target transcripts while F7-2 shows high selectivity for the target transcript under various experimental conditions. The 3′-UTRs of all F7-1 off-target genes show statistically significant enrichment of the reverse complement of the F7-1 siRNA seed region located in the guide strand. Seed region enrichment was confirmed in off-target transcripts modulated by siRNA targeting the glucocorticoid receptor. To investigate how these sites contribute to off-target recognition of F7-1, we employed CXCL5 transcript as model system because it contains five F7-1 seed sequence motifs with single base mismatches. We show by transient transfection of reporter gene constructs into HEK293 cells that three out of five sites located in the 3′-UTR region are required for F7-1 off-target activity. For further mechanistic dissection, the sequences of these sites were synthesized and inserted either individually or joined in dimeric or trimeric constructs. Only the fusion constructs were silenced by F7-1 while the individual sites had no off-target activity. Based on F7-1 as a model, a single mismatch between the siRNA seed region and mRNA target sites is tolerated for target recognition and the CXCL5 data suggest a requirement for binding to multiple target sites in off-target transcripts.
Keywords: off-target effects, seed region, single base mismatch, siRNA
Introduction
RNA interference (RNAi) is a naturally occurring mechanism in higher eukaryotes that degrades foreign RNA introduced by viral invaders and other pathogens. The RNAi machinery is well-conserved from plants to animals and allows in mammalian cells not only cell defense mechanisms but also endogenous micro-RNAs (miRNAs) controlled gene regulation. miRNAs are short intracellular RNA duplexes that bind in a sequence-dependent manner to target transcripts, resulting in reduced transcript abundance either by Ago-2 dependent mRNA degradation or by mRNA destabilization.1 Shortly after discovery of the RNAi mechanism in nematodes by Fire et al.,2 Elbashir et al.3 demonstrated that transfection of short double-stranded RNAs (siRNAs) can trigger in mammalian cells RNAi-mediated decay of specific transcripts. This mechanism, also known as exogenous RNAi pathway, has led siRNA to become an indispensable and valuable research tool for functional genomics in cell lines and rodent animal models.4
Based on the possibility of targeting the transcript of virtually any gene in the cell, significant interest in exploring RNAi as a novel treatment modality for human diseases was elicited.5,6 However, this approach suffers from a lack of efficient and specific delivery vehicles to mediate transport of the drug to the target organ. In vivo activity of siRNA delivered to the liver by liposome-based nanoparticles was first demonstrated in primates where down-modulation of Apolipoprotein B resulted in significant lowering of blood cholesterol levels.7 This study raised further hope for the application of RNAi therapy to humans although the toxicity of the delivery vehicle is currently a serious concern for advancement into the clinic. The short siRNA length of 21 base pairs on one hand and the complexity of eukaryotic genomes on the other hand limit the design of target-specific siRNAs with unique binding sites. Sequence-independent responses like activation of the innate immune system can largely be circumvented by chemical modifications.8 Today, it is still challenging to predict off-target effects based on computational algorithms since the exact molecular determinants that direct on- and off-target recognition of transcripts are unclear.9-11 Several studies have shown that target complementarity to the 7-nucleotide seed region of siRNA (positions 2–8 from the 5′ end on the guide strand) is sufficient for entry into the RNA-induced silencing complex (RISC) and subsequent mRNA cleavage and degradation by a microRNA-like mechanism.12-14 In order to minimize seed sequence-driven off-target binding, siRNA design rules have been developed that take thermodynamic stability, balanced GC content as well as structural and positional parameters into account.8 Furthermore understanding of the rules that drive miRNA binding to specific mRNA targets is likely to improve siRNA target selectivity which is highly desirable for therapeutic applications in humans.
Here we analyze the off-target activity of two synthetic siRNAs that target human blood coagulation factor 7 (F7) mRNA in a panel of human cell lines as model. Based on in silico prediction these siRNAs, termed F7-1 and F7-2, are devoid of significant sequence complementarity to any other human transcript. We applied genome-wide microarray profiling to experimentally address this prediction and we showed that F7-1 modulates a significant number of off-target transcripts while F7-2 is virtually mono-specific for F7. We used the F7-1 off-target CXCL5 as model system to analyze the mechanism of off-target recognition by transient transfection of appropriately designed reporter constructs. This analysis showed that cooperative binding of F7-1 siRNA to at least two sites with seed sequence similarity is required to induce off-target activity. Possible relevance of our findings for the development of RNAi therapeutics is discussed.
Results
On-target activity of F7-1 and F7-2
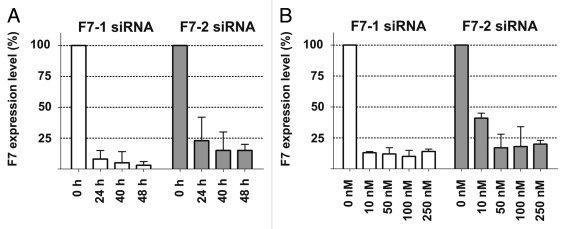
F7-1 and F7-2 siRNAs target human blood coagulation factor 7 either in the coding region or in the 3′-UTR respectively. Both siRNAs were considered for development of RNAi therapeutics for treatment of blood clotting disorders. Therefore it is critical to test whether these molecules have off-target activity although complementarity searches in the human transcriptome did not yield any significant hits. Here we use a panel of human cell lines under various experimental conditions to investigate the specificity of both siRNAs. To validate the experimental system, we transfected the human hepatoma cell line Hep3B with F7-1 or F7-2 and measured knock-down of factor 7 mRNA at various time points after transfection (Fig. 1A) or by applying a concentration range from 10 nM to 250 nM (Fig. 1B). Both siRNAs show comparable on-target knock-down activity between 75% (F7-2) and 85% (F7-1) 24 h after transfection at a concentration of 10 nM (Fig. 1A). Both siRNAs show maximal knock-down activity at 50 nM and further increase beyond this point had no significant effect (Fig. 1B).
Figure 1. Time- and concentration-dependent knock-down of F7 mRNA by F7–1 and F7–2 siRNA in Hep3B cells. (A) Cells were transfected with each siRNA at 10 nM and knock-down efficiency was measured followed 24h, 40h and 48h incubation relative to mRNA levels of lipofectamine transfected cells set to 100%. (B) Cells were transfected with increasing amounts of siRNA and F7 knock-down was measured 24 h after transfection with lipofectamine transfected cells as reference. All quantifications were performed with a commercial branched-DNA assay (Panomics). For details see Materials and Methods.
Off-target activity of F7-1 and F7-2
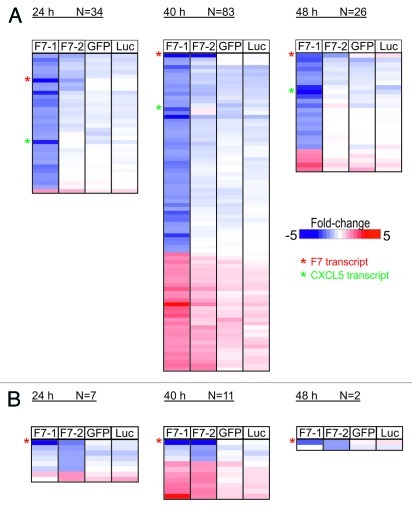
We next analyzed whether target knock-down of F7 mRNA by F7-1 or F7-2 targets additional transcripts. We repeated the time course experiment shown above and analyzed the responses to F7-1 and F7-2 using whole-genome transcript imaging together with green-fluorescence protein (GFP) or luciferase (Luc) siRNA as non-targeting controls in Hep3B cells (Fig. 2). We applied a fold-change of 2 (FC > 2 or FC < -2) and a p value < 0.05 as parameters to call off-target genes for each experimental condition. Twenty-four hours after transfection F7-1 yields 34 transcripts fulfilling our selection criteria and this set of genes is only marginally modulated by F7-2 or by established controls targeting GFP and luciferase (Fig. 2A).11 At 40 h the expression of 83 genes is affected by F7-1 and the number of modulated off-target genes declines after 48 h to levels similar to the 24 h time point. In contrast to F7-1, F7-2 siRNA modulates significantly fewer transcripts with a maximum of 11 genes at 40 h and only two genes are affected 48 h after transfection (Fig. 2B). As expected, the genuine target F7 is down-modulated under F7-1 and F7-2 conditions with similar efficiency.
Figure 2. Time-dependent induction of off-target effects by F7-1 (A) and F7-2 (B) siRNAs in Hep3B cells measured by genome wide microarray profiling. Hep3B cells were transfected with F7-1, F7-2, GFP or luciferase siRNA at 10 nM for the time points indicated above the heatmaps. (A) shows transcripts modulated by F7-1 and the fold change of the same set of transcripts is shown in cells transfected with F7-2, GFP or luciferase siRNA to illustrate specificity for F7-1. (B) shows the equivalent data for F7-2. Expression fold change is displayed as a color gradient from blue (downregulation) to red (upregulation) as indicated by the reference bar. Each line in the heatmap display corresponds to a gene and gene symbols were omitted for clarity. Red asterisk in the heatmap indicate expression levels of F7 for the different conditions while green asterisk indicate expression levels of CXCL5.
siRNA concentration-dependent off-target activity
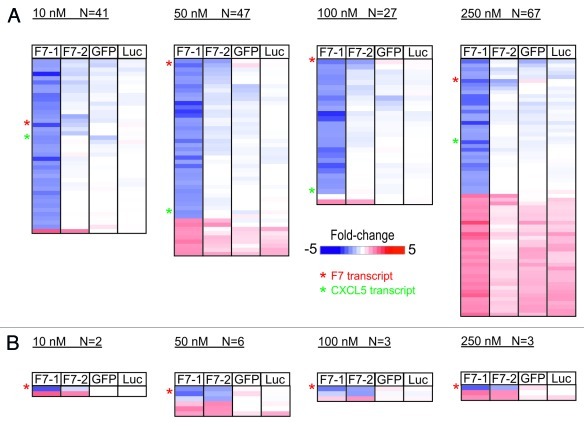
We next asked whether an increase in the siRNA concentration far above effective levels would raise the number of co-modulated transcripts. From DNA gel blotting experiments it has long been known that an excess of probe leads to unspecific hybridization and high background. Related to RNAi, overloading of the intracellular RISC machinery with exogenous siRNAs could relax the control of gene expression by the miRNA system resulting in upregulation of genes that are normally repressed by miRNAs.15 We transfected Hep3B cells for 24 h with the same set of siRNAs used above (F7-1, F7-2, GFP and Luc) at 10 nM, 50 nM, 100 nM and 250 nM. For F7-1 the number of down-modulated genes remained in the same order of magnitude across the entire concentration range (Fig. 3A) while F7-2 has only marginal off-target activity (Fig. 3B). At high F7-1 concentration (250 nM) about 30 genes are significantly upregulated. The same set is modulated to some extent by F7-2, GFP or luciferase siRNAs. Based on the intracellular RISC machinery overloading model,15 we would expect significant upregulation of numerous genes by F7-2 which was not the case. Hence, the induction of mRNA expression is at least partially related to the F7-1 siRNA sequence.
Figure 3. siRNA concentration-dependent induction of off-target effects by F7–1 (A) and F7–2 (B) siRNA in Hep3B cells measured by genome wide microarray profiling after 24h treatment. For details see legend of Figure 2.
Off-target activity of F7-1 in other cell lines
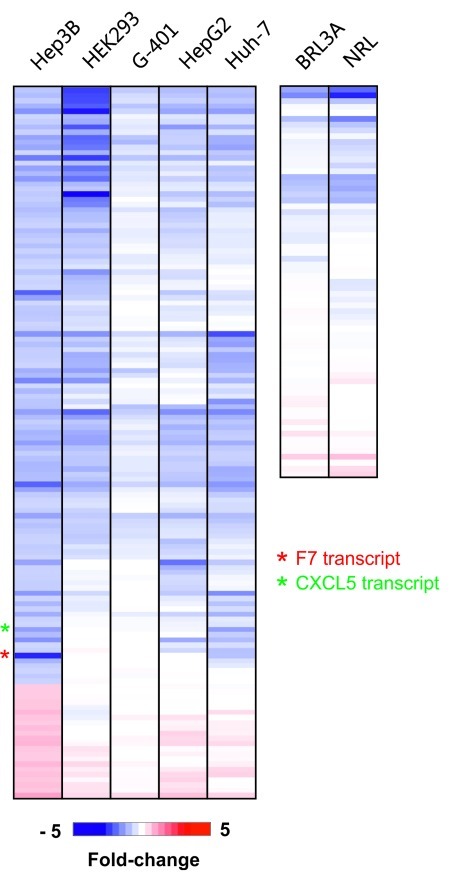
The RNAi host defense mechanism is considered ubiquitous and operates in virtually all vertebrate tissues and cell lines. We tested whether host cell specific factors such as miRNA content influence off-target activity of siRNA. In addition to our standard cell line Hep3B, we transfected the human hepatoma cell lines HepG2 and Huh-7 together with the human kidney cell lines G-401 and HEK293 with F7-1 for 24 h at 10 nM followed by microarray-based off-target analysis as above. In addition, we included the rat liver cell lines BRL3A and NRL for the assessment of species restriction of RNAi as shown by Burchard et al.16 We used the gene panel modulated by F7-1 in Hep3B as reference (fold-change > 1.5 or \ < -1.5 and p value < 0.05) and retrieved the corresponding expression data from the other cell lines. Based on this comparison, virtually all genes that are downregulated in Hep3B are also modulated in at least one of the added cell lines, provided the transcript is expressed (Fig. 4; left panel). In the rat cell lines only a small portion (< 10) of the human F7-1 off-target orthologs is down-modulated (Fig. 4; right panel) which is consistent with species restriction of RNAi.16
Figure 4. Detection of F7-1 off-target transcripts in additional human and rat cell lines. The human hepatoma cell lines Hep3B, HepG2 and Huh-7 and the human kidney lines G-401 and HEK293 were transfected with 10 nM F7-1 siRNA for 24 h followed by transcript profiling. In addition, the rat cell lines BRL3A and NRL cells were included to demonstrate species specificity of siRNA off-target effects. Analogous to the experiments shown in Figures 2and3, the transcripts modulated by F7-1 in Hep3B cells were used as reference and the fold change-factor of these genes in the other cell lines is shown in a gradient from blue (downregulation) to red (upregulation) as in Figures 2and3. For the rat cell lines the fold change of the human orthologs present on the rat array is shown. Note that virtually all genes modulated in Hep3B are found in at least one of the other human lines with exception of the upregulated genes.
Enrichment analysis of seed region matches in F7-1 off-target genes
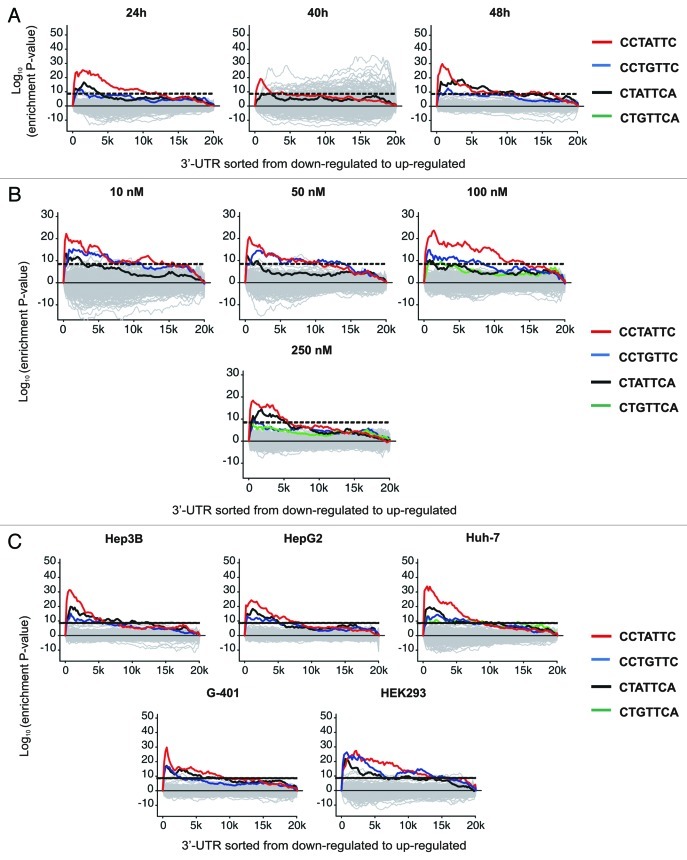
The microarray-based off-target analysis suggests a sequence-dependent mechanism of transcript recognition by F7-1 since a BLAST-based search of all known human transcripts with both full-length sequences of F7-1 and F7-2 did not generate any significant matches. Therefore, we had to consider the possibility that seed sequence similarity to F7-1 (5′-end positions 2–8 of the antisense guide strand) triggers recognition and subsequent down-modulation of off-target transcripts.14 To address this possibility, we performed a word enrichment analysis based on the Sylamer program which was originally developed for the in silico detection of potential miRNA targets.17 We scanned the 3′-UTRs of all 20,000 transcripts detected by the microarray under all conditions (time, dose and cell type) for enrichment of unique heptamer motifs after purging repetitive stretches and poly-A tails from the database entries. According to van Dongen et al., a word becomes significant when the log10 (enrichment p value) is higher than 8. The most over-represented word fulfilling these criteria across the entire data set is the reverse complement of the F7-1 seed region 5′ CCTATTC 3′, followed by the single mismatch version 5′ CCTGTTC 3′ (Fig. 5; A, time course; B, concentration; C, human cell lines). Strikingly, significant enrichment of the single mismatch version of 5′ CCTATTC 3′ occurred specifically in the 3′-UTRs of the most downregulated F7-1 off-target transcripts. This finding suggests that even imperfect matching to the siRNA seed region can be sufficient for off-target recognition and entry into the RISC pathway. This result further supports our previous notion that target recognition by F7-1 is sequence driven and we consider perfect or imperfect seed region similarity as a pre-requisite for off-target recognition. As control, we scanned the 5′-UTRs of the same gene set for heptamer enrichment which did not yield any significant enrichment data. Furthermore, the ‘Sylamer’ analysis reveals that more than half of the down-modulated transcripts have multiple perfect or partial F7-1 seed region matches. A positive correlation between the number of seed sequence matches in the 3′-UTR and siRNA silencing potency has been shown by others.14 We therefore sorted all F7-1 off-target transcripts according to seed sequence frequency but could not confirm the reported association between seed region frequency and knock-down efficiency in our data set.14 The adenylate kinase 4 (AK4) transcript for instance contains 11 imperfect copies of the F7-1 seed motif in the 3′-UTR and yet it is only moderately down-modulated by F7-1 (data not shown). Interestingly, the F7-1 seed motif was not recovered in any of the genes that are upregulated at high siRNA concentrations or prolonged exposure (Figs. 2 and 3) suggesting an RNAi-independent mechanism. In addition, microarray profiling can only detect changes in transcript abundance and changes of protein abundance caused by translational inhibition require proteomic approaches.18
Figure 5. Enrichment analysis of the F7-1 seed region matches in the 3′-UTR sequence of all expressed genes considering all experimental conditions (A, dose, B, time course, C, human cell lines panel). The ‘Sylamer’ algorithm scans the 3′-UTRs for enrichment of random heptamer motifs. The reverse complement of the F7-1 seed region (red) is significantly enriched in the panel of modulated genes followed by the single mismatch version (5′ CCTGTTC 3′; blue) in most conditions. Enrichment of the 5′ CTATTCA 3′ (black) or 5′ CTGTTCA 3′ (green) heptamers was considered less significant by the program. In the graphic display, the most downregulated genes appear on the left side of the x-axis and induced genes are on the right side of each window. The curve indicates the frequency of each motif and the statistical cut-off is marked by a dotted line. The log10 of the enrichment p value is plotted on the y-axis. Motifs without any significant enrichment in either condition are shown in gray.
Seed sequence enrichment in siRNAs targeting the glucocorticoid receptor
To further support the conclusions drawn above, we assessed the off-target activity of two additional siRNAs (GCR-1 and GCR-2) targeting the human glucocorticoid receptor (GR). Both siRNAs were considered as candidates for clinical development because they had the lowest number of predicted off-targets in the human transcriptome based on in silico screening. Similar to F7, GCR-1 targets the coding region and GCR-2 the 3′-UTR of the glucocorticoid receptor transcript. To keep the study in a manageable scope we performed transcript profiling at a concentration of 10 nM for 24 h followed by transcript profiling and ‘Sylamer’ analysis as above. To avoid mRNA target modulation by GR signaling we have chosen Hep3B cells for the analysis because these cells do not express the glucocorticoid receptor. Under these conditions, complement factor B (CFB) was the only GCR-1 off-target gene (data not shown) while GCR-2 targeted a total of 38 genes fulfilling our selection criteria (Fig. S1, A). We used all transcripts detected in the data set for GCR-1 and GCR-2 for a heptamer motif enrichment analysis using the Sylamer algorithm as above. No significant motif was enriched in the GCR-1 data set (Fig. S1, B) while significant enrichment of the reverse complement of the GCR-2 seed region (5′ ATATAGA 3′) occurred in genes down-modulated by GCR-2 (Fig. 1C). We conclude that seed sequence homology is required for entry into the RISC mediated RNAi pathway.
Mechanism of F7-1 mediated off-target recognition of CXCL5 transcripts
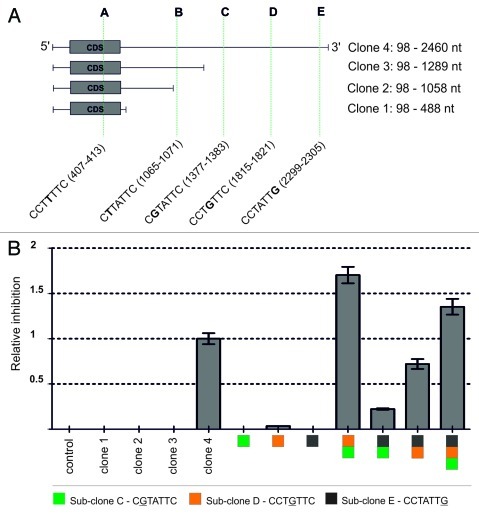
Next we analyzed the requirement for seed sequence based off-target recognition. In particular, we tested whether binding of siRNA to multiple sites in the 3′-UTR as suggested by Lin et al.14 is necessary or whether binding of F7-1 to a single site is sufficient for target knock-down. We chose the chemokine gene CXCL5 as model because it is consistently silenced by F7-1 under all experimental conditions and the microarray data were confirmed using branched-DNA assay (data not shown; Figs. 2 and 3). In addition, CXCL5 3′-UTR region contains five evenly spaced seed motifs (named A, B, C, D and E) which allows experimental dissection of the role of each heptamer motif by mutagenesis (Fig. 6). To identify the target sites of F7–1 experimentally we employed the commercial pMIR-REPORT luciferase reporter gene system originally designed to detect short miRNA target sites in 3′-UTRs. We cloned the full-length cDNA of CXCL5 transcript (2,460 base pairs) into pMIR-REPORT (clone 4) and three truncated versions lacking sites C, D and E (Fig. 6A). These constructs were transfected into CXCL5 negative HEK293 cells, together with F7-1 (10 nM, 24 h) and the β-galactosidase signal normalization vector. Using this assay, only clone 4 showed significant inhibition (Fig. 6B) suggesting that the heptamer sites C, D and E in the CXCL5 transcript are preferred targets of F7-1. It further implies that single mismatches are tolerated in the process of mRNA target recognition.
Figure 6. Schematic outline of the CXCL5 cloning strategy used for F7–1 binding site mapping. (A) Clone 4 contains the full-length CXCL5 cDNA sequence without poly-A tail. Clones 1–3 represent truncated versions of CXCL5 sequence containing seed motifs A–E with the exact positions indicated in brackets. The coding region of CXCL5 is shown as a gray box. (B) Detection of F7-1 predicted binding sites in CXCL5 cDNA clones by reporter gene assays. The CXCL5 pMIR_REPORT sub-clones 1–4 were transiently co-transfected into HEK293 cells with siRNA and β-galactosidase plasmid for calibration. The data represent the average of three independent experiments with three biological replicates each.
To further dissect the roles of binding sites C, D and E we generated additional reporter constructs containing the C, D and E seed region complement sites including 25 nucleotides upstream of the predicted binding motif. In addition to monomer constructs, we also assembled clones containing all possible dimer configurations and a single construct combing all three binding sites. The resulting plasmids were co-transfected into HEK293 cells together with F7-1 and the β-galactosidase normalization plasmid. None of the monomer sub-clones C (green), D (orange) or E (black) responded to F7-1 while the dimers responded in the order C-D, D-E and C-E (Fig. 6B). The triple construct C-D-E showed inhibition in the range of C-D. These results provide first experimental evidence that a single mismatch in a discontinuous stretch of the seed sequence is tolerated to trigger RNAi as suggested based on isothermal titration calorimetry of siRNA/RISC complexes.19 The functional dissection of the F7-1 binding sites suggests that off-target activity of siRNA requires annealing to at least two seed complement sites on the target strand.
Discussion
We have analyzed the off-target activity of a pair of siRNAs (F7-1 and F7-2) that target human blood coagulation factor 7 with high efficiency. We used microarray profiling to detect co-regulated genes under various experimental conditions and in multiple cell lines. Based on this analysis, F7-1 suppresses about 40 off-target transcripts greater than 2-fold while F7-2 is essentially mono-specific for its target. All transcripts targeted by F7-1 contain between one and 11 heptamer motifs complementary to the seed region of F7-1 with single base mismatches ranging from 1 to 10 occurrences. Seed sequence enrichment in off-target transcripts was confirmed independently for GCR-2 which targets the glucocorticoid receptor and has similar off-target activity as F7-1. Using the chemokine gene CXCL5 as an off-target model, we have identified by transient transfection of reporter gene constructs three preferred target sites for F7-1 in the 3′-UTR. Recognition of at least two of these sites is required for F7-1 off-target activity in our experimental model.
F7-1 and F7-2 were considered as candidates for the development of RNAi based therapeutics. The sequences of both constructs have no significant matches in the human transcriptome based on refined BLAST searches (Pamela Tan, personal communication). However the experimental assessment of siRNA specificity showed that F7-1 modulates a significant number of transcripts under various conditions while F7-2 has negligible off-target activity (Figs. 2 and 3). Based on transcriptional responses in HepG2 cells that do not express F7 we can eliminate the possibility that off-target modulation is a response to F7 depletion: the number of transcripts down-modulated by F7-1 was in the same order of magnitude as in Hep3B.
Several groups have shown independently that 7-nucleotide seed region sequence complementarily is sufficient for RISC binding and mRNA knock-down.12-14;20 This provides a plausible explanation why none the off-targets found experimentally was detected in silico. The word enrichment analysis further supports seed sequence-mediated target recognition because all genes affected by F7-1 or GCR-2 contain perfect or partial matches to the reverse complement of the siRNA seed region with no obvious preference for the CDS or 3′-UTR. Likewise, we detected seed sequence matches in all genes down-modulated by F7-2 siRNA (data not shown). In at least half of the F7-1 affected genes we found multiple copies of the seed heptamer motifs but we could not demonstrate a positive correlation between the prevalence of the seed motifs and knock-down efficiency as shown by Lin et al.14 Furthermore, we failed to detect any heptamer sequence enrichment in the genes upregulated by F7-1 indicating a seed sequence independent mechanism. We also considered the possibility that a miRNA controlling expression of the upregulated genes was down-modulated by F7-1 which cannot be detected mRNA microarrays. However, according to the TARbase5.0 database not a single gene of the upregulated set is a validated miRNA target. Overloading of the intracellular RISC machinery by transfected siRNA at high concentration was suggested as an alternative mechanism to activate expression of genes normally silenced by miRNAs.15 According to this model, we would also expect upregulation of genes in the F7-2 transfection experiments at high concentration or prolonged siRNA exposure which was not the case.
The CXCL5 transfection experiments demonstrate a direct role of the heptamer seed motifs in gene silencing by F7-1 together with the tolerance for single mismatches provided that cooperative binding to two sites occurs. However, not all available motifs are recognized and it is currently unclear which factors drive targeting of three out of five motifs in the CXCL5 sequence by F7-1. The regular spacing of about 300 nucleotides between the individual binding sites rules out preferred binding to the C-D-E sites due to steric hindrance. In addition, these sites are recognized and active in the synthetic dimer and trimer constructs where spacing is reduced to about 30 nucleotides. The close proximity of the sites in the recombinant constructs eliminates to a large extent a prominent role of RNA folding and other structural features of the native transcript. All F7-1 target sites contain single base mismatches at different positions. This could lead to site preference and cooperative binding since the mismatch position can impact heteroduplex stability between the siRNA seed region and the target mRNA.19 In fact, the mismatch position is at the 5′ end in C, centered in D and 3′ in E (Fig. 6). Maximal knock-down activity in the range of clone 4 requires binding of F7-1 to the D site in conjunction with the C site located upstream of D. Cooperative binding to D and E induces only partial knock-down and the presence of all sites results in the triple construct displays similar activity like the C-D pair and clone 4. We noted that the mismatch of the critical D site is G:U and it has been shown that this wobble mismatch leads to formation of a stable RNA duplex.19 When G:U is located at position 4 or 5 of the seed region, as in D, RISC loading is efficient and only marginally reduced compared with a perfect match control.19 In contrast, the mismatch in the C and E sites is located at terminal positions 7 or 2 of F7-1 seed region respectively. This configuration leads to substantial reduction of target affinity and RISC loading.19 In addition, the C and E sites flank the dominant D site which may further favor cooperative binding and recognition of these binding sites in CXCL5.21;22 Thus, we propose that binding of F7-1 to the dominant D site transforms the target mRNA into a stable state which enables cooperative binding to the C or E sites. We consider RNA folding and secondary structure as less important since the native CXCL5 mRNA and the mono-cistronic, artificial transcript generated by pMIR-REPORT luciferase vector exhibit comparable knock-down activity despite different sequence context. At this point it is unclear why the A and B sites are not targeted by F7-1. The mismatch nucleotide in site A is at the favorable position 5 of F7-1 seed region which leads to formation of a stable heteroduplex. However, localization of this site in the coding sequence of CXCL5 might disable F7-1 binding (Fig. 6). The mismatch in B occurs at position 7 of F7-1 seed region and this localization is likely to reduce the binding affinity of F7-1. The considerations above suggest that the position of the mismatch in the heptamer target site of F7-1 siRNA is a major determinant for preferred recognition in our F7-1/CXCL5 model system. Finally, the partial usage of seed sites in our model may explain the lack of correlation between knock-down efficiency and seed motif prevalence in our studies. The requirement for at least two seed sequence motifs in a favorable configuration may explain the on-target specificity of F7-2 and the lack of off-target effects for the GFP and luciferase controls. The occurrence of multiple seed matches in all down-modulated F7-1 off-target transcripts supports this view.
Experimental RNAi in cell lines and animal models has become an indispensable tool for drug target validation and other biomedical applications in the last decade. Currently, ongoing pharmaceutical research explores RNAi as a novel therapeutic modality due to the fascinating opportunity of targeting any transcript.23 Advancement in this field is currently limited by the lack of efficient and safe delivery vehicles and the inability to design mono-specific siRNAs to avoid off-target activity. The short length of the seed sequence in the siRNA guide strand together with the high prevalence of heptamer motifs in any genome make the design of mono-specific siRNA highly challenging. The seed region motifs of F7-1 (5′ CCTATTC 3′) and F7-2 (5′ CTTCGAT 3′) for instance have 3,260 or 1,320 hits in all RefSeq annotated transcripts and randomly generated heptamer sequences occur at frequencies ranging from 1,200 to 3,700 hits in the RefSeq database. Therefore, we consider experimental off-target detection in human cell lines currently as a favorable approach to assess off-target activity of siRNAs. The high on-target specificity of F7-2 proves that it should be possible to design siRNAs with a favorable off-target profile. A recent report24 describes a new 2-stage algorithm termed SVMicrO for the prediction of miRNA targets based on training of support vector machine with data available in miRecords or with data from published microarray experiments. Once the rules for accurate target recognition by miRNAs are understood, these principles may be translated to siRNA design since the RISC processing pathway is shared.
Currently, new generations of siRNAs are under development aimed at reducing the off-target activity seen in standard ribonucleotide constructs. Asymmetric siRNAs for instance contain a full-length antisense strand and a truncated passenger strand which results in a significant reduction of off-target activity.25 Another strategy employs the replacement of the RNA seed region by DNA which diminishes thermodynamic stability of duplexes with single nucleotide mismatches of the DNA seed region binding and the RNA target.26 An alternative, is perhaps the use of antisense oligonucleotides as drugs which is currently revisited because their stability and specificity have undergone significant improvements in the last decade.27 At the time of writing, 35 clinical studies with antisense nucleotides are ongoing compared with five phase 1 studies with siRNA. Target recognition of antisense oligonucleotides depends solely on proper Watson and Crick base-pairing to the target which is entirely RISC-independent. Consequently, off-target effects due to seed sequence matches are not an issue, implying that design of mono-specific oligonucleotides is less problematic.
In summary, our study further reduces the stringency for primary sequence based off-target recognition. Further studies are necessary to show general validity of our conclusions for other siRNAs with off-target activity. Based on the high statistical occurrence of heptamer motifs in nucleic acids it is obvious to postulate existence of additional rules and mechanisms contributing to naturally occurring target specificity of miRNAs. Based on our current knowledge it remains challenging to design highly mono-specific siRNAs such as F7-2 using in silico prediction algorithms.
Materials and Methods
siRNA constructs
Two siRNAs, named F7-1 and F7-2 siRNA, were designed to target human blood coagulation factor 7 (F7). F7-1 siRNA (5′ ATGTGGAAAA ATACCTATTC T 3′) targets the coding region of F7 mRNA whereas F7-2 siRNA (5′ GATATGCACA CACACGGATG C 3′) targets the 3′-UTR. As a control for gene expression microarray experiments two additional non-targeting siRNAs were used; the green-fluorescence protein, GFP (5′ CCACATGAAG CAGCACGACT T 3′) and the firefly luciferase, Luc (5′ CTTACGCTGA GTACTTCGAT T 3′). GCR-1 (5′ TGGTCGAACA GTTTTTTCTdT dT 3′) and GCR-2 (5′ TGCTTAACTA CATATAGATdT dT 3′) siRNAs were designed to target the coding region and the 3′-UTR of the human glucocorticoid receptor respectively.
Cell culture, siRNA transfection and RNA isolation
Cell lines were obtained from Roche Non-Clinical Biorepository group. They were cultured under standard conditions (37°C, 5% CO2) with the following media: DMEM with 4.5 g/L glucose (Invitrogen, 41965–039) and 10% FBS for Huh-7, EMEM with GlutaMAX-I (Invitrogen, 41090–028) and 10% FBS for HepG2 and Hep3B, McCoy’s 5A medium with L-Glutamine (Invitrogen, 26600–023) and 10% FBS for G-401, DMEM (Invitrogen, 41090–028) with 10% FBS for HEK293 and Ham’s F12 (Invitrogen, 21127–022) with 10% FBS for BRL3A and NRL. Hep3B and Huh-7 cell lines express F7 while the other cell lines do not. Two hundred fifty thousand cells per well were seeded in a 6-well plate 14 h before transfection. The cells were transfected with 10 nM, 50 nM, 100 nM or 250 nM siRNA using LipofectamineTM 2000 reagent (Invitrogen, 11668–019). Each transfection was performed in triplicate and the cells were harvested with 1 mL of TRIzol reagent (Invitrogen, 15596–026) 24 h, 40 h or 48 h after transfection. Total RNA isolation was performed using Invitrogen’s standard protocol. RNA quality was assessed by agarose gel electrophoresis and quantification was performed using the RiboGreen® RNA quantitation kit (Invitrogen, R-11490).
Quantigene branched-DNA assay
Quantification of F7 and CXCL5 mRNAs was performed using QuantiGene2.0 branched-DNA assay (b-DNA) from Panomics (QS0008). Total RNA was isolated from siRNA-treated cells and hybridized to specific b-DNA probes according to the manufacturer’s instructions and raw data were collected by measuring luminescence in a microplate reader (Victor3 multilabel counter, Perkin Elmer). Data were normalized by calculating the ratio between the signal intensity of the target mRNA and the signal intensity of the housekeeping gene GAPDH.
Microarray experiments and gene expression data analysis
Gene expression microarray experiments were conducted on HumanWG-6.V3 (BD101–0603, 48,804 probes derived from human genes in the NCBI RefSeq and UniGene databases) and RatRef-12.V1 (BD27–302, 22,519 probes derived from rat genes in the NCBI RefSeq and UniGene databases) Illumina Beadchips. 500 ng of total RNA was amplified and biotinylated using the TotalPrep RNA amplification kit (Ambion, AMIL1791). For each sample, 1,500 ng or 750 ng of labeled cRNAs were hybridized onto human or rat beadchips, respectively, using the Illumina whole genome gene expression direct hybridization protocol. The beadchips were scanned using Illumina iScan and the raw data were generated by GenomeScan software (Illumina). The raw data were then log2 transformed and quantile normalized without background subtraction. ANOVA statistical tests were performed in order to calculate samples’ p value and fold-change, the fold-change being a number describing how much the expression value of a gene changes upon treatment. Pairwise comparisons were performed between the treated samples and the control and changes were considered significant for fold-change > 2 or < -2 and a p value < 0.05.
Word enrichment analysis
mRNA sequences were retrieved from the RefSeq database28 and only 3′-UTR regions were included in the analysis. Redundant sequences were masked using the RSAT algorithm.29 Finally, the poly-A tails were removed from the 3′-end. Sequences were sorted by ascending experimental fold-change. Enrichment or depletion for 7-nucleotide word in this ranked gene list was analyzed using the ‘Sylamer’ program.17
Cloning and plasmid/siRNA co-transfection
Full-length and truncated cDNA fragments of CXCL5 were amplified from Hep3B total RNA using Titan One Tube RT-PCR System kit (Roche, 11939823001) using a common 5′-end primer (5′ GGCGTTTAAA CCGCAGCGCT CTCTTGACCA CT 3′) and truncation-specific 3′-end primers (clone 1: 5′ GCGGTTTAAA CTTCCATGCG TGCTCATTTC TC 3′; clone 2: 5′ CGGGTTTAAA CATTCTACAT AGATAAGGTG TG 3′; clone 3: 5′ CGGGTTTAAA CTCAGGAATT TGCCTGTTTT C 3′; clone 4: 5′ CGGGTTTAAA C GTTTAAACTT TTTTTTTTTT TTTTTTGA 3′) (Fig. 6). The corresponding PCR products were cloned into pCR2.1-TOPO vector (Invitrogen, K4500–40) and after sequence verification cDNA fragments were then sub-cloned into the unique PmeI site of the commercial vector pMIR_REPORT luciferase (Apply Biosystems, AM5795). The resulting clones were referred as clone 4 for the full-length sequence and clones 1–3 for the truncated sequences (Fig. 6). The sequences corresponding to the F7-1 target sites C, D, E, the dimer constructs C-D, C-E or D-E and the trimer construct C-D-E were chemically synthesized and sub-cloned into pMIR_REPORT Luciferase vector in a 5′-3′ orientation using SpeI (5′ end) and HindIII (3′ end) overhangs (Table S1). ApaI restriction site was used for clone screening.
Dual-light luciferase assay
We used the Dual-light® system from Applied Biosystems (T1005) as readout for transcript regulation after 10 nM F7–1, CXCL5 and luciferase siRNA treatment. Luciferase siRNA from Microsynth (5′ CaaCACCCCA ACAUCUUCGTT 3′) and CXCL5 siRNA from Dharmacon (5′ CCGCUGCUGU GUUGAGAGA 3′) target respectively the coding region of the firefly luciferase and CXCL5. 0.045 ng/µL of pMIR_REPORT luciferase vector was co-transfected with 0.01 ng/µL of β-galactosidase control vector into HEK293 cells in order to normalize the luciferase signal intensity across the samples. Firefly luciferase and β-galactosidase signal intensity was measured using a microplate reader combined with a dispenser (Victor3 multilabel counter, Perkin Elmer).
Supplementary Material
Supplementary PDF file supplied by authors.
Acknowledgments
We would like to thank Drs Guido Steiner, Laura Badi and Gonzalo Duran Pacheco for their support in the statistical analysis of microarray data and support in bioinformatics. We are grateful to Drs Rainer Constien and Pamela Tan for stimulating discussions and supply of siRNA constructs. We thank Prof P. Jacquart for stimulating discussions prior to submission of the paper. Finally, we thank the entire siRNA safety team, in particular Dr Thomas Weiser and Dr Thomas Singer for continued interest and project support.
Glossary
Abbreviations:
- siRNA
small interfering RNA
- miRNA
micro-RNA
- RISC
RNA-induced silencing complex
- FC
fold-change
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/18121
References
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of mirnas and sirnas. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded rna in caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide rnas mediate rna interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Blagbrough IS, Zara C. Animal models for target diseases in gene therapy using DNA and sirna delivery strategies. Pharm Res. 2009;26:1–18. doi: 10.1007/s11095-008-9646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanotto D, Rossi JJ. The promises and pitfalls of rna-interference-based therapeutics. Nature. 2009;457:426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: A progress report on sirna-based therapeutics. Nat Rev Drug Discov. 2007;6:443–53. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. Rnai-mediated gene silencing in non-human primates. Nature. 2006;441:111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 8.Hajeri PB, Singh SK. Sirnas: Their potential as therapeutic agents - part i. Designing of sirnas. Drug Discov Today. 2009;14:851–8. doi: 10.1016/j.drudis.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. Expression profiling reveals off-target gene regulation by rnai. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AL, Linsley PS. Noise amidst the silence: Off-target effects of sirnas? Trends Genet. 2004;20:521–4. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagenblatt A, et al. Off-target effects of sirna specific for gfp. BMC Mol Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, et al. Experimental validation of the importance of seed complement frequency to sirna specificity. RNA. 2008;14:853–61. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, et al. 3′ utr seed matches, but not overall identity, are associated with rnai off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 14.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, et al. Sirna-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–35. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small rnas globally perturbs gene regulation by endogenous micrornas. Nat Biotechnol. 2009;27:549–55. doi: 10.1038/nbt0709-671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, et al. Microrna-like off-target transcript regulation by sirnas is species specific. RNA. 2009;15:308–15. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microrna binding and sirna off-target effects from expression data. Nat Methods. 2008;5:1023–5. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena S, Jonsson ZO, Dutta A. Small rnas with imperfect match to endogenous mrna repress translation. Implications for off-target activity of small inhibitory rna in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 19.Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in rna silencing by a piwi/mid domain protein. Mol Cell. 2009;33:204–14. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread sirna “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–87. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microrna target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 22.Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human risc. Nat Struct Mol Biol. 2005;12:469–70. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 23.Sibley CR, Seow Y, Wood MJ. Novel rna-based strategies for therapeutic gene silencing. Mol Ther. 2010;18:466–76. doi: 10.1038/mt.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Yue D, Chen Y, Gao SJ, Huang Y. Improving performance of mammalian microrna target prediction. BMC Bioinformatics. 2010;11:476. doi: 10.1186/1471-2105-11-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Rogoff HA, Li CJ. Asymmetric rna duplexes mediate rna interference in mammalian cells. Nat Biotechnol. 2008;26:1379–82. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 26.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, et al. Functional dissection of sirna sequence by systematic DNA substitution: Modified sirna with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–51. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett CF, Swayze EE. Rna targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–93. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. Ncbi reference sequences (refseq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, et al. Rsat: Regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119-27. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.