Abstract
The cellular and molecular mechanisms underlying the pathogenesis of cataracts leading to visual impairment remain poorly understood. In recent studies, several mutations in the cytoplasmic sterile-α-motif (SAM) domain of human EPHA2 on chromosome 1p36 have been associated with hereditary cataracts in several families. Here, we have investigated how these SAM domain mutations affect EPHA2 activity. We showed that the SAM domain mutations dramatically destabilized the EPHA2 protein in a proteasome-dependent pathway, as evidenced by the increase of EPHA2 receptor levels in the presence of the proteasome inhibitor MG132. In addition, the expression of wild-type EPHA2 promoted the migration of the mouse lens epithelial αTN4-1 cells in the absence of ligand stimulation, whereas the mutants exhibited significantly reduced activity. In contrast, stimulation of EPHA2 with its ligand ephrin-A5 eradicates the enhancement of cell migration accompanied by Akt activation. Taken together, our studies suggest that the SAM domain of the EPHA2 protein plays critical roles in enhancing the stability of EPHA2 by modulating the proteasome-dependent process. Furthermore, activation of Akt switches EPHA2 from promoting to inhibiting cell migration upon ephrin-A5 binding. Our results provide the first report of multiple EPHA2 cataract mutations contributing to the destabilization of the receptor and causing the loss of cell migration activity.
Introduction
Cataract, the lens opacity disease, is the leading cause of blindness in the world, accounting for 48% of the cases [1]. Congenital cataract (CC) is one of the common causes of visual impairment in infants up to 25% [2]. Recent studies have examined the excess clustering of the disease in families with a high risk for cataract developments [3]. In addition, as much as 40% of early-onset cataracts may have a genetic basis [4]. Genetic studies have identified numerous underlying mutations including crystalline genes (CRYAA, CRYAB, CRYBB1, CRYBB2, CRYBB3, CRYBA3/A1, CRYBA4, CRYGC, GRYGD, and CRYGS) [5]–[13], connexin genes (GJA3, GJA8) [14], [15], and intermediate-filament-like factors (VIM) [16]. Recent genetic analyses revealed an additional novel pathway for cataract formation, mediated by mutations in the Eph receptor tyrosine kinase-type A2 (EPHA2) [17]–[20].
Human EPHA2 resides within the critical region on chromosome 1p36 that was previously defined in an Australian family with autosomal dominant total congenital cataracts [21], [22]. A recent study on the variations in the EPHA2 receptor tyrosine kinase gene within this region has identified a missense mutation c.2842G>T which substitutes an amino-acid from glycine to tryptophan at codon 948 (GGG>TGG: p.G948W) for autosomal dominant posterior polar cataracts in Caucasians [20]. In addition, other recent findings identified missense [c.2819C>T (p.T940I) in a Chinese family], frameshift [c.2915_2916delTG (p.V972GfsX39) in a British family] and splicing (c.2826-9G>A in an Australian family) mutations in EPHA2 in three independent families developing CC from different ancestral groups [19]. All of these mutations are located in the cytoplasmic sterile-α-motif (SAM) domain at the C-terminus of EPHA2 [20], [23], [24], suggesting that the SAM domain of EPHA2 may have an important role in the regulation of EPHA2 function and lens development.
The SAM domain is a conserved protein module in many key regulatory proteins, scaffolding proteins, and transcription factors. Mutations in the SAM domain have been observed to cause several human diseases [19], [20], [25]–[34]. For example, SAM domain mutations in the TP63 have been shown to affect SUMO-1-mediated regulation which would influence the protein stability causing ectodermal dysplasia syndromes [31], [32]. These defects are derived from increased TP63 ubiquitination as a result of the SAM domain mutation [29]. The 12p13 ETV6 (TEL: translocation ETS leukemia) SAM domain mutations block polymerization of ETV6-NTRK3 (EN) and transformation activity [26], [33]. The importance of this domain has led to numerous studies on the structure and stoichiometry of SAM domain complexes [19]. However, although SAM domains are capable of forming both homo- and hetero-oligomers in vitro, it remains unclear how SAM domains mediate protein interactions and what mechanisms regulate its association in vitro or in vivo. The presence of a conserved SAM domain within the cytoplasmic region of all Eph receptors indicates that it may play a role in regulating Eph receptor signaling. Since SAM domains facilitate protein-protein interactions [24] through homo- and hetero-oligomerization with other SAM domains, it is possible that EphA2 SAM domain mutations interfere with receptor oligomerization or clustering into higher-order complexes essential for physiologic signaling [23].
In the human genome, there are 14 Eph-coding genes (9 EPHAs and 5 EPHBs) and 8 ephrin ligand-coding genes (5 EFNAs and 3 EFNBs) [35]. Eph-related receptor tyrosine kinases (RTKs) have been implicated in the control of axon guidance, cell migration, angiogenesis, and patterning of the nervous system. Our previous studies showed that ephrin-A5 acts as a ligand for EphA2 in the lens, and the loss of ephrin-A5 function leads to cataracts in mice [36]. Additionally, ephrin-A5 interacts with the EphA2 receptor to regulate the adherens junction complex by enhancing recruitment of β-catenin to N-cadherin [36]. However, a molecular mechanism of EPHA2 signaling through the SAM domain that regulates lens development remains unknown. Here, we show that the EPHA2 SAM domain is required for protein stability and that the receptor utilizes both ligand-dependent and independent mechanisms to regulate lens epithelial cell biology.
Results
Mutations in the SAM domain of EPHA2 gene reduce protein levels
Our previous observations on the role of the ephrinA5/EphA2 molecules on lens development [36] suggest that EphA2 may act as a critical mediator in lens function. Consistent with our hypothesis, it has been shown that mutations in the EPHA2 gene within human chromosome 1p36 region lead to cataracts [17]–[20], [37]. Interestingly, four of the known mutations within EPHA2 are located in the SAM domain of the C-terminal region of EPHA2 (Figure 1A) that serves as a potential protein interaction site [19], [20], [23], [24]. To examine the consequences of these mutations, we generated four mutant EPHA2 genes: the missense mutants c.2819C>T (p.T940I) and c.2842G>T (p.G948W), the frameshift mutant c.2915_2916delTG (p.V972GfsX39), and the splicing mutant c.2826-9G>A (Figure 1A). In the c.2819C>T EPHA2 mutant, isoleucine replaces the wild-type threonine at residue 940 between H-3 and H-4 segments in the SAM domain [19]. The missense mutant c.2842G>T has a G T mutation of codon 948 (GGG>TGG) resulting in the missense substitution of glycine by tryptophan [20]. The c.2915_2916delTG mutant has a deletion of 2 bp in exon 17 resulting in a mutant EPHA2 protein with a novel C-terminal polypeptide of 39 amino acid residues. The c.2826-9G>A substitution creates a novel splice acceptor site which adds an intronic sequence into the mRNA generating a novel 71 amino acid residues at the C-terminus, of which the last 39 residues are identical to that of the novel polypeptide produced by the c.2915_2916delTG frameshift mutation [19].
T mutation of codon 948 (GGG>TGG) resulting in the missense substitution of glycine by tryptophan [20]. The c.2915_2916delTG mutant has a deletion of 2 bp in exon 17 resulting in a mutant EPHA2 protein with a novel C-terminal polypeptide of 39 amino acid residues. The c.2826-9G>A substitution creates a novel splice acceptor site which adds an intronic sequence into the mRNA generating a novel 71 amino acid residues at the C-terminus, of which the last 39 residues are identical to that of the novel polypeptide produced by the c.2915_2916delTG frameshift mutation [19].
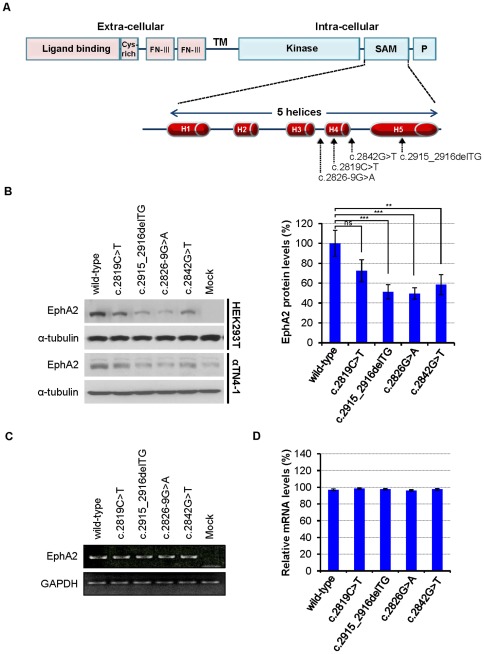
Figure 1. EPHA2 cataract mutations in the SAM domain.
(A) Schematic diagram showing the domains of EPHA2 receptor and the locations of four SAM domain mutations found in human cataracts (c.2819C>T; c.2915_2916delTG; c.2826-9G>A; and c.2842G>T) in the EPHA2 gene. FN-III: fibronectin type-III domain; TM: transmembrane domain; Kinase: protein tyrosine kinase domain; SAM: sterile-α-motif domain; P: PDZ-binding motif. The SAM domain comprises 5 α-hecices (H1–5). (B) Reduction of mutant EPHA2 protein levels in transfected cells expressing EPHA2 mutants. Protein levels of EPHA2 mutants are decreased in both HEK293T and αTN4-1 cells. The blot was reprobed with anti-α-tubulin as a loading control. The graphs represent the quantification of relative band intensity of EphA2 as connected by the levels of α-tubulin from three independent experiments. Total EphA2 protein band intensity was determined using ImageJ software. Mean values are presented with  S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant. (C, D) No difference between wild-type and mutant EPHA2 genes in transcription levels. (C) Semi-quantitative RT-PCR and (D) Real-time PCR for wild-type and mutant EPHA2 genes were performed using total RNA, isolated from transfected HEK293T cells. GAPDH transcript levels are used as controls. The graphs represent the quantification of western blots from three independent experiments.
S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant. (C, D) No difference between wild-type and mutant EPHA2 genes in transcription levels. (C) Semi-quantitative RT-PCR and (D) Real-time PCR for wild-type and mutant EPHA2 genes were performed using total RNA, isolated from transfected HEK293T cells. GAPDH transcript levels are used as controls. The graphs represent the quantification of western blots from three independent experiments.
To investigate whether the EPHA2 SAM domain mutations affect EPHA2 expression, we examined EPHA2 protein levels. Wild-type and mutant EPHA2 genes were transfected into HEK293T and mouse lens epithelial αTN4-1 cells. Wild-type EPHA2 is expressed at high levels in both HEK293T and αTN4-1 cells, while the mutant EPHA2 genes, c.2915_2916delTG, c.2826G>A and c. 2842G>T, showed low levels compared to the wild-type (Figure 1B). However, one of the mutant proteins, c.2819C>T, did not show a significantly lower level of expression from that of the wild-type protein, although it appeared to be somewhat reduced. We next examined whether these differences were due to the differences in transcription. Semi-quantitative (Figure 1C) and real-time RT-PCR (Figure 1D) reactions were used to investigate the mRNA levels. After transfection into HEK293T cells, PCR reactions for wild-type and mutant EPHA2 genes were performed using total RNA of the transfected cells, and EPHA2 PCR products were normalized to GAPDH transcript levels. No differences were found in mRNA level between the wild-type and mutants (Figure 1C,D), suggesting that these SAM domain mutations affect EPHA2 protein levels posttranscriptionally. We also expressed the wild-type and mutant EPHA2 SAM domain constructs as GST-fusion proteins in E. coli and found that the solubility of the mutant proteins was significantly reduced (Figure S1A,B) indicating an alteration of protein conformation. These results together suggest that mutations in the SAM domain of EPHA2 receptor reduce protein stability, and the reduced mutant protein levels may also in part, be a result of defective protein synthesis or maturation related to their altered conformation.
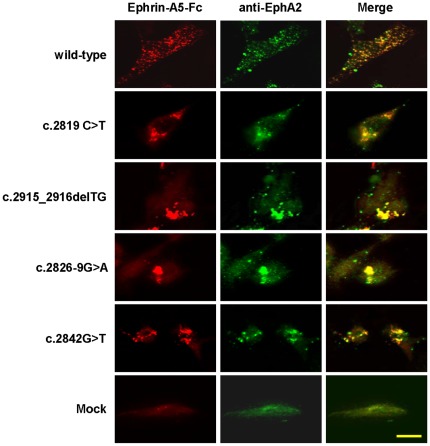
We also investigated EPHA2 protein expression using ligand-mediated immunofluore- scence staining. Detectable binding of ephrin-A5 was observed in EphA2−/− MEF cells expressing wild-type and mutant EPHA2 genes (Figure 2). Wild-type EPHA2 showed evenly distributed small protein aggregates throughout the cells (Figure 2). In contrast, EPHA2 mutants exhibited large protein aggregates, suggesting that the mutations in the SAM domain lead to enhanced protein aggregation, consistent with decreased GST-SAM domain fusion protein solubility in E. coli (Figure S1B).
Figure 2. Subcellular localization of the wild-type and mutant EPHA2 receptors on transfected EphA2−/− MEF (E13.5) cells.
EphA2 knockout MEF cells expressing wild-type or mutant EPHA2 were incubated with clustered ephrin-A5-Fc before fixation and treated with anti-Fc antibodies (red). After washing with PBS, cells were counterstained with anti-EphA2 antibody (green). Images were captured using a Nikon Eclipse C1 confocal microscope. Scale bar, 50 µm.
The ubiquitin-proteasome pathway mediates degradation of EPHA2 mutants
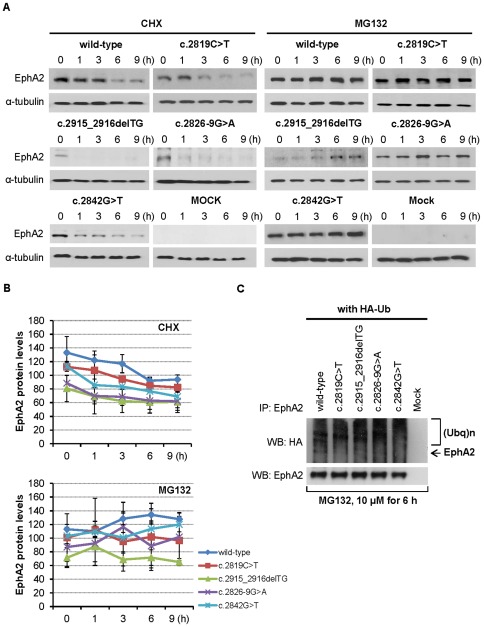
To determine whether the SAM domain mutations reduce the half-life of EPHA2 protein, we examined effects of cycloheximide (CHX), a protein synthesis inhibitor. HEK293T cells expressing wild-type and mutant EPHA2 genes were treated with 50 µg/mL CHX to block new protein synthesis. EPHA2 proteins with mutations in the SAM domain showed more rapid degradation kinetics compared to the wild-type (Figure 3A). The half-life of wild-type EPHA2 is approximately 3 hours (Figure 3B), whereas the half-life of the mutant proteins has been reduced to less than 1 hour (Figure 3A,B). These results demonstrate that mutations in the SAM domain result in rapid EPHA2 proteolysis.
Figure 3. EPHA2 degradation is mediated by proteasomal pathway.
(A) Mutant EPHA2 proteins have a reduced half-life. HEK293T cells were treated for indicated time with the protein biosynthesis inhibitor CHX (50 µg/mL) or the proteasome inhibitor MG132 (10 µM). Cell lysates were immunoblotted with anti-EphA2 antibody. Lysates were resolved by SDS-PAGE and western blot analysis was performed using indicated antibodies as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control. (B) Quantification of EphA2 protein levels over time. Mean values are presented with  S.D as indicated. (C) EPHA2 mutants have increased ubiquitination. Cells transfected with EPHA2 and HA-tagged ubiquitin were treated with 10 µM MG132 for 6 hours, and were then lysed. Immunoprecipitated EphA2 was further analyzed with western blotting using anti-HA antibodies to detect ubiquitinated EphA2 as described in the Materials and Methods. The smear band is characteristic ubiquitin immunoreactivity. The amount of total EphA2 is shown as a loading control.
S.D as indicated. (C) EPHA2 mutants have increased ubiquitination. Cells transfected with EPHA2 and HA-tagged ubiquitin were treated with 10 µM MG132 for 6 hours, and were then lysed. Immunoprecipitated EphA2 was further analyzed with western blotting using anti-HA antibodies to detect ubiquitinated EphA2 as described in the Materials and Methods. The smear band is characteristic ubiquitin immunoreactivity. The amount of total EphA2 is shown as a loading control.
To investigate whether EPHA2 degradation is mediated through proteasomal and lysosomal pathways, cells were treated with either the specific proteasome inhibitor MG132 or the lysosomal proton pump inhibitor bafilomycin A1. In the presence of 10 µM MG132, the expression levels of mutant EPHA2 proteins gradually increased over time (Figure 3A,B). Co-treatment with CHX and MG132 also largely prevented degradation of EPHA2 proteins (Figure S2A,B). In contrast, bafilomycin A1 did not affect the levels of EPHA2 proteins (Figure S3). These results indicate that EPHA2 proteins are degraded by proteasomes rather than lysosomes, and that the SAM domain is critical in modulating the degradation.
Proteasomal inhibition is normally associated with the accumulation of polyubiquitinylation on proteins. To confirm EPHA2 is degraded via the ubiquitin-mediated proteasomal pathway, wild-type and mutant EPHA2 genes were cotransfected with HA-tagged ubiquitin (HA-Ub) in the presense of MG132. The EPHA2 proteins were then immunoprecipitated with anti-EphA2 antibody and analyzed with western blotting for the presence of ubiquitin using an anti-HA antibody. The anti-HA antibody detected an increase in the intensity of high-molecular-mass EPHA2 proteins (Figure 3C), suggesting that the mutant proteins had increased polyubiquitination. Taken together, our data indicate that these particular mutations in the SAM domain of the EPHA2 receptor enhance proteasome-mediated EPHA2 protein degradation.
Ephrin-A5 stimulation induces tyrosine phosphorylation of EPHA2 mutants
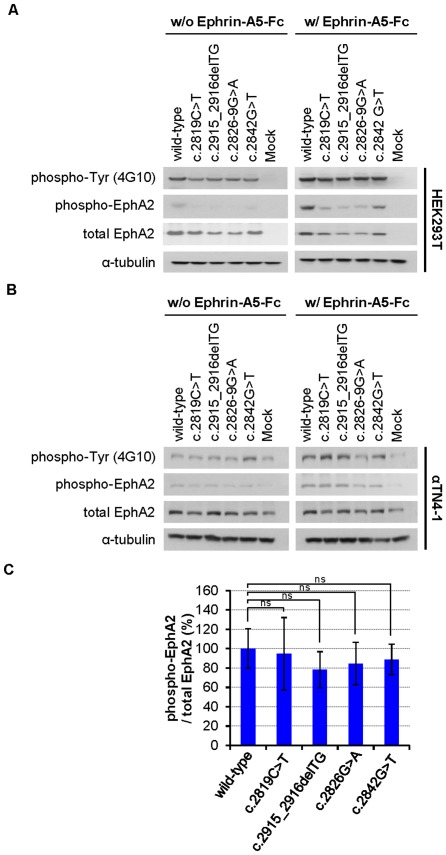
One important question is whether the EPHA2 mutants can still be activated by ephrin-A5. To address this question, EPHA2 wild-type and mutant genes were transfected into HEK293T and αTN4-1 cells, and the cells were stimulated with clustered recombinant ephrin-A5-Fc at 37°C for 30 minutes. The cell lysates were analysed with two phospho-specific antibodies: anti-phospho-Tyrosine (4G10) and anti-phospho-EphA2. EPHA2 wild-type and mutants showed similar tyrosine phosphorylation in response of ephrin-A5 ligand (Figure 4). This analysis showed that all mutants were autophosphorylated, although the levels were lower than the wild-type due to reduced protein concentration (Figure 4A,B). Ephrin-A5 stimulation further enhanced EPHA2 tyrosine phosphorylation. To confirm that EPHA2 is specifically phosphorylated, we probed the cell lysates with an anti-phospho-EphA2 (Tyr594) antibody, which detects transfected levels of EPHA2 proteins only when phosphorylated on Tyr594 and does not cross-reacted with other activated protein tyrosine kinases [38]. As shown in Figure 4A and Figure 4B, ephrin-A5 also enhanced EPHA2 Y594 phosphorylation in both HEK293T and αTN4-1. Quantification of intensity of the phosphorylated EPHA2 after protein level corrections showed similar activity between the wild-type and mutant EPHA2 proteins, although the mutants had reduced protein expression level (Figure 4C).
Figure 4. Tyrosine phosphorylation of EPHA2 receptor by ephrin-A5 is not affected by SAM domain mutations.
(A, B) Ephrin-A5 ligand stimulates EPHA2 phosphorylation. HEK293T (A) and αTN4-1 (B) cells were grown to confluence and growth factor-starved for 24 hours. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media and cell lysates were immunoblotted with indicated antibodies. Western blot analysis was performed as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control. (C) The ratios of levels of phospho-EphA2 to total EphA2 are similar between the wild-type and mutant EPHA2 proteins. The graphs show total band intensity of anti-phospho-EphA2 immunoblot to total EphA2 and represent the average of three independent experiments. Quantification of phospho-EphA2 protein/total EphA2 protein levels was performed using ImageJ software. Mean values are presented with  S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups.
S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups.
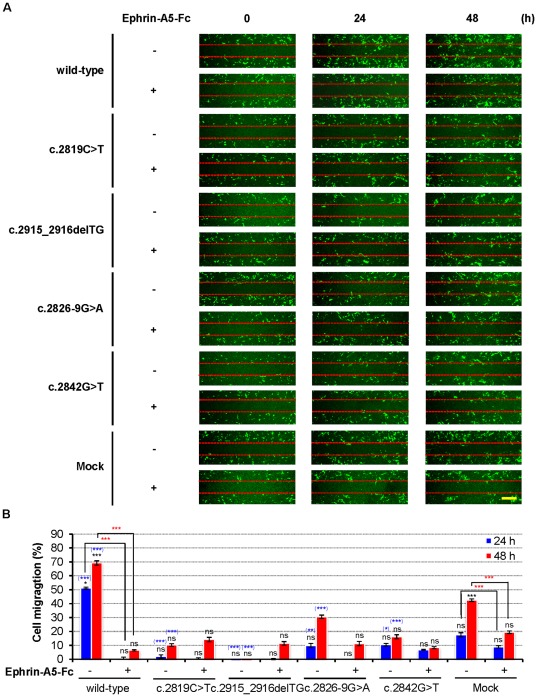
EPHA2 cataract mutations result in the loss of the ability to promote cell migration
To examine how the cataract mutations affect EPHA2 function, we investigated EPHA2 regulation on cell migration using the wound-healing assay, a common method in analyzing cell migration. A confluent cell monolayer was wounded using a pipette tip, introducing a cell-free area, and the migration of cells into the wound was monitored by capturing images at the indicated time points. In the absence of ephrin-A5 stimulation, wild-type EPHA2 promoted cell migration, while EPHA2 mutations in the SAM domain greatly reduced this ability (Figure 5A). Cells expressing wild-type EPHA2 began to migrate into the wound at 24 hours, while ephrin-A5 treatment significantly impaired the EPHA2-induced migration. Wound-healing assays were also quantified by measuring the distance by which transfected cells migrated into the cell-free region. Cells expressing wild-type EPHA2 migrated markedly faster than the mutants, occupying 51.25% (3.075 mm/6 mm) of the cell-free area after 24 hours and to 69.17% (4.15 mm/6 mm) after 48 hours (Figure 5B). The cataract mutations reduced the ability of EPHA2 to promote cell migration (Figure 5A,B). Although the wild-type EPHA2 promoted cell migration, treatment with ephrin-A5 impaired EPHA2-mediated cell migration (Figure 5A,B). Similar results were obtained using HEK293A cells (Figure S4A,B). In addition, statistical analysis showed there were significant differences between wild-type EPHA2 and the mock control groups (Figure 5B and Figure S4). The effects of mutant EPHA2 genes were also statistically different compared to that of mock-transfected αTN4-1 cells (Figure 5B).
Figure 5. SAM domain of EPHA2 is essential for ligand-independent promotion of cell migration.
(A) Mutant EPHA2 genes fail to promote αTN4-1 cell migration. αTN4-1 cells were grown to confluency and serum-starved for 24 hours. A scratch wound was made with a micropipette tip and the edge of cells was marked. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media, and cells were allowed to migrate toward the center of the wound and photographed at the indicated times (representative figure of three independent experiments). The position of the initial scratch is indicated by dotted lines. Scale bar, 500 µm. (B) Quantification of the effects of EPHA2 genes on αTN4-1 cell migration. The graphs represent the measurement of migration distance from three independent experiments. Mean values are presented with  S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
These observations demonstrate that EPHA2 promotes cell migration in the absence of ligand. However, ligand stimulation resulted in a switch of EPHA2 function, turning promotion to inhibition of cell migration.
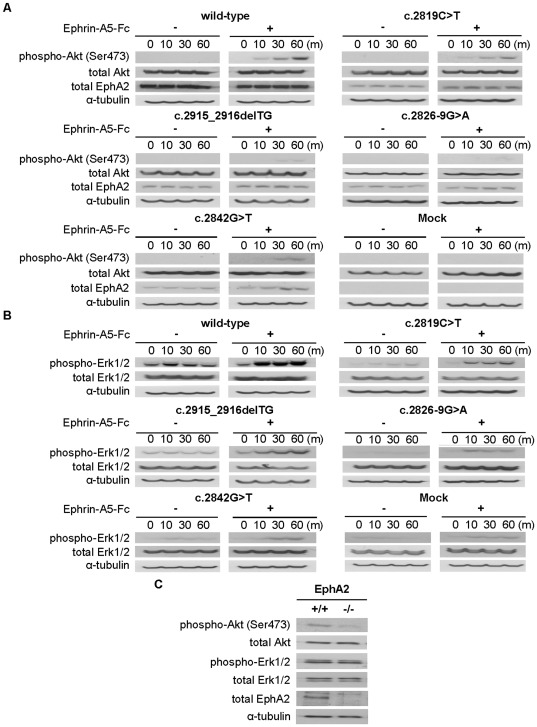
EPHA2 SAM domain mutations reduce Akt activation
To assess whether the SAM domain mutations affect EPHA2 biochemical functions, we determined the effects of EPHA2 on Akt and extracellular signal-regulated kinase (Erk) 1/2 signaling [18], [39], [40]. HEK293T cells transfected with wild-type and mutant EPHA2 genes were serum-starved for 24 hours, then treated with 2 µg/mL ephrin-A5-Fc at the indicated time points. Stimulated cells were lysed and examined for Akt and Erk activation. Ephrin-A5 stimulation of HEK293T cells expressing wild-type EPHA2 resulted in an increase of Akt phosphorylation at Ser473 in a time-dependent manner. In contrast, ephrin-A5-induced Akt phosphorylation was severely reduced in EPHA2 mutant-transfected cells after ephrin-A5 stimulation (Figure 6A). Akt phosphorylation was almost undetectable in cells expressing c.2915_2916delTG, c.2826-9G>A and c.2842G>T. Similar results were obtained using αTN4-1 cells (Figure S5A,B). The ability of the various EphA2 genes to activate Akt closely correlated with the receptor protein levels, as evidenced by the similar ratios of phospho-Akt (Ser473) to total EphA2 protein signals (data not shown).
Figure 6. Ligand-stimulated EPHA2 activation regulates Akt and Erk activation.
(A, B) Mutant EPHA2 proteins exhibit reduced activation of Akt and Erk by ephrin-A5. HEK293T cells were grown to confluence and serum-starved for 24 hours. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media and cell lysates were immunoblotted with anti-phospho-Akt (Ser473) or anti-phospho-Erk (1/2), and then reprobed with anti-α-tubulin as a loading control. (C) Inactivation of EphA2 gene leads to reduction of Akt activity in mouse lenses. Each lens was prepared from 22 days old mice and extracted with lysis buffer. Total lens proteins were resolved by SDS-PAGE and western blot analysis was performed using indicated antibodies as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control.
To determine whether the SAM domain mutations affect p44/p42 mitogen-activated protein kinase (MAPK) phosphorylation, lysates of HEK293T cell transfected with various EPHA2 genes were probed with anti-phospho-Thr202/Tyr204 Erk1/2 antibody. Ephrin-A5 stimulation of HEK293T cells expressing wild-type EPHA2 resulted in a robust increase in Erk phosphorylation, while cells expressing the EPHA2 mutant genes showed little or no activation (Figure 6B). In addition, the intensity of the phosphorylated Erk1/2 showed also a dependence on EPHA2 protein expression levels, similar to Akt activity (data not shown).
To examine whether EphA2 inactivation affects activation of Akt and Erk1/2 in vivo, we analyzed the lenses of postnatal day 22 (P22) EphA2+/+ and EphA2−/− mice. Akt activation was clearly detected in the wild-type EphA2 mouse lens (Figure 6C). However, Akt phosphorylation was almost undetectable in EphA2−/− mice. In contrast, levels of phospho-Erk1/2 did not show any decrease (Figure 6C).
Discussion
EPHA2 is a member of the Eph family receptor tyrosine kinases, and is strongly expressed in the cortical lens fiber cells. Recent studies have shown that mutations or deletion of EphA2 gene lead to cataracts in humans and mice [18]–[20]. Complementing these observations, our previous study identified that the loss of ephrin-A5 also leads to cataracts in mice [36], indicating that EphA2 serves as a receptor for the ligand in maintaining the clarity of the crystalline lens. The current study aims to define the nature of the EPHA2 SAM domain mutations. Previous studies showed that the SAM domains in Eph receptors may have multiple functions [38], [41]–[46]. The SAM domain of EphB2 receptor can self-associate and forms oligomers [42], [44], although the exact function is not known at present. In contrast, the SAM domain of EphA4 receptor has been shown to negatively regulate receptor kinase activity [43], but is not required for topographic mapping in the brain [41]. This domain has also been shown to mediate recruitment of downstream signaling molecules [45], [46]. A recent study by Fang et al. showed that the tyrosine 929 in the SAM domain of EphA2 is required for Ephrin-A1-induced vascular assembly [38]. Our analysis revealed an additional function in the maintenance of receptor stability.
EPHA2 SAM domain mutations cause increased receptor proteolysis
Eph receptors have an extracellular region consisting of an ephrin-binding domain and two fibronecin type III repeats, and an intracellular region comprised of a regulatory juxtamembrane domain, a tyrosine kinase domain, a SAM domain and a PDZ-binding motif [23]. SAM domains mediate important protein-protein interactions and are found in a variety of signaling molecules that exert diverse cellular functions [25]. A highly conserved SAM domain in the cytoplasmic region of all Eph receptors is located at the C-terminal region of the receptors and contain five α helical domains [25], [47]. Our studies suggest that the SAM domain of the EPHA2 protein modulates ubiquitinylation and regulates the stability of the receptor. As a first step to examine the specific effects of EPHA2 SAM domain mutations, we investigated the stability of the mutant EPHA2 proteins in two different cell lines, HEK293T and αTN4-1. Our results show that the EPHA2 proteins with SAM domain mutations exhibit increased degradation in a proteasome-dependent pathway, as treatment with the proteasome inhibitor MG132 enhanced EPHA2 protein levels. Ubiquitin-mediated proteasomal degradation pathway plays an important role in regulating a wide variety of cellular processes, since many proteins are degraded through this pathway [48], [49]. Thus we conclude that human EPHA2 SAM domain mutations cause cataracts through the reduction of EPHA2 protein levels.
Although the SAM domain modulates EPHA2 stability and protein solubility, it cannot be ruled out that the PDZ-binding motif may also contribute to EPHA2 stability, since the two mutants without the PDZ-binding domain, c.2915_2916delTG and c.2826-9G>A, showed lower steady state protein levels.
Mutations in the SAM domain affect EPHA2 protein solubility and subcellular localization
Many human diseases such as Alzheimer's and Parkinson's diseases as well as cataracts, have been demonstrated as “protein condensation diseases”, in which the pathogenic proteins form insoluble aggregates [50]. It has been shown also previously that the change of sequence parameters by mutations can affect protein expression and solubility in vivo [50]. To gain insights into the effects of the cataract mutations on protein solubility, we generated SAM domain fusion proteins from the wild-type and mutant EPHA2 receptors. Interestingly, all the GST-SAM domain mutant fusion proteins were insoluble in contrast to the wild-type GST-SAM domain fusion protein. These results indicate that SAM domain mutations influence protein stability and degradation rate through changes of solubility and folding efficiency. Several pathogenic proteins have been reported to have mutations in specific domains which induce aggregate formation and deplete the proteins from their normal cellular environment due to incorrect protein folding [51], [52]. Indeed we also observed that the mutations in the SAM domain of EPHA2 alter the patterns of subcellular distribution. The mutant proteins form much bigger aggregates than the wild-type. These observations support the notion that the EPHA2 cataract mutations induce protein misfolding leading to instability, which may lead to cellular disorganization and eventual lens opacity. Recent studies have shown that the solubility of several cataract-linked mutants of human γD-crystallin is severely compromised as a result of the mutations [50]. Well known examples are the R36S and the P23T mutants of human γD-crystallin, which can spontaneously crystallized at very low concentrations in vitro as a result of the lowered solubility of the mutant protein [50]. Similarly, mutant EPHA2 receptors form large protein aggregates, consistent with an insolubility issue.
EPHA2 regulates cell migration
It has been well established that wild-type EphA2 regulates cell migration, proliferation and invasion in a number of cell types [53]–[59]. Given the wealth of information linking EphA2 to cell migration, we evaluated the functional effects of the EPHA2 SAM domain mutations using the wound-healing assay. We showed that EPHA2 promotes cell migration in the absence of ligand stimulation, and that the SAM domain mutations diminish this activity, possibly due to the reduction of EPHA2 protein levels. Statistical analysis showed that there were significant differences between the wild-type and mutant EPHA2 genes. Similar enhancement effects in cell migration were reported following EphA2 expression in other cell types, including MDA-MB-231 breast cancer cells [59], PC3 carcinoma cells [60], U373 glioblastoma cells, and U87 glioblastoma cells [57]. Inactivation of EphA2 has also been shown to impair cell migration [58], [61]. For example, EphA2-deficient murine pulmonary microvascular (MPMEC) endothelial cells have impaired angiogenesis [58], [61]. Ligand-independent promotion of cell migration by EphA2 is likely mediated through interaction with Ephexin4 and the eventual activation of Rac1 [59].
Ephrin ligand binding induces Eph receptor phosphorylation and activation [53], [56], [57], [61]. The activation of EphA2 has been shown to negatively regulate cell migration [57], [60], [62], proliferation [53] and invasion [54]–[56] in a number of cell lines including U373, U87, A172, PC3, MDA-MB-231, MDA-MB-435, MCF10A, Capan2, G48a, U87 and U251. We also observed that stimulation of EPHA2 with ephrin-A5 resulted in the inhibition of migration of HEK293T and αTN4-1 cells. Although EphA2 activation inhibits cell migration in these studies, the opposite effects have also been reported [58], [61], [63]. Brantley-Sieders et al., showed that stimulation of lung microvascular endothelial cells (MPMEC) and bovine pulmonary microvascular endothelial cells (BPMEC) with ephrin-A1 induces cell migration [58], [61]. In human cardiac stem cells (CSC), ephrin-A1 promotes cell migration and enhances cardiac repair [63]. Taken together, our results are consistent with previous studies showing that EPHA2 is a major regulator of cell migration, and that the effects depend on whether the ligand is present and the cellular context.
Akt activation serves as a switch for EPHA2 function in cell migration
Mechanism underlying EPHA2 suppression of cell migration after ligand stimulation remains incompletely defined. The phosphoinositide 3-kinase (PI3K) signaling pathway has been shown to regulate cell growth, proliferation and migration [62], [64]. Akt and Akt-related serine-threonine kinases are activated by ligand stimulated growth factor receptor signaling in a PI3K-dependent manner [64]. We have shown here that activation of EPHA2 by ephrin-A5 in both αTN4-1 and HEK293 cells resulted in Akt activation, which correlates with inhibition of cell migration. Akt activation has been shown to inhibit migration and invasion of MDA-MB-231, MDA-MB-435 and SUM-159-PT breast cancer cells [65]. Another recent study has shown that down-regulation of Akt1 enhanced epidermal growth factor (EGF)-stimulated cell migration in MCF-10A breast cancer cells [66]. These results are consistent with our observation that activation of Akt mediates the inhibitory effects on migration. Thus, Akt activation serves as molecular switch for EphA2 function in cell migration.
The effects of EphA2 activation on Akt activity can vary depending on the cellular context. A number of studies have shown that stimulation of EphA2 by ephrin-A1 resulted in inhibition of Akt activity in certain cell lines (U87, U251, A172, MCF7, G48a, ES2, HEYA8 and corneal epithelial cells) [39], [54], [57], [62]. However, Akt is activated following ephrin-A1 stimulation in a number of other cell lines (B16, LNCaP, BxPC-3, PANC-1 and MIA PaCa-2) [39], [67]. Consistent with these latter studies, we observed Akt activation by ephrin-A5 stimulation, and found that Akt (Ser473) became highly phosphorylated in response to the addition of the ligand. The molecular mechanisms underlying this activation need further investigation. We also showed Akt activation depends on EphA2 expression levels, and reduced Akt activation may be responsible at least in part for cataract development in patients with EphA2 SAM domain mutations, since our in vivo studies reveal that Akt activation was significantly reduced in EphA2 knockout mouse lens. Thus, the ability of EPHA2 to cause Akt dephosphorylation or phosphorylation appears to depend on cell specific environment, and there may be kinase-dependent and -independent pathways that regulate Akt activity.
In sum, our studies show that mutations in the SAM domain of EPHA2 receptor induce EPHA2 protein instability. In addition, ephrin-A5 stimulation induces Akt activation, which in turn suppresses cell migration. These observations provide new insights in the mechanism by which defects in to EPHA2 signaling causes human cataracts.
Materials and Methods
Ethics statement
Animal studies were performed under standard conditions and treated in accordance with the Guidelines for the Animal Care and Use Committee at Rutgers University and ARVO Statement for the Use of Animals in Opthalmic and Vision Research (Rutgers approval ID number #93-052).
Mice and tissue extraction
The EphA2−/− [68] mice were kindly provided by Dr. Bingcheng Wang (Case Western Reserve University School of Medicine, Cleveland, Ohio, USA). All animals used were 22 days old in this study. For tissue extraction, lenses were dissected from mouse eyes and homogenized in ice-cold lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 100 µg/mL phenylmethylsulfonyl fluoride (PMSF), 1 µg/mL aprotinin, 10 µg/mL leupeptin, and 1 mM Na3VO4. The samples were cleared by centrifugation at 13,000 g for 2 minutes and used in western blot analysis.
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) cells obtained from American Type Culture Collection (ATCC), and mouse lens epithelial αTN4-1 cells [69] were generously provided by Dr. Bingcheng Wang (Case Western Reserve University School of Medicine, Cleveland, Ohio, USA). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich, USA) containing 4500 mg/L glucose/L, 584 mg/L L-glutamine/L with 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin solution (10000 units penicillin and 10 mg streptomycin/mL in 0.9% NaCl, Sigma-Aldrich, USA) at 37°C. Primary mouse embryonic fibroblasts (MEFs) were isolated from EphA2−/− E13.5 embryos and cultured in DMEM supplemented with 10% FBS [70]. Cells from passage 2 were used for transfection. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Protein levels were evaluated by immunoblotting 2 days after transfection.
Expression of EPHA2 genes
The human EPHA2 wild-type (GenBank NM_004431.3) and c.2842G>T were cloned using PCR previously [19]. Three other mutant cDNAs (c.2819C>T; c.2915_2916delTG; c.2826-9G>A) were generated by DNA synthesis (GeneScript USA Inc.). All cDNAs were cloned into the eukaryotic expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). All mutations were verified by DNA sequencing.
Antibodies and reagents
Antibodies used for immunoblot are from the following sources: anti-phospho-EphA2 (Tyr594) (1∶500, #3970), anti-Akt (1∶1000, #4691S), anti-phospho-Akt (Ser473) (1∶1000, #9271S), anti-Erk1/2 (1∶1000, #4695S), anti-phospho-Erk1/2 (1∶1000, #9101S) and anti-HA-Tag (1∶1000, #2367) from Cell Signaling Technology (Beverly, MA, USA); anti-EphA2 (1∶500, #E1026), anti-α-tubulin (1∶5000, #T6074), cycloheximide (CHX) (#C4859) and MG132 (Z-Leu-Leu-Leu-al) (#C2211) from Sigma-Aldrich (USA); bafilomycin A1 (#ab120497) and anti-EphA2 (1∶1000, #ab5386) from Abcam (Cambridge, MA) and anti-phospho-tyrosine (4G10) (1∶3000, #16-316) from Milipore Corporation (Billerica, MA).
Ephrin-A5 preparation and treatment
Recombinant ephrin-A5-Fc protein was purchased from R&D Systems (Minneapolis, MN, USA; #374-EA). To form clustered ephrin-A5, ephrin-A5-Fc (2 µg/mL) was cross-linked with anti-human Fc IgG (Jackson Immuno-Research, Immuno-Research, West Grove, PA, USA) at a 5∶1 ratio in µgs for 2 hours at 37°C as described in our previous studies [71]. Transfected cells were stimulated by ephrin-A5-Fc after serum starvation at 37°C overnight.
Western blot analysis
Tissue and cells were lysed in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 100 µg/mL phenylmethylsulfonyl fluoride (PMSF), 1 µg/mL aprotinin, 10 µg/mL leupeptin, and 1 mM Na3VO4 for 30 minutes at 4°C. Lysates were cleared by centrifugation at 13,000 g for 2 minutes. 30 µg of protein samples were boiled in 2 SDS-PAGE loading buffer and fractionated on 7.5% (w/v) SDS-PAGE gels and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). After blocking with 5% (w/v) dried skim milk in PBST (PBS with 0.1% Tween 20) for 1 hour, the membranes were probed with the indicated antibodies, coupled with a HRP-conjugated secondary antibody. Bands were visualized with chemiluminescence using ECL western blotting detection reagents (Amersham Pharmacia Biotech, UK) according to the manufacturer's instructions. Protein band intensities were quantified using NIH ImageJ software.
SDS-PAGE loading buffer and fractionated on 7.5% (w/v) SDS-PAGE gels and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). After blocking with 5% (w/v) dried skim milk in PBST (PBS with 0.1% Tween 20) for 1 hour, the membranes were probed with the indicated antibodies, coupled with a HRP-conjugated secondary antibody. Bands were visualized with chemiluminescence using ECL western blotting detection reagents (Amersham Pharmacia Biotech, UK) according to the manufacturer's instructions. Protein band intensities were quantified using NIH ImageJ software.
Immunoprecipitation
Briefly, various antibodies were added to the cell lysate, and incubated at 4°C overnight. The immunocomplex was recovered by using protein A-agarose beads (Roche Molecular Biochemicals, Indianapolis, IN, USA; #1719408) and centrifugation. After washing the protein A-Agarose beads five times with lysis buffer, the precipitated proteins were recovered by boiling in 40 µL 2 SDS-PAGE loading buffer for 5 minutes.
SDS-PAGE loading buffer for 5 minutes.
Detection of ubiquitination
HEK293T cells at 80% confluence were co-transfected with 2 µg of EPHA2 plasmid DNA and 1 µg of HA-tagged ubiquitin (HA-Ub) using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). At 48 hours post-transfection, cells were treated with 10 µM MG132 for 6 hours, and then were lysed with cell lysis Buffer. Cell lysates were clarified at 13,000 g for 2 minutes. Immunoprecipitation was carried out using an EphA2 antibody at 1 µg/mg of total protein at 4°C for 2 hours. Immune complexes were collected using protein A agarose (Millipore, Billerica, MA, USA) at 4°C for 1 hour. The beads were then washed with immunoprecipitation wash buffer containing 20 mM Tris-HCl (pH 7.4), 10% glycerol, 50 mM NaCl, 0.2% NP-40, 0.5 mM PMSF and 0.5 mM Na3VO4. The samples were re-suspended in SDS sample buffer and fractionated on 7.5% polyacrylamide gel. Immunoprecipitates or total cell lysates were analysed by western blotting as described above and probed with the anti-HA-Tag antibody.
Inhibition of protein synthesis and degradation
HEK293T cells were transfected with the EPHA2 wild-type and four mutants, c.2819C>T, c.2915_2916delTG, c.2826-9G>A, and c.2842G>T. At 48 hours post-transfection, the culture media were replaced with fresh DMEM+10% FBS and cells were either treated with the protein biosynthesis inhibitor CHX at 50 µg/mL, the proteasome inhibitor MG132 at 10 µM or the lysosomal inhibitor bafilomycin A1 at 100 nM, incubated for various times, then lysed. Total cell lysates were analysed by western blotting as described above.
Expression of glutathione S-transferase (GST) fusion protein and GST pull-down assay
The following primer pairs containing Sma I for forward primer and Not I for reverse primer were used to generate GST-tagged recombinant plasmids: wild-type EPHA2 SAM domain, Forward primer 5′-tgttgcccgggattccgcacggtgtccgagtg-3′ and reverse primer 5′-ccttctcgagtcaagtgttcacctggtcctt-3′; and mutant EPHA2 SAM domains, forward primer 5′-tgttgcccgggattccgcacggtgtccgagtg-3′ and reverse primer 5′-ccttctcgagtcagaaataaataaagtcccc-3′. PCRs were performed with the following cycle conditions: 95°C for 30 sec, 54°C for 30 sec, and 72°C for 30 sec for 25-cycles. PCR products were cloned into pGEX5X-1 expression vector. All GST-tagged SAM domain constructs were transformed into BL21 (DE3) E. coli, the proteins were induced with 1 mM isopropyl-β-D-thiogalactoside (IPTG) at 37°C for 4 hours. Whole-cell extracts were prepared and separated into soluble and insoluble fractions, and then the amount of recombinant GST fusion proteins were determined by coomassie or silver staining. For GST-wild-type SAM, GST-c.2819C>T SAM and GST-c.28442G>T SAM, 66 amino acids from the EphA2 SAM domain were fused to GST in frame at the 3′ end, resulting fusion proteins with the molecular weight of 33.6 kDa. For GST-c.2915_2916delTG SAM and GST-c.2826-G>A SAM, the mutated SAM domains contain 105 amino acids and 108 amino acids, respectively, resulting in molecular weights of 38 and 38.2 kDa.
RNA preparation, reverse transcription (RT)-PCR and real-time PCR
To study the transcription of the EPHA2 plasmids using semi-quantitative RT-PCR, total RNA was isolated from transfected cells using a QIAGEN total RNA isolation kit (Valencia, CA, U.S.A.). cDNA was generated by reverse transcription reaction using 5 µg of total RNA per sample with random primers and the SuperScript II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). The following primer pairs were used for the analysis of EPHA2 levels using PCR, generating a product of 248 bp: EPHA2, forward primer 5′-ttgtcatgtgggaggtgatg-3′ and reverse primer 5′-aaagtcagccagggtcttg-3′; and GAPDH, forward primer 5′-ttgccatcaatgaccccttca-3′ and reverse primer 5′-cgccccacttgattttgga-3′. PCRs were performed with the cycle conditions of 95°C for 30 sec, 60°C for 30 sec, and 68°C for 30 sec for 25 cycles.
Real-time PCR was performed using the ABI PRISM® 7000 sequence detection system and PCR reactions including SYBR Green dye. Results for target genes were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression for each sample, and relative expression was calculated using the comparative threshold cycle method [72].
Immunofluorescence staining
EphA2−/− MEF (E13.5) cells expressing wild-type and mutant EPHA2 genes were serum-starved overnight and then incubated with 2 µg/mL cross-linked ephrin-A5-Fc for 30 minutes. Cells were washed twice with phosphate buffered saline (PBS) at room temperature, and fixed in 4% paraformaldehyde (PFA) on ice for 30 minutes. Cells were permeablized with 0.3% PBS including Tween-20, and then blocked with 5% goat serum at room temperature for 2 hours. Secondary antibody Biotin-SP-AffiniPure goat anti-human IgG (Jackson Immuno-Research, Immuno-Research, West Grove, PA, USA) incubations were performed at room temperature for 1 hour. After extensive washing with PBS, CY3-conjugated streptavidin (Jackson Immuno-Research, Immuno-Research, West Grove, PA, USA) was added for 2 hours. Subsequently, samples were washed again with PBS and then incubated with anti-EphA2 antibody (1∶200, Abcam, Cambridge, MA), and images of stained cells were captured with a Nikon Eclipse C1 confocal microscope with 200 magnification.
magnification.
Cell migration assay
HEK293A cells were seeded on collagen-coated dishes and cultured for 24 hours. Cells expressing wild-type or mutant EPHA2 constructs were serum-starved overnight. GFP-expressing vector, pEGFP-N1 was cotransfected with the EPHA2 clones to identify transfected cells in the wound-healing assay. For ligand treatments, the culture media were replaced with fresh DMEM+10% FBS prior to stimulation. Cells were stimulated with 2 µg/mL cross-linked ephrin-A5-Fc. The migration distances of cells were monitored at the indicated time points (0, 24, and 48 hours) after wounding and quantified as described [73]. Images were captured using a Nikon Eclipse C1 confocal microscope with 40 magnification. The relative migration distance of GFP-positive cells into wound was determined using NIH ImageJ and Adobe Photoshop CS3 software.
magnification. The relative migration distance of GFP-positive cells into wound was determined using NIH ImageJ and Adobe Photoshop CS3 software.
Statistical analysis
Statistical analyses were performed using the Prism® software (GraphPad Software, La Jolla, CA). Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). A two-tailed student t-test was used to analyze statistical significance between two groups. All values are presented as the standard deviation of the mean ( S.D.) from at least three independent experiments. Value of P<0.05 was considered to be statistically significant.
S.D.) from at least three independent experiments. Value of P<0.05 was considered to be statistically significant.
Supporting Information
EPHA2 cataract mutations reduce SAM domain solubility in E. coli . GST alone and GST-fusion proteins containing either wild-type SAM domain, two missense mutations c.2819C>T SAM and c.2842G>T SAM, the frameshift mutation c.2916_2916delTG SAM, and the splicing mutation c.2826-9G>A SAM were overexpressed in BL21 (DE3) E. coli with 1 mM IPTG at 37°C for 4 hours. (A) GST-fusion proteins were highly induced in bacterial cells by IPTG treatment. After induction with 1 mM IPTG, whole-cell extracts were prepared fractionated with SDS-PAGE, and stained with Coomassie Blue. (B) The solubility of the EPHA2 mutant proteins was significantly reduced. Whole-cell extracts were separated into soluble (S) and insoluble (I) fractions, and then the amount of soluble and insoluble recombinant GST fusion proteins were determined by silver staining. Arrowheads indicate the position of the fusion proteins.
(TIF)
EPHA2 degradation is mediated by proteasomal pathway. (A) MG132 prevents degradation of EPHA2 protein. HEK293T cells were treated for indicated time with the protein biosynthesis inhibitor CHX (50 µg/mL) and the proteasome inhibitor MG132 (10 µM). Cell lysates were immunoblotted with anti-EphA2 antibody. Lysates were resolved by SDS-PAGE and western blot analysis was performed using indicated antibodies as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control. (B) Graphs show EphA2 protein levels over time. Mean values are presented with  S.D as indicated.
S.D as indicated.
(TIF)
Bafilomycin A1 does not stabilize EPHA2 protein. HEK293T cells were transfected and treated for indicated time with the lysosomal inhibitor bafilomycin A1 (100 nM). Cell lysates were immunoblotted with anti-EphA2 antibody. Lysates were resolved by SDS-PAGE and western blot analysis was performed as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control.
(TIF)
SAM domain of
EPHA2
is essential for ligand-independent promotion of cell migration of HEK293A cells. (A) EPHA2 SAM domain mutants lack migration promoting activity in HEK293A cells. HEK293A cells were grown to confluence and serum-starved for 24 hours. A scratch wound was made with a micropipette tip and the edge of cells as marked. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media, and cells were allowed to migrate toward the center of the wound and photographed at the indicated times (representative figure of three independent experiments). The position of the initial scratch is indicated by dotted lines. Scale bar, 500 µm. (B) Quantification of EPHA2 genes on HEK293A cell migration. The graphs represent the measurement of migration distance from three independent experiments. Mean values are presented with  S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
(TIF)
Ligand-stimulated EPHA2 activation regulates Akt activation in αTN4-1 cells. (A) Mutant EPHA2 genes have reduced ability to activate Akt. αTN4-1 cells were grown to near confluence, and growth factor-starved for 24 hours. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media and cell lysates were immunoblotted with indicated antibodies. The blot was probed with anti-phospho-Akt (Ser473), and then reprobed with anti-α-tubulin as a loading control. (B) Wild-type and mutant EPHA2 genes have similar activity in Akt activation when corrected for EPHA2 protein levels. Graphs show ratio of phosphor-Akt to total EPHA2. Quantification of phospho-Akt protein/total EPHA2 protein was determined using ImageJ software. Mean values are presented with  S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups. Data for the other two mutants were not quantified, due to the very low levels of the signals.
S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups. Data for the other two mutants were not quantified, due to the very low levels of the signals.
(TIF)
Acknowledgments
We thank Dr. Joo-young Im for generously providing the HA-tagged ubiquitin clone and Dr. Hyuck Kim for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported in part by the National Institutes of Health (NIH) grant (1RO1EY019012) to Dr. Zhou, and the National Natural Science Foundation of China (NSFC) grant (30973276) to Dr. Zhang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Foster A, Resnikoff S. The impact of Vision 2020 on global blindness. Eye. 2005;19:1133–1135. doi: 10.1038/sj.eye.6701973. [DOI] [PubMed] [Google Scholar]
- 2.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–149. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty CA, Taylor HR. The genetics of cataract. Invest Ophthalmol Vis Sci. 2001;42:1677–1678. [PubMed] [Google Scholar]
- 4.SanGiovanni JP, Chew EY, Reed GF, Remaley NA, Bateman JB, et al. Infantile cataract in the collaborative perinatal project: prevalence and risk factors. Arch Ophthalmol. 2002;120:1559–1565. doi: 10.1001/archopht.120.11.1559. [DOI] [PubMed] [Google Scholar]
- 5.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, et al. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 6.Berry V, Francis P, Reddy MA, Collyer D, Vithana E, et al. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–1145. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay DS, Boskovska OB, Knopf HLS, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–1221. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, et al. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human β-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–668. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 9.Riazuddin S, Yasmeen A, Yao W, Sergeev YV, Zhang Q, et al. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–2106. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- 10.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, et al. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 11.Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, et al. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–709. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H, Ma Z, Li Y, Liu B, Li Z, et al. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–710. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, et al. The [gamma]-crystallins and human cataracts: a puzzle made clearer. The American Journal of Human Genetics. 1999;65:1261–1267. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiels A, Mackay D, Ionides A, Berry V, Moore A, et al. A Missense Mutation in the Human Connexin50 Gene (GJA8) Underlies Autosomal Dominant. Am J Hum Genet. 1998;62:526–532. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackay D, Ionides A, Kibar Z, Rouleau G, Berry V, et al. Connexin46 mutations in autosomal dominant congenital cataract. Am J Hum Genet. 1999;64:1357–1364. doi: 10.1086/302383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller M, Bhattacharya SS, Moore T, Prescott Q, Wedig T, et al. Dominant cataract formation in association with a vimentin assembly disrupting mutation. Hum Mol Genet. 2009;18:1052–1057. doi: 10.1093/hmg/ddn440. [DOI] [PubMed] [Google Scholar]
- 17.Kaul H, Riazuddin SA, Shahid M, Kousar S, Butt NH, et al. Autosomal recessive congenital cataract linked to EPHA2 in a consanguineous Pakistani family. Mol Vis. 2010;16:511–517. [PMC free article] [PubMed] [Google Scholar]
- 18.Jun G, Guo H, Klein BEK, Klein R, Wang JJ, et al. EPHA2 is associated with age-related cortical cataract in mice and humans. PLoS Genet. 2009;5:e1000584. doi: 10.1371/journal.pgen.1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Hua R, Xiao W, Burdon KP, Bhattacharya SS, et al. Mutations of the EPHA2 receptor tyrosine kinase gene cause autosomal dominant congenital cataract. Hum Mutat. 2009;30:E603–E611. doi: 10.1002/humu.20995. [DOI] [PubMed] [Google Scholar]
- 20.Shiels A, Bennett TM, Knopf HLS, Maraini G, Li A, et al. The EPHA2 gene is associated with cataracts linked to chromosome 1p. Mol Vis. 2008;14:2042–2055. [PMC free article] [PubMed] [Google Scholar]
- 21.McKay JD, Patterson B, Craig JE, Russell-Eggitt IM, Wirth MG, et al. The telomere of human chromosome 1p contains at least two independent autosomal dominant congenital cataract genes. British journal of ophthalmology. 2005;89:831. doi: 10.1136/bjo.2004.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond CJ, Snieder H, Spector TD, Gilbert CE. Genetic and environmental factors in age-related nuclear cataracts in monozygotic and dizygotic twins. N Engl J Med. 2000;342:1786–1790. doi: 10.1056/NEJM200006153422404. [DOI] [PubMed] [Google Scholar]
- 23.Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 24.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 25.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:286. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 26.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cicero DO, Falconi M, Candi E, Mele S, Cadot B, et al. NMR structure of the p63 SAM domain and dynamical properties of G534V and T537P pathological mutants, identified in the AEC syndrome. Cell Biochem Biophys. 2006;44:475–489. doi: 10.1385/CBB:44:3:475. [DOI] [PubMed] [Google Scholar]
- 28.Kantaputra PN, Hamada T, Kumchai T, McGrath JA. Heterozygous mutation in the SAM domain of p63 underlies Rapp-Hodgkin ectodermal dysplasia. J Dent Res. 2003;82:433–437. doi: 10.1177/154405910308200606. [DOI] [PubMed] [Google Scholar]
- 29.Kantaputra PN, Malaivijitnond S, Vieira AR, Heering J, Dötsch V, et al. Mutation in SAM domain of TP63 is associated with nonsyndromic cleft lip and palate and cleft palate. Am J Med Genet A. 2011;155:1432–1436. doi: 10.1002/ajmg.a.34011. [DOI] [PubMed] [Google Scholar]
- 30.McGrath JA, Duijf PHG, Doetsch V, Irvine AD, Waal R, et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- 31.Ghioni P, Bolognese F, Duijf PHG, Van Bokhoven H, Mantovani R, et al. Complex Transcriptional Effects of p63 Isoforms: Identification of Novel Activation and Repression Domains {dagger}. Mol Cell Biol. 2002;22:8659–8668. doi: 10.1128/MCB.22.24.8659-8668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang WC, Ko TP, Li SSL, Wang AHJ. Crystal structures of the human SUMO 2 protein at 1.6 Å and 1.2 Å resolution. Eur J Biochem. 2004;271:4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 33.Tognon CE, Mackereth CD, Somasiri AM, McIntosh LP, Sorensen PHB. Mutations in the SAM domain of the ETV6-NTRK3 chimeric tyrosine kinase block polymerization and transformation activity. Mol Cell Biol. 2004;24:4636–4650. doi: 10.1128/MCB.24.11.4636-4650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin MT, Turel-Ermertcan A, Chan I, McGrath JA, Oztürkcan S. Ectodermal dysplasia showing clinical overlap between AEC, Rapp-Hodgkin and CHAND syndromes. Clin Exp Dermatol. 2004;29:486–488. doi: 10.1111/j.1365-2230.2004.01584.x. [DOI] [PubMed] [Google Scholar]
- 35.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Cooper MA, Son AI, Komlos D, Sun Y, Kleiman NJ, et al. Loss of ephrin-A5 function disrupts lens fiber cell packing and leads to cataract. Proc Natl Acad Sci U S A. 2008;105:16620–16625. doi: 10.1073/pnas.0808987105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan W, Hou S, Jiang Z, Hu Z, Yang P, et al. Association of EPHA2 polymorphisms and age-related cortical cataract in a Han Chinese population. Mol Vis. 2011;17:1553–1558. [PMC free article] [PubMed] [Google Scholar]
- 38.Fang WB, Brantley-Sieders DM, Hwang Y, Ham AJL, Chen J. Identification and functional analysis of phosphorylated tyrosine residues within EphA2 receptor tyrosine kinase. J Biol Chem. 2008;283:16017–16026. doi: 10.1074/jbc.M709934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang NY, Fernandez C, Richter M, Xiao Z, Valencia F, et al. Crosstalk of the EphA2 receptor with a serine/threonine phosphatase suppresses the Akt-mTORC1 pathway in cancer cells. Cellular Signal. 2011;23:201–212. doi: 10.1016/j.cellsig.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brannan JM, Sen B, Saigal B, Prudkin L, Behrens C, et al. EphA2 in the early pathogenesis and progression of non-small cell lung cancer. Cancer Prev Res (Phila) 2009;2:1039–1049. doi: 10.1158/1940-6207.CAPR-09-0212. [DOI] [PubMed] [Google Scholar]
- 41.Dufour A, Egea J, Kullander K, Klein R, Vanderhaeghen P. Genetic analysis of EphA-dependent signaling mechanisms controlling topographic mapping in vivo. Development. 2006;133:4415–4420. doi: 10.1242/dev.02623. [DOI] [PubMed] [Google Scholar]
- 42.Smalla M, Schmieder P, Kelly M, Ter Laak A, Krause G, et al. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 1999;8:1954–1961. doi: 10.1110/ps.8.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park EK, Warner N, Bong YS, Stapleton D, Maeda R, et al. Ectopic EphA4 receptor induces posterior protrusions via FGF signaling in Xenopus embryos. Mol Biol Cell. 2004;15:1647–1655. doi: 10.1091/mbc.E03-09-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang G, Hunter S, Hwang Y, Chen J. Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-Kinase-dependent Rac1 activation. J Biol Chem. 2007;282:2683–2694. doi: 10.1074/jbc.M608509200. [DOI] [PubMed] [Google Scholar]
- 46.Yamazaki T, Masuda J, Omori T, Usui R, Akiyama H, et al. EphA1 interacts with integrin-linked kinase and regulates cell morphology and motility. J Cell Sci. 2009;122:243–255. doi: 10.1242/jcs.036467. [DOI] [PubMed] [Google Scholar]
- 47.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nature structural biology. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 48.Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 49.Winer IS, Bommer GT, Gonik N, Fearon ER. Lysine residues Lys-19 and Lys-49 of β-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem. 2006;281:26181–26187. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- 50.Pande A, Ghosh KS, Banerjee PR, Pande J. Increase in surface hydrophobicity of the cataract-associated, P23T mutant of human gammaD-crystallin is responsible for its dramatically lower, retrograde solubility. Biochemistry. 2010;49:6122–6129. doi: 10.1021/bi100664s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai YC, Weissman AM. The Unfolded Protein Response, Degradation from the Endoplasmic Reticulum, and Cancer. Genes Cancer. 2010;1:764–778. doi: 10.1177/1947601910383011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chai A, Withers J, Koh YH, Parry K, Bao H, et al. hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum Mol Genet. 2008;17:266–280. doi: 10.1093/hmg/ddm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zantek ND, Azimi M, Fedor-Chaiken M, Wang B, Brackenbury R, et al. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 1999;10:629–638. [PubMed] [Google Scholar]
- 54.Gopal U, Bohonowych JE, Lema-Tome C, Liu A, Garrett-Mayer E, et al. A novel extracellular Hsp90 mediated co-receptor function for LRP1 regulates EphA2 dependent glioblastoma cell invasion. PLoS One. 2011;6:e17649. doi: 10.1371/journal.pone.0017649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem Biophys Res Commun. 2004;320:1096–1102. doi: 10.1016/j.bbrc.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 56.Wykosky J, Gibo DM, Stanton C, Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol Cancer Res. 2005;3:541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 57.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brantley-Sieders DM, Fang WB, Hicks DJ, Zhuang G, Shyr Y, et al. Impaired tumor microenvironment in EphA2-deficient mice inhibits tumor angiogenesis and metastatic progression. FASEB J. 2005;19:1884–1886. doi: 10.1096/fj.05-4038fje. [DOI] [PubMed] [Google Scholar]
- 59.Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, et al. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol. 2010;190:461–477. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parri M, Buricchi F, Taddei ML, Giannoni E, Raugei G, et al. Ephrin-A1 repulsive response is regulated by an EphA2 tyrosine phosphatase. J Biol Chem. 2005;280:34008–34018. doi: 10.1074/jbc.M502879200. [DOI] [PubMed] [Google Scholar]
- 61.Brantley-Sieders DM, Caughron J, Hicks D, Pozzi A, Ruiz JC, et al. EphA2 receptor tyrosine kinase regulates endothelial cell migration and vascular assembly through phosphoinositide 3-kinase-mediated Rac1 GTPase activation. J Cell Sci. 2004;117:2037–2049. doi: 10.1242/jcs.01061. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan N, Fatima A, Peng H, Bryar PJ, Lavker RM, et al. EphA2/Ephrin-A1 signaling complexes restrict corneal epithelial cell migration. Invest Ophthalmol Vis Sci. 2012;53:936–945. doi: 10.1167/iovs.11-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goichberg P, Bai Y, D'Amario D, Ferreira-Martins J, Fiorini C, et al. The Ephrin A1-EphA2 system promotes cardiac stem cell migration after infarction. Circ Res. 2011;108:1071–1083. doi: 10.1161/CIRCRESAHA.110.239459. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 65.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, et al. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 66.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang Q, Jorgensen C, Pawson T, Hedley DW. Effects of dasatinib on EphA2 receptor tyrosine kinase activity and downstream signalling in pancreatic cancer. Br J Cancer. 2008;99:1074–1082. doi: 10.1038/sj.bjc.6604676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, et al. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 69.Yamada T, Nakamura T, Westphal H, Russell P. Synthesis of alpha-crystallin by a cell line derived from the lens of a transgenic animal. Curr Eye Res. 1990;9:31–37. doi: 10.3109/02713689009000052. [DOI] [PubMed] [Google Scholar]
- 70.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005:28.21.21–28.21.28. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 71.Yue X, Dreyfus C, Kong TAN, Zhou R. A subset of signal transduction pathways is required for hippocampal growth cone collapse induced by ephrin A5. Dev Neurobiol. 2008;68:1269–1286. doi: 10.1002/dneu.20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liliental J, Moon SY, Lesche R, Mamillapalli R, Li D, et al. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EPHA2 cataract mutations reduce SAM domain solubility in E. coli . GST alone and GST-fusion proteins containing either wild-type SAM domain, two missense mutations c.2819C>T SAM and c.2842G>T SAM, the frameshift mutation c.2916_2916delTG SAM, and the splicing mutation c.2826-9G>A SAM were overexpressed in BL21 (DE3) E. coli with 1 mM IPTG at 37°C for 4 hours. (A) GST-fusion proteins were highly induced in bacterial cells by IPTG treatment. After induction with 1 mM IPTG, whole-cell extracts were prepared fractionated with SDS-PAGE, and stained with Coomassie Blue. (B) The solubility of the EPHA2 mutant proteins was significantly reduced. Whole-cell extracts were separated into soluble (S) and insoluble (I) fractions, and then the amount of soluble and insoluble recombinant GST fusion proteins were determined by silver staining. Arrowheads indicate the position of the fusion proteins.
(TIF)
EPHA2 degradation is mediated by proteasomal pathway. (A) MG132 prevents degradation of EPHA2 protein. HEK293T cells were treated for indicated time with the protein biosynthesis inhibitor CHX (50 µg/mL) and the proteasome inhibitor MG132 (10 µM). Cell lysates were immunoblotted with anti-EphA2 antibody. Lysates were resolved by SDS-PAGE and western blot analysis was performed using indicated antibodies as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control. (B) Graphs show EphA2 protein levels over time. Mean values are presented with  S.D as indicated.
S.D as indicated.
(TIF)
Bafilomycin A1 does not stabilize EPHA2 protein. HEK293T cells were transfected and treated for indicated time with the lysosomal inhibitor bafilomycin A1 (100 nM). Cell lysates were immunoblotted with anti-EphA2 antibody. Lysates were resolved by SDS-PAGE and western blot analysis was performed as described in the Materials and Methods. The blot was reprobed with anti-α-tubulin as a loading control.
(TIF)
SAM domain of
EPHA2
is essential for ligand-independent promotion of cell migration of HEK293A cells. (A) EPHA2 SAM domain mutants lack migration promoting activity in HEK293A cells. HEK293A cells were grown to confluence and serum-starved for 24 hours. A scratch wound was made with a micropipette tip and the edge of cells as marked. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media, and cells were allowed to migrate toward the center of the wound and photographed at the indicated times (representative figure of three independent experiments). The position of the initial scratch is indicated by dotted lines. Scale bar, 500 µm. (B) Quantification of EPHA2 genes on HEK293A cell migration. The graphs represent the measurement of migration distance from three independent experiments. Mean values are presented with  S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
S.D as indicated. Statistical differences were analyzed using one-way analysis of variance (ANOVA) or calculated by a two-tailed student t-test. Black asterisks
, comparison between time 0 and 24 hours and time 0 and 48 hours; Blue asterisks
, comparison between the mock groups and the listed wild-type or mutant EPHA2 genes at 24 or 48 hours; Red asterisks
, comparison between untreated and treated conditions at 24 or 48 hours. ***, P<0.001; **, P<0.01; *, P<0.05; and ns, not significant. Values of P<0.05 were considered to be statistically significant.
(TIF)
Ligand-stimulated EPHA2 activation regulates Akt activation in αTN4-1 cells. (A) Mutant EPHA2 genes have reduced ability to activate Akt. αTN4-1 cells were grown to near confluence, and growth factor-starved for 24 hours. 2 µg/mL cross-linked ephrin-A5-Fc was then added to the starvation media and cell lysates were immunoblotted with indicated antibodies. The blot was probed with anti-phospho-Akt (Ser473), and then reprobed with anti-α-tubulin as a loading control. (B) Wild-type and mutant EPHA2 genes have similar activity in Akt activation when corrected for EPHA2 protein levels. Graphs show ratio of phosphor-Akt to total EPHA2. Quantification of phospho-Akt protein/total EPHA2 protein was determined using ImageJ software. Mean values are presented with  S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups. Data for the other two mutants were not quantified, due to the very low levels of the signals.
S.D as indicated. Statistical differences between multiple groups were analyzed using one-way analysis of variance (ANOVA). Values of P<0.05 were considered to be statistically significant. ns: No statistically significant difference between the two groups. Data for the other two mutants were not quantified, due to the very low levels of the signals.
(TIF)