Abstract
The ribosomal P proteins are located on the stalk of the ribosomal large subunit and play a critical role during the elongation step of protein synthesis. The single chain recombinant antibody C5 (scFv C5) directed against the C-terminal region of the Trypanosoma cruzi P2β protein (TcP2β) recognizes the conserved C-terminal end of all T. cruzi ribosomal P proteins. Although this region is highly conserved among different species, surface plasmon resonance analysis showed that the scFv C5 possesses very low affinity for the corresponding mammalian epitope, despite having only one single amino-acid change. Crystallographic analysis, in silico modelization and NMR assays support the analysis, increasing our understanding on the structural basis of epitope specificity. In vitro protein synthesis experiments showed that scFv C5 was able to specifically block translation by T. cruzi and Crithidia fasciculata ribosomes, but virtually had no effect on Rattus norvegicus ribosomes. Therefore, we used the scFv C5 coding sequence to make inducible intrabodies in Trypanosoma brucei. Transgenic parasites showed a strong decrease in their growth rate after induction. These results strengthen the importance of the P protein C terminal regions for ribosomal translation activity and suggest that trypanosomatid ribosomal P proteins could be a possible target for selective therapeutic agents that could be derived from structural analysis of the scFv C5 antibody paratope.
Introduction
Trypanosoma cruzi is a protozoan parasite responsible for Chagas' disease. This is an endemic disease in Latin America that affects 18–20 million people. No vaccines are available at present and drugs used for treatment show undesirable side effects. The identification of new targets for chemotherapy is a major challenge in the control of the disease and the protein synthesis machinery has been proven to be such a target in other species. Insight into the mechanism capable of selectively blocking protein synthesis could thus lead to the discovery of new therapeutic agents.
The large subunit of the eukaryotic ribosome possesses a long and protruding stalk formed by the ribosomal P proteins. These proteins include P0, an approximately 34 kDa polypeptide, and two distinct, but closely related peptides of about 11 kDa, P1 and P2. All of them share a conserved, highly acidic motif at its C-terminal end. An additional P protein, named P3, has been described in plants [1]. The number of P1/P2 subtypes varies among species. In higher eukaryotes, the P1 and P2 families have only one member. In Saccharomyces cerevisiae, these families have two members, P1α/P1β and P2α/P2β [2]. Trypanosoma cruzi also possesses two different P1 and P2 proteins [3], [4]. Interestingly, the T. cruzi P0 protein has a C-terminal end that deviates from the eukaryotic P consensus and bears similarity to that of the L10 protein of Archaea [5]. The GTPase activity of the eukaryotic elongation factor 2 (eEF-2), which catalyses the translocation of peptidyl-tRNA from the A to the P site of the ribosome, is dependent on the presence of P proteins on the large ribosomal subunit [6]. Specifically, the C-terminal region of the ribosomal P proteins was shown to be essential during this step [7], [8]. Thus, the ribosomal stalk is directly involved in the translocation step of protein synthesis [9]. It has been previously shown that antibodies against the C-terminal region of ribosomal P proteins (markers of systemic lupus erythematosus in humans) and their scFv recombinant forms posses the ability to block in vitro translation in a rabbit reticulocyte lysate system [10], [11]. In chronic Chagas' heart disease, antibodies against the C-terminal region of T. cruzi ribosomal P proteins have been also detected [12], [13]. However, fine epitope mapping demonstrated that the specificity of the antibodies induced in these two pathological disorders is different [14], [15]. The single chain recombinant antibody (scFv) C5 directed against the C-terminal region of the ribosomal P2β protein of T. cruzi (R13 epitope), targets the five P proteins that constitute the stalk [16], [17]. Four of them (P1α, P1β, P2α, P2β) contain the same C-terminal epitope, R13 (Figure 1A); and the fifth, P0, has a closely related epitope called P015 (Figure 1A) [3], [16], [18]. This antibody however, as shown in this work, possesses very low affinity for the corresponding mammalian epitope (H13) that has one single non-conservative amino acid change in the third residue. We found that the scFv C5 was able to specifically block in vitro protein synthesis by trypanosomatid ribosomes, but had virtually no effect on translation by mammalian ribosomes. We expressed for the first time an intrabody (intracellular antibody), derived from scFv C5, in trypanosomatid cells resulting in growth arrest. Therefore, we propose the ribosomal stalk as a novel potential chemotherapeutic target, and the scFv C5 paratope as a model for peptide mimetics synthesis for selective blocking of the parasite protein synthesis apparatus.
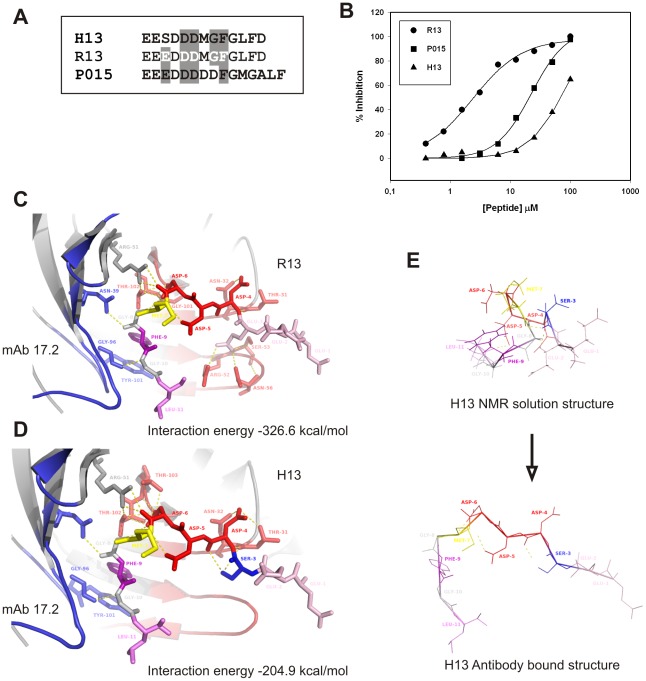
Figure 1. scFv C5 Epitope specificity.
A. Sequence aligment of T. cruzi R13 and P015 peptides with the mammalian counterpart H13 peptide. White letters correspond to residues necessary for antibody recognition, as identified by alanine scanning. Grey background corresponds to those residues conserved in the other two peptides. B. Inhibition of the interaction between scFv C5 and TcP2β protein by R13, P015 and H13 peptides, using surface plasmon analysis. The figure corresponds to one representative result out of 3 independent assays. C. Crystal structure of the complex mAb 17.2-R13 (PDB 3SGE). D. Model of the interaction of mAb 17.2 with the H13 peptide. The interaction energy is indicated on each case. E. NMR solution structure of H13 peptide (PDB 1S4J) in comparison with the antibody-bound peptide structure modeled.
Materials and Methods
Synthetic peptides
Peptides were prepared by the solid-phase method of Merrifield as was previously described [19], using a semiautomatic multisynthesizer NPS 4000 (NeoMPS SA, Strasbourg, France).
Surface Plasmon Resonance
The BIACORE 3000 system, sensor chip CM5, surfactant P20, amine coupling kit containing N-hydroxysuccinimide (NHS) and N-Ethyl-N′-dimethylaminopropyl carbodiimide (EDC), ethanolamine were from BIACORE (Uppsala, Sweden). Biosensor assays were performed with HBS-EP buffer as running buffer (10 mM HEPES, 150 mM sodium chloride, 3 mM EDTA, 0.005% (v/v) surfactant P20, pH 7.4). The low carboxylated dextran matrix (B1) was activated with 35 µl of a mixture 0.2 M N-ethyl-N-dimethylaminopropyl carbodiimide and 0.05 M N-hydroxysuccinimide at 5 µl/min. TcP2β-GST fusion protein and GST (as control) were immobilized with the standard BIACORE protocol at a density of 0.05 pmol/mm2 [4]. The scFv C5 was then pre-incubated with different concentrations of P015, R13 or H13 peptides for 30 min and then injected on the sensor chip for 2 min, followed by a dissociation phase of 3 min at a flow rate of 30 µl/min. The sensor chip surface was regenerated after each experiment by injecting 20 µL of 3 M MgCl2. From the decrease in the initial linear kinetics of the interaction in the presence of increasing amounts of inhibitor, the IC50 could be determined for each inhibitor as previously described [16].
ScFv C5 DNA constructs and intrabody expression
Expression of scFv C5 in E. coli and purification was performed as previously described [16]. For in vivo expression, the DNA encoding for scFv C5 was amplified by PCR using the following oligonucleotides carrying HindIII restriction site (C5ForHIND: 5′-GT AAGCTT GCC ATG GCC GAA GTG CAG C-3′) and BamHI restriction site (C5RevBAM: 5′-GT GGATCC CCG TTT TAT TTC CAG CTT GGT CC-3′) and inserted into pHD1700 vector as previously described [20]. The N-terminal myc-tagged scFv C5 construct was transfected into blood stream 1313-514 T. brucei inducible cell line [21], expressed and detected by Western blot using peroxidase anti-myc monoclonal antibody (1∶50,000) (Santa cruz biotechnology). Monoclonal antibody to aldolase [22] (1∶4000) was used to detect aldolase, used as the loading control. For growth rate analysis, parasites were seeded at 2×105 cells/ml and counted every 24 hours. The concentration was adjusted (by diluting appropriately) every 24 hours back to 2×105 cells/ml. The cumulative number of parasites corresponds to the number of parasites per milliliter multiplied by the dilution factor for each day (day after day).
Molecular modeling
All procedures were performed with Discovery Studio 2.5 software from Accelrys (San Diego, CA, USA). The scFv C5 model derives from the parental mAb 17.2 crystal structure Apo (3SGD) or in complex with R13 peptide (3SGE). The former structure was used as template to build a model antibody-H13 interaction. For this purpose, we mutated the aspartic acid 3 of R13 by serine and optimized the conformation of both the mutated residues and any surrounding residues that lay within a cut-off radius of 2 Å. Five models thus obtained were scored by the Discrete Optimized Protein Energy (DOPE). We continued analysis using the lowest energy model (DOPE = −96583.96 kcal/mol). Finally, the complex was subjected to a minimization step (max 500), RMS gradient 0.1 Kcal/mol by conjugated gradient.
Gel electrophoresis and Western blot analysis
SDS-PAGE analysis was performed as a standard procedure using 12% acrylamide gels followed by immunoblotting. Proteins were transferred from gels onto a Hybond ECL nitrocellulose transfer membrane (Amersham Pharmacia, UK) using a mini trans-blot system (Bio-Rad, Hercules, CA, USA) in transfer buffer (25 mM Tris-HCl, 190 mM glycine, 20% (v/v) methanol, pH 8.3). Membranes were soaked in PBS-T (20 mM Na2HPO4, 1.8 mM K2HPO4, 150 mM NaCl, 2.7 mM KCl, 0.1% Tween 20, pH 7.4) supplemented with 5% (p/v) non-fat milk powder. This was followed by incubation with scFv C5 200 nM together with peroxidase-conjugated anti-His Ab (∼350 nM) (Sigma, St. Louis, MO, USA), for detection of ribosomal P proteins and with peroxidase-conjugated anti-His Ab (∼350 nM) alone for intrabody detection. The Ab was diluted in the blocking solution PBS-T 1% (p/v) non-fat milk powder. Proteins on transferred membranes were revealed with tetramethyl-benzidine (TMB) (Sigma, St. Louis, MO, USA) after a brief wash with dextran sulphate 1% (Sigma, St. Louis, MO, USA).
Ribosome purification
Purification of ribosomes from different sources and in vitro protein synthesis assays were performed as described [23]. Briefly, mammalian ribosomes were obtained from rat liver (20 g), which was washed in sucrose 0.25 M and then homogenized in buffer containing 50 mM Tris–HCl, pH 7.5; 250 mM KCl; 5 mM magnesium acetate; sucrose 0.25 M and supplemented with 10% of S150 fraction [23]. The homogenate was treated with 10 U/ml of a-amylase and 0.1 mM CaCl2 for 15 min at 4°C, and then centrifuged for 4 min at low speed. The supernatant was again centrifuged for 20 min at 23,000 g. The pellet was discarded and the supernatant containing polysomes was supplemented with Triton X-100 1%, deoxycholate 0.5% and centrifuged for 5 min at 16,000 g. The new supernatant fluid (around 12 ml) was layered onto two layers of 2 M and 1.5 M sucrose made up in Buffer A (50 mM Tris–HCl, pH 7.5; 5 mM magnesium acetate; 250 mM KCl, dithiothreitol 1 mM and 10% of S150 fraction) and centrifuged 16 h at 140,000 g. The pellet corresponding to polysomes was rinsed and resuspended in a buffer containing 10 mM Tris–HCl, pH 7.5; 10 mM KCl and 1.5 mM magnesium acetate. Ribosome concentration was determined by optical density at 260 nm.
Polysomes from trypanosomatids were obtained from cultures at the exponential phase of growing. Cultures were treated with cycloheximide (50 ug/ml) for 10 min before harvesting by centrifugation at 4°C. Cells were washed twice with PBS containing 50 ug/ml cycloheximide. The cells were resuspended in Lysis Buffer (20 mM Tris–HCl, pH 7.5; 1 mM MgCl2; 5 mM KCl; 3 mM CaCl2; 5 mM 2-merchaptoethanol and 250 mM sucrose) and lysed with 0.2–0.4% of Nonidet P40 at 4°C. The homogenate was centrifuged 2–3 times for 20 min at 1,000 g and the final supernatant fraction containing ribosomes was layered onto a discontinuous gradient of 2 and 1.5 M of sucrose made up in the following buffer: 10 mM Tris–HCl, pH 7.5; 1 mM magnesium acetate; and 100 mM potasium acetate. The gradient was centrifuged for 16 h at 140,000 g. The supernatant was discarded and the pellet carefully rinsed and resuspended as described above.
In vitro protein synthesis assays
The reaction mixtures were prepared on ice and contained: 19 amino acids 50 uM each (excepting Met); 2 mM dithiothreitol; 100 mM potassium acetate; 3.5 mM magnesium acetate; 75 ug/ml wheat germ tRNA; 18 mM Hepes/KOH, pH 7.5; 1 mM ATP; 0.5 mM GTP; 7.5 mM creatine phosphate; 37.5 ug/ml creatine phosphokinase; rat liver S150 fraction (24 ug of protein); 0.3 A260U of ribosomes and 2 uCi of [35S] methionine in a final volume of 30 ul. Reactions were performed at 30°C during 60 min and stopped by adding 150 ul of 1.5 M NaOH; 1 mM Met, 170 ug/ml BSA. After incubation for 30 min at 37°C, proteins were precipitated with 1 ml of cold TCA 25%. After 60 min on ice, the samples were filtered and washed with TCA 10% and ethanol using glass fiber filters. Radioactivity retained in the filters was measured by liquid scintillation counting. For inhibition assays, reaction mixtures were incubated on ice with the scFv C5 antibody. For inhibition reversion, the scFv C5 was preincubated with the H13/R13 peptides for 20 min. After that, reactions were initiated by incubation at 30°C.
Trypanosome growth and transfection
Bloodstream form of Trypanosoma brucei was cultured in HMI-9 medium [24] supplemented with 10% (v/v) fetal calf serum (Sigma-Aldrich), and transfected as described previously [25]. Transfected cells were then cloned by serial dilution in a 24 well microtiter plate. For induction, cells were cultured in the medium above containing 0,1 mg/ml tetracycline and monitored for phenotypic changes (96 hours) or expression levels (72 hours) post induction.
Data mining and phylogenetic analysis
Ribosomal P protein sequences of Trypanosoma cruzi, Saccharomyces cerevisiae and Homo sapiens were used to perform BLAST searches [26] against NCBI-GenBank and GeneDB databases, using the tblastn and blastp algorithms. All protein sequences used for the phylogenetic analysis are listed in Table S1. Sequences were aligned with MEGA 4.0 software [27] using the Clustal W algorithm [28] with the PAM protein weight matrix, and all parameters with default settings. A phylogenetic tree was modeled using the minimum evolution method with the pairwise deletion option, and all parameters with default settings. Support for the branching order was determined by 1,000 bootstrap replicates.
Statistical analysis
Translation inhibition studies were analyzed by two-way ANOVA with Bonferroni's post test. Densitometry was analyzed by one-way ANOVA with Tukey's Multiple Comparison Test. T. brucei growth rate was analyzed by two-way ANOVA with Bonferroni's post test. All analysis were done using GraphPad Prism version 5.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com.
Results
Epitope Specificity
As it was previously reported the scFv C5 (that derives from the monoclonal antibody 17.2) recognizes the C-terminal region of T. cruzi ribosomal P2β protein (TcP2β, R13 epitope), and is able to recognize all five T. cruzi ribosomal P proteins [16], [29]. Four of them (TcP1α, TcP1β, TcP2α and TcP2β) contain the R13 epitope ( Figure 1A ); and the fifth, TcP0, contains a homologous C-terminal region called P015 ( Figure 1A ). Alanine replacement of R13 peptide showed as important residues for antibody interaction the motif 3-EdDDmGF-9 [29] ( Figure 1A , white letters). The mammalian counterpart epitope (H13) possesses one single, non-conservative amino acid change in the first key glutamic acid position ( Figure 1A ). Therefore, using surface plasmon resonance (SPR) analysis, we tested the ability of the three peptides to inhibit the interaction between the scFv C5 and TcP2β. Inhibition curves were performed using recombinant TcP2β protein fixed to a sensor chip and the scFv C5 in solution together with increasing concentrations of R13, P015 and H13 peptides ( Figure 1B ). Complete inhibition curves could be only obtained with R13 (IC50 = 2,37±0.23 µM) (Rsqr = 0.992) and P015 (IC50 = 22,64±2.03 µM) (Rsqr = 0.997) peptides but not with the H13 peptide. These results demonstrated the low affinity of scFv C5 for the mammal epitope. Interestingly, both P015 and H13 peptides possess one amino acid change in this motif but the replacement of glycine 8 by aspartic acid in P015 peptide seemed to be less essential than the replacement of glutamic 3 by serine in H13 peptide for scFv C5 interaction.
The crystallographic structure of mAb 17.2 in complex with R13 peptide has been recently solved (PDB 3SGE) [17]. In silico replacement of GLU3 by SER showed a strong reduction of the interaction energies that fall from −326.6 kcal/mol (R13 complex) to −204.9 kcal/mol (H13 complex) ( Figure 1C and D ). Particularly, there were three hydrogen bonds established between GLU3 and CDR-H2 residues (ARG-52, SER-53 and ASN-56) that were completely lost by the replacement by SER. In addition H13 peptide, but not R13, was able to acquire a stable conformation in solution using NMR [30], indicating that a strong conformational change is needed for the transition from the free solution form of H13 to the antibody bound conformation ( Figure 1E ) been energetically more expensive than binding to R13 peptide.
Translation inhibition
We analyzed the ability of scFv C5 to detect the ribosomal P proteins of T. cruzi, T. brucei, C. fasciculata and R. norvegicus by western blot. As it was previously shown [16] the scFv C5 strongly reacted with the T. cruzi P0, P1 and P2 proteins ( Figure 2A , lane 1). The sequence of the C-terminal peptide of T. brucei shows a conservative change of Glu3 by Asp in three of the low molecular weight P proteins (Table S2), and the sequences of C. fasciculata P proteins are not known. Figure 2A shows that scFv C5 clearly detected the P proteins on T. brucei and C. fasciculata extracts ( Figure 2A , lanes 2 and 3 respectively). Similar results were obtained analyzing equal amounts of purified ribosomes from these different organisms (not shown). In contrast, no protein bands were detected using rat extracts ( Figure 2A lane 4), as was expected from SPR analysis.
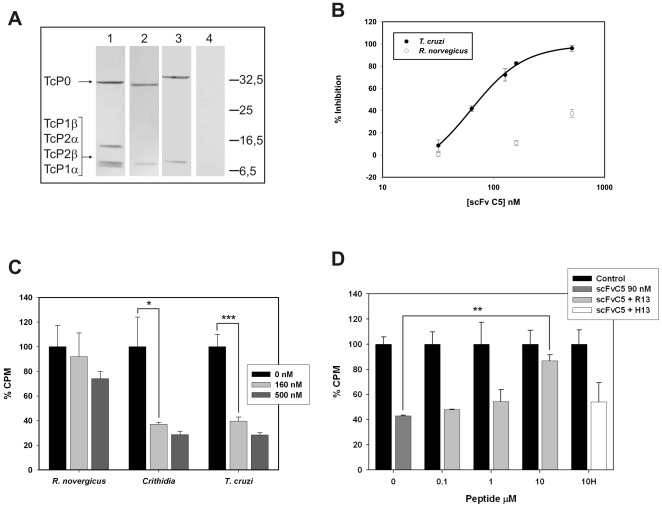
Figure 2. Translation inhibition.
A. Crude extracts from T. cruzi (lane 1), T. brucei (lane 2), C. fasciculata (lane 3) and R. norvegicus (lane 4) were analyzed by SDS-PAGE and Western blot with scFv C5. The position of previously characterized P proteins from T. cruzi (Tc) is shown on the left. The image is representative of three independent assays. B. Dose-response effect of scFv C5 on protein synthesis by ribosome extracts from R. norvegicus and T. cruzi. C. Effect of scFv C5 160 nM on in vitro protein synthesis in ribosome extracts from R. norvegicus, C. fasciculata and T. cruzi compared with the translation inhibitor emetine at 0.1 mg/mg. Average values for control assays were 80,000 cpm, 48,000 cpm and 43,000 cpm for R. norvegicus, C. fasciculata and T. cruzi, respectively. The scFv C5 significantly inhibited protein synthesis by C. fasciculata and T. cruzi ribosomes (*, p<0.05; ***, p<0.001). D. Effect of the preincubation with R13 or H13 peptide on the translation inhibition by scFv C5 (**; p<0.01).
Because scFv C5 reacts with the functional domain of the ribosomal P proteins, we used an in vitro cell-free translation system to determine the ability of this antibody to inhibit protein synthesis. Background values of radioactivity incorporation in the absence of protein synthesis were obtained in the presence of emetine (a well-characterized elongation inhibitor). It should be mentioned that the only difference among the translation mixtures was the ribosome source, ruling out a possible non-specific effect of scFv C5 on processes upstream from translation like ATP regeneration or amino acid activation. Our ribosome preparations lack initiation activity, as we have verified using the recently characterized IF4A inhibitor hippuristanol [23]. The ribosome concentration was estimated around 130 nM (∼10 units of absorbance at 260 nm per ml). Under these conditions, inhibition of protein synthesis on T. cruzi ribosomes by scFv C5 was dose-dependent with an IC50 of around 62,4 nM ( Figure 2B ). When rat ribosomes were used, it was not possible to obtain a complete translation inhibition curve even with 520 nM scFv C5 ( Figure 2B ). Moreover, scFv C5 up to 500 nM strongly inhibited the incorporation of [35S]-methionine on T. cruzi and C. fasciculata protein synthesis but had no effect on radioactivity incorporation when rat ribosomes were used ( Figure 2C ). Under the same conditions, it was possible to displace the scFv C5 translation inhibition activity by addition of R13 peptide with a maximal effect at 10 µM. However, peptide H13 did not affect scFv C5 inhibition activity at the same concentration ( Figure 2D ). These observations demonstrate that scFv C5 effectively inhibits the elongation step of protein synthesis as a consequence of its interaction with the C-terminal region of parasite ribosomal P proteins. Moreover, differences in epitope affinity correlate with the selective functional effect observed.
Intrabody expression in T. brucei
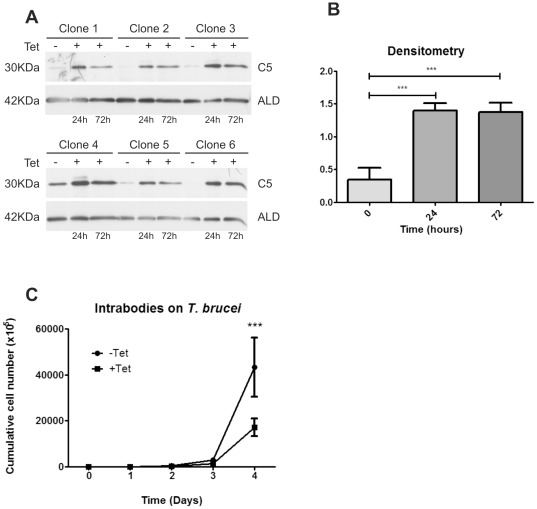
At present, there are no confident inducible expression systems available in T. cruzi and intrabody transfection using classical T. cruzi constitutive expression vectors (pTREX and pRIBOTEX) give no viable clones. Taking advantage from a tightly regulated, inducible expression system in T. brucei, we assessed intrabody expression in this organism. We subcloned the scFvC5 coding sequence into the pHD 1700 c-myc vector and transfected it into the T. brucei cell line 1313-514. Stable clones were obtained by serial dilution. Tetracycline induction of six different clones gives detectable protein at 24 hours post-induction and almost no detectable protein in the absence of inducer ( Figure 3A ). Intrabody expression levels remained stable until 72 h as can be seen in western blot analysis and the average densitometry analysis ( Figure 3B ). All clones were then subjected to a four day tetracycline induction period comparing the growth rate of induced and uninduced parasites. The averaged growth rate of the six different clones is shown in figure 3C . There was a detectable delay in the growth rate starting at day 2 that became statistically different (p<0.001) at day 4, indicating a detrimental effect of the intrabody expression on the logarithmic growing phase as can be seen in figure 3C . Out of the six clones assessed, clones 2 and 6 that showed no intrabody background expression without tetracycline, were analyzed three times showing reproducible growth rate inhibition after tetracycline induction (Figure S1). In addition, luciferase expression or a control intrabody did not affect the growing rate of induced parasites (Figure S2).
Figure 3. scFv C5 intrabody expression in T. brucei.
A. Western blot detection of C5 intrabody expression at 0, 24 and 72 hours in six different clones. C5 was detected by an anti-Myc antibody. The loading control ALD was detected by an anti-aldolase antibody. B. Averaged densitometry analysis of intrabody expression (***; p<0.001). C. Averaged growth rate curve of the six different clones induced and un-induced C5 intrabody parasites (***; p<0.001).
Stalk organization and epitope analysis
To better understand the organization of the stalk components in different protozoan we generated a sequence alignment (using MEGA 4.0) of the ribosomal P proteins of all genome sequenced protozoan. We searched by homology all ribosomal P proteins (putative and previously reported ones) from the following organisms: Trypanosoma cruzi (Tc), Trypanosoma brucei (Tb), Trypanosoma vivax (Tv), Trypanosoma congolense (To), Leishmania infantum (Li), Leishmania braziliensis (Lb), Leishmania major (Lm), Plasmodium knowlesi (Pk), Plasmodium falciparum (Pf), Plasmodium berghei (Pb), Plasmodium chabaudi (Pc), Dictyostelium discoideum (Dd), Eimeria tenella (Et), Theileria annulata (Ta), Theileria parva (Tp), Entamoeba histolytica (Eh) and Cryptosporidium parvum (Cp). We also included the ribosomal P proteins of Homo sapiens (Hs), Arabidopsis thaliana (At) and Saccharomyces cerevisiae (Sc) as key markers (Table S1). It is important to note that two independent gene duplication events yielded P1 and P2 subtypes (α and β) in trypanosomatids and S. cerevisiae. The clusters belonging to trypanosomatids P proteins P0, P1α, P1β, P2α and P2β are all supported by high bootstrap values (Figure 4A). Therefore it is possible that trypanosomatids possess common stalk architecture, probably different from the other analyzed organisms.
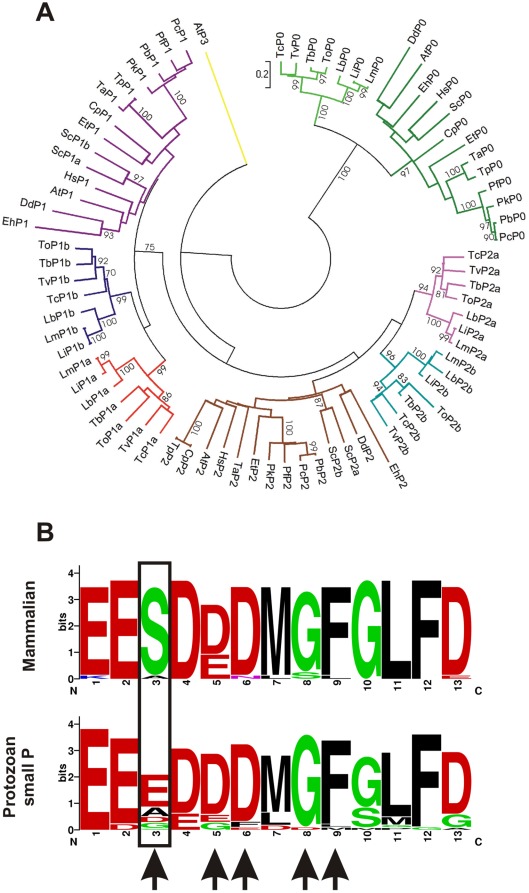
Figure 4. P proteins phylogeny and epitope analysis.
A. Phylogeny inference of ribosomal P proteins. Red, branch of trypanosomatid P1α proteins. Blue, branch of trypanosomatid P1β proteins. Magenta, branch of trypanosomatid P2α proteins. Light blue, branch of trypanosomatid P2β proteins. Brown, branch of non-trypanosomatid P2 proteins. Violet, branch of non-trypanosomatid P1 proteins. Yellow, Arabidopsis thaliana P3. Green, branch of P0 where has been grouped separated the trypanosomatids P0 (light green). Bootstrap values above 70 are shown. B. The sequences of the C-terminal residues of mammalian P-proteins and the C-terminal region of the small protozoan P-proteins were aligned using WebLogo [31]. Arrows indicate critical residues for scFv C5 recognition. The region boxed corresponds to the key difference between mammal and protozoan epitopes.
Thereafter we generated a sequence alignment of the C-terminal epitope corresponding to different protozoan and mammal organisms (Table S2) using weblogo [31]. This allowed a graphical representation of an amino acid multiple sequence alignment showing that in mammals the main amino acid at position 3 is a serine in contrast to protozoan where this position is occupied mainly by glutamic acid, followed by alanine, aspartic acid and finaly glycine ( Figure 4B ). Noteworthy, alanine scanning replacement of R13 peptide showed the importance of glutamic acid 3 for recognition of the antibody [29]. Since the C-terminal region of Leishmania P proteins posses an alanine in position 3, it rules out the possibility of an effect of the C5 antibody on this organism.
Discussion
In the present work, we demonstrated a strong correlation between the affinity of scFv C5 for the C-terminal end of ribosomal P proteins from different species and its ability to inhibit translation in a cell-free system. The difference on IC50 observed by SPR (R13 vs H13) together with in silico structural analysis based on the recently solved crystallographic structure of the complex Fab 17.2-R13 peptide; clarify how the scFv C5 selectively blocks protein synthesis by trypanosomatid ribosomes. Western blot and sequence data analysis showed that different trypanosomatids posses the same C-terminal epitope, all being potentially susceptible to scFv C5 activity. When T.cruzi cells were transfected for constitutive expression of scFv C5 intrabody it was not possible to complete the selection period because all cells died. Profiting from the availability of inducible vectors for T. brucei and that scFvC5 recognizes T. brucei P proteins in Western Blot, we tested the effect of scFv C5 intrabody induction on these cells. The scFv C5 intrabody could be detected at protein level using its myc tag at 24 hs post-induction. Induced parasites showed a delay in growth rate, consistent with a blockade of ribosomal translation activity. As can be seen on figure S1, it is difficult to establish a relationship between intrabody expression level and functional effect. The inhibition mechanism can be resumed as a protein (scFvC5) that needs to be translated before binding the ribosomal P proteins and thus inhibiting protein synthesis. It means that it is really difficult to obtain total inhibition because the synthesis of the intrabody will also be blocked. We think that the most probable scenario could be interpreted as an equilibrium between scFv translation, protein activity and growth inhibition. In addition, the scFv C5 was able to inhibit in vitro protein synthesis in ribosome extracts from T. brucei at a similar extent than T. cruzi ribosome extracts (Figure S3). Intrabodies have been successfully expressed in different cell types [32], [33], [34], [35]. To our knowledge, in this work we have described for the first time the expression of an intrabody in trypanosomatid cells. This knock-down strategy has some potential advantages over RNAi for studying genes that have very high mRNA-turnover rates or whose protein products have very low turnover rates, because they directly interact with the target protein, being less affected by target dynamics. Because intrabodies act at the protein level, they will be of value to specifically block the function of protein complexes and to model drug–target interactions [36].
Sequence analysis showed that mammalian ribosomal P proteins possess a conserved C-terminal region slightly different from the protozoan one, being the main difference the presence of a serine in position 3 ( Figure 4 ). This residue showed to be a key residue in scFv C5 recognition, as has been shown all along this work. If this is so, this antibody can potentially inhibit translation activity in different parasitic protozoan cells without affecting the host cell. Moreover, these results reveal the functional C-terminal end of trypanosomatid ribosomal P proteins as a possible specific target for chemotherapy. In addition, the paratope-forming residues from mAb 17.2 recently crystallized (PDB 3SGE) [17] could be an interesting starting point for designing peptide mimetics [37], [38], [39], [40] as specific inhibitors of trypanosomatid translation.
Supporting Information
Growth curves of all clones assessed for intrabody expression. The growth curves and expression profile of each individual clone are shown in every row. On the left side we present the western blots of scFvC5 expression performed at 0 and 24 hours post-induction. The averaged growth curve and densitometry is shown at the bottom and corresponds to the results showed in figure 3. The growth curves of three replicates of clones 2 and 6 and the average growth curve are also shown. Red squares correspond to averaged growth curves. (***; p<0.001).
(TIF)
Control intrabody expression in T. brucei . Growth rate of T. brucei transfected with the control scFv S3, scFv C5 or Luciferase in presence of tetracycline using the pLew inducible system.
(TIF)
Translation inhibition. Effect of scFv C5 50 nM on in vitro protein synthesis in ribosome extracts from T. cruzi and T. brucei compared with the translation inhibitor emetine at 0.1 mg/mg. Average values for control assays were 6,000 cpm and 19,000 cpm for T. cruzi and T. brucei, respectively.
(TIF)
C-terminal region of the ribosomal P-proteins analyzed in Figure 4B . A. Different mammalian P-proteins C-terminal sequences potentially hosts of parasitic infections. B. Protozoan P1/P2 C-terminal sequences.
(DOC)
Acknowledgments
We thank Dr. Sylviane Muller and Prof. Sylvie Fournel (UPR 9021 Strasbourg, France) for helpful collaboration in the publication of this article as well as Dr. I.D. Algranati for his advice about in vitro translation experiments and helpful discussions about the manuscript. We sincerely thank Prof. Dr. Christine Clayton (ZMBH, Heidelberg, Germany) for facilitating the expressions in T. brucei. M.J.A., S.A.L. and K.A.G. are members of scientific career of CONICET. L.S. and T.D. have fellowships from CONICET.
In memory of Dr. Mariano J. Levin (1951–2010), Director of the Laboratory of Molecular Biology of Chagas' disease, INGEBI-CONICET, Buenos Aires, Argentina, from 1985 to 2010.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research has been initially supported by grants from the World Health Organization/Special Program for Research and Training in Tropical Diseases, Universidad de Buenos Aires, Ministerio de Salud y Acción Social-Beca Ramón Carrillo-Arturo Oñativia, and the National Agency of Scientific and Technological Promotion (FONCYT BID 1201/OC-AR 01–14389). Support of ECOS-SECyT project “Anticorps anti-proteines ribosomales P de T. cruzi comme inhibiteur specifique de la traduction” (France-Argentine, 2005–2008) is also acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bailey-Serres J, Vangala S, Szick K, Lee CH. Acidic phosphoprotein complex of the 60S ribosomal subunit of maize seedling roots. Components and changes in response to flooding. Plant Physiol. 1997;114:1293–1305. doi: 10.1104/pp.114.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 3.Juri Ayub M, Smulski CR, Nyambega B, Bercovich N, Masiga D, et al. Protein-protein interaction map of the Trypanosoma cruzi ribosomal P protein complex. Gene. 2005;357:129–136. doi: 10.1016/j.gene.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Smulski CR, Longhi SA, Ayub MJ, Edreira MM, Simonetti L, et al. Interaction map of the Trypanosoma cruzi ribosomal P protein complex (stalk) and the elongation factor 2. J Mol Recognit. 2010;24:359–370. doi: 10.1002/jmr.1089. [DOI] [PubMed] [Google Scholar]
- 5.Levin MJ, Vazquez M, Kaplan D, Schijman AG. The Trypanosoma cruzi ribosomal P protein family: classification and antigenicity. Parasitol Today. 1993;9:381–384. doi: 10.1016/0169-4758(93)90088-w. [DOI] [PubMed] [Google Scholar]
- 6.Lavergne JP, Conquet F, Reboud JP, Reboud AM. Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 1987;216:83–88. doi: 10.1016/0014-5793(87)80761-5. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 8.Bargis-Surgey P, Lavergne JP, Gonzalo P, Vard C, Filhol-Cochet O, et al. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur J Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 9.Ballesta JP, Rodriguez-Gabriel MA, Bou G, Briones E, Zambrano R, et al. Phosphorylation of the yeast ribosomal stalk. Functional effects and enzymes involved in the process. FEMS Microbiol Rev. 1999;23:537–550. doi: 10.1111/j.1574-6976.1999.tb00412.x. [DOI] [PubMed] [Google Scholar]
- 10.Zampieri S, Mahler M, Bluthner M, Qiu Z, Malmegrim K, et al. Recombinant anti-P protein autoantibodies isolated from a human autoimmune library: reactivity, specificity and epitope recognition. Cell Mol Life Sci. 2003;60:588–598. doi: 10.1007/s000180300050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stacey DW, Skelly S, Watson T, Elkon K, Weissbach H, et al. The inhibition of protein synthesis by IgG containing anti-ribosome P autoantibodies from systemic lupus erythematosus patients. Arch Biochem Biophys. 1988;267:398–403. doi: 10.1016/0003-9861(88)90045-8. [DOI] [PubMed] [Google Scholar]
- 12.Levin MJ, Mesri E, Benarous R, Levitus G, Schijman A, et al. Identification of major Trypanosoma cruzi antigenic determinants in chronic Chagas' heart disease. Am J Trop Med Hyg. 1989;41:530–538. doi: 10.4269/ajtmh.1989.41.530. [DOI] [PubMed] [Google Scholar]
- 13.Mesri EA, Levitus G, Hontebeyrie-Joskowicz M, Dighiero G, Van Regenmortel MH, et al. Major Trypanosoma cruzi antigenic determinant in Chagas' heart disease shares homology with the systemic lupus erythematosus ribosomal P protein epitope. J Clin Microbiol. 1990;28:1219–1224. doi: 10.1128/jcm.28.6.1219-1224.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan D, Ferrari I, Bergami PL, Mahler E, Levitus G, et al. Antibodies to ribosomal P proteins of Trypanosoma cruzi in Chagas disease possess functional autoreactivity with heart tissue and differ from anti-P autoantibodies in lupus. Proc Natl Acad Sci U S A. 1997;94:10301–10306. doi: 10.1073/pnas.94.19.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez Bergami P, Mateos P, Hoebeke J, Levin MJ, Baldi A. Sequence analysis, expression, and paratope characterization of a single-chain Fv fragment for the eukaryote ribosomal P proteins. Biochem Biophys Res Commun. 2003;301:819–824. doi: 10.1016/s0006-291x(02)03074-7. [DOI] [PubMed] [Google Scholar]
- 16.Smulski C, Labovsky V, Levy G, Hontebeyrie M, Hoebeke J, et al. Structural basis of the cross-reaction between an antibody to the Trypanosoma cruzi ribosomal P2beta protein and the human beta1 adrenergic receptor. FASEB J. 2006;20:1396–1406. doi: 10.1096/fj.05-5699com. [DOI] [PubMed] [Google Scholar]
- 17.Pizarro JC, Boulot G, Bentley GA, Gómez KA, Hoebeke J, et al. Crystal Structure of the Complex mAb 17.2 and the C-Terminal Region of Trypanosoma cruzi P2β Protein: Implications in Cross-Reactivity. PLoS Negl Trop Dis. 2011;5:e1375. doi: 10.1371/journal.pntd.0001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez Bergami P, Scaglione J, Levin MJ. Antibodies against the carboxyl-terminal end of the Trypanosoma cruzi ribosomal P proteins are pathogenic. FASEB J. 2001;15:2602–2612. doi: 10.1096/fj.01-0132com. [DOI] [PubMed] [Google Scholar]
- 19.Muller S, Couppez M, Briand JP, Gordon J, Sautiere P, et al. Antigenic structure of histone H2B. Biochim Biophys Acta. 1985;827:235–246. doi: 10.1016/0167-4838(85)90208-0. [DOI] [PubMed] [Google Scholar]
- 20.Manful T, Cristodero M, Clayton C. DRBD1 is the Trypanosoma brucei homologue of the spliceosome-associated protein 49. Mol Biochem Parasitol. 2009;166:186–189. doi: 10.1016/j.molbiopara.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Alibu VP, Storm L, Haile S, Clayton C, Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Molecular and Biochemical Parasitology. 2005;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Clayton CE. Import of fructose bisphosphate aldolase into the glycosomes of Trypanosoma brucei. J Cell Biol. 1987;105:2649–2654. doi: 10.1083/jcb.105.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juri Ayub M, Ma KW, Shaw PC, Wong KB. Trypanosoma cruzi: high ribosomal resistance to trichosanthin inactivation. Exp Parasitol. 2008;118:442–447. doi: 10.1016/j.exppara.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 25.Biebinger S, Wirtz LE, Lorenz P, Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol Biochem Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahler E, Sepulveda P, Jeannequin O, Liegeard P, Gounon P, et al. A monoclonal antibody against the immunodominant epitope of the ribosomal P2beta protein of Trypanosoma cruzi interacts with the human beta 1-adrenergic receptor. Eur J Immunol. 2001;31:2210–2216. doi: 10.1002/1521-4141(200107)31:7<2210::aid-immu2210>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 30.Soares MR, Bisch PM, Campos de Carvalho AC, Valente AP, Almeida FC. Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis. FEBS Lett. 2004;560:134–140. doi: 10.1016/S0014-5793(04)00088-2. [DOI] [PubMed] [Google Scholar]
- 31.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boldicke T, Weber H, Mueller PP, Barleon B, Bernal M. Novel highly efficient intrabody mediates complete inhibition of cell surface expression of the human vascular endothelial growth factor receptor-2 (VEGFR-2/KDR). J Immunol Methods. 2005;300:146–159. doi: 10.1016/j.jim.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Wolfgang WJ, Miller TW, Webster JM, Huston JS, Thompson LM, et al. Suppression of Huntington's disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci U S A. 2005;102:11563–11568. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer's Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kontermann RE. Intrabodies as therapeutic agents. Methods. 2004;34:163–170. doi: 10.1016/j.ymeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Stocks MR. Intrabodies: production and promise. Drug Discov Today. 2004;9:960–966. doi: 10.1016/S1359-6446(04)03269-6. [DOI] [PubMed] [Google Scholar]
- 37.Saragovi HU, Fitzpatrick D, Raktabutr A, Nakanishi H, Kahn M, et al. Design and synthesis of a mimetic from an antibody complementarity-determining region. Science. 1991;253:792–795. doi: 10.1126/science.1876837. [DOI] [PubMed] [Google Scholar]
- 38.Takasaki W, Kajino Y, Kajino K, Murali R, Greene MI. Structure-based design and characterization of exocyclic peptidomimetics that inhibit TNF alpha binding to its receptor. Nat Biotechnol. 1997;15:1266–1270. doi: 10.1038/nbt1197-1266. [DOI] [PubMed] [Google Scholar]
- 39.Berezov A, Greene MI, Murali R. Structure-based approaches to inhibition of erbB receptors with peptide mimetics. Immunol Res. 2003;27:303–308. doi: 10.1385/IR:27:2-3:303. [DOI] [PubMed] [Google Scholar]
- 40.Casset F, Roux F, Mouchet P, Bes C, Chardes T, et al. A peptide mimetic of an anti-CD4 monoclonal antibody by rational design. Biochem Biophys Res Commun. 2003;307:198–205. doi: 10.1016/s0006-291x(03)01131-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves of all clones assessed for intrabody expression. The growth curves and expression profile of each individual clone are shown in every row. On the left side we present the western blots of scFvC5 expression performed at 0 and 24 hours post-induction. The averaged growth curve and densitometry is shown at the bottom and corresponds to the results showed in figure 3. The growth curves of three replicates of clones 2 and 6 and the average growth curve are also shown. Red squares correspond to averaged growth curves. (***; p<0.001).
(TIF)
Control intrabody expression in T. brucei . Growth rate of T. brucei transfected with the control scFv S3, scFv C5 or Luciferase in presence of tetracycline using the pLew inducible system.
(TIF)
Translation inhibition. Effect of scFv C5 50 nM on in vitro protein synthesis in ribosome extracts from T. cruzi and T. brucei compared with the translation inhibitor emetine at 0.1 mg/mg. Average values for control assays were 6,000 cpm and 19,000 cpm for T. cruzi and T. brucei, respectively.
(TIF)
C-terminal region of the ribosomal P-proteins analyzed in Figure 4B . A. Different mammalian P-proteins C-terminal sequences potentially hosts of parasitic infections. B. Protozoan P1/P2 C-terminal sequences.
(DOC)