Abstract
A proper immune response ensures survival in a hostile environment and promotes longevity. Recent evidence indicates that innate immunity, beyond antimicrobial effectors, also relies on host-defensive mechanisms. The Caenorhabditis elegans transcription factor SKN-1 regulates xenobiotic and oxidative stress responses and contributes to longevity, however, its role in immune defense is unknown. Here we show that SKN-1 is required for C. elegans pathogen resistance against both Gram-negative Pseudomonas aeruginosa and Gram-positive Enterococcus faecalis bacteria. Exposure to P. aeruginosa leads to SKN-1 accumulation in intestinal nuclei and transcriptional activation of two SKN-1 target genes, gcs-1 and gst-4. Both the Toll/IL-1 Receptor domain protein TIR-1 and the p38 MAPK PMK-1 are required for SKN-1 activation by PA14 exposure. We demonstrate an early onset of immunosenescence with a concomitant age-dependent decline in SKN-1-dependent target gene activation, and a requirement of SKN-1 to enhance pathogen resistance in response to longevity-promoting interventions, such as reduced insulin/IGF-like signaling and preconditioning H2O2 treatment. Finally, we find that wdr-23(RNAi)-mediated constitutive SKN-1 activation results in excessive transcription of target genes, confers oxidative stress tolerance, but impairs pathogen resistance. Our findings identify SKN-1 as a novel regulator of innate immunity, suggests its involvement in immunosenescence and provide an important crosstalk between pathogenic stress signaling and the xenobiotic/oxidative stress response.
Author Summary
Innate immunity promotes survival by combating pathogenic threat. During infection, tissue damage is induced both by invading pathogens and immune effectors such as toxins and free radicals. Therefore, it is important to elucidate by what self-protective mechanisms the host defends itself against pathogenic stress. The conserved SKN-1 protein of the roundworm Caenorhabditis elegans directs a detoxification response neutralizing harmful compounds as well as confers tolerance to oxidative stress. Here we identify SKN-1 as a novel regulator of C. elegans innate immunity. We show that SKN-1 contributes to resistance against infection caused by two bacterial pathogens. Components of a pathogen-responsive signaling pathway are required to activate SKN-1 in intestinal cells at the site of infection. Moreover, the SKN-1-dependent response to pathogen exposure declines during aging, whereas mild metabolic and oxidative stresses, known to extend lifespan, evoke a SKN-1-dependent boosting of immunity. Finally, we find that elimination of an inhibitory protein leads to an excessive activation of SKN-1 and impairs pathogen resistance. Thus, SKN-1 integrates various chemical, metabolic and microbial signals to elicit a self-protective detoxification response, which promotes innate immunity and may be relevant to diseases and aging of the human immune system.
Introduction
A proper immune response ensures survival in a hostile environment and contributes to longevity. The nematode Caenorhabditis elegans provides a valuable genetic tool for studying innate immunity and various aspects of host-pathogen interactions. During infection, both bacterial virulence factors and host antimicrobial defense mechanisms present oxidative and proteotoxic noxae inducing tissue-damage, especially in the intestine [1]–[5]. Accordingly, several self-protective stress-response regulators including the forkhead transcription factor DAF-16/FOXO [6], the heat shock transcription factor HSF-1 [7] and the X-box binding protein 1 (XBP-1) [8] are required for robust immunity. Moreover, the DAF-16-regulated antioxidant enzymes SOD-3 and CTL-2 contribute to immunity by protecting intestinal cells from reactive oxygen species during exposure to Enterococcus faecalis [9]. Strikingly, hyper-activation of DAF-16 enhances susceptibility to bacterial infection [10].
These data illustrate a critical role of stress response in innate immunity, and raise questions about the co-ordination of antimicrobial and host-defense mechanisms. Antimicrobial responses are mediated by a canonical p38 mitogen-activated protein kinase (MAPK) pathway, which is conserved from nematodes to humans [11], [12]. Besides, the insulin/IGF-like signaling (IIS) and TGF-β pathways are also involved in the regulation of the pathogen-specific immune response in C. elegans [13], [14]. Both p38 MAPK and IIS pathways regulate the Nrf1/2/3 ortholog SKN-1, a transcription factor that orchestrates both oxidative and xenobiotic stress responses in C. elegans [15], [16]. However, the involvement of SKN-1 in the regulation of pathogen stress response is unknown.
In nematodes, three SKN-1 isoforms exist. While the function of SKN-1A has not been elucidated yet, SKN-1B and C provide distinct biological functions. SKN-1B is expressed in the ASI neurons, and mediates lifespan extension in response to dietary restriction [17]. In contrast, intestinal SKN-1C is required for oxidative stress resistance and contributes to longevity by reduced IIS [15]. SKN-1 activity is regulated by phosphorylation and degradation. Under normal conditions, inhibitory phosphorylations by GSK-3 and IIS kinases, AKT-1/2 and SGK-1, retain SKN-1 in the cytosol [15], [18], where it is rapidly targeted to proteasomal degradation by the WD40 repeat protein WDR-23 [19], [20]. In response to oxidative stress, the p38 MAPK ortholog PMK-1 phosphorylates SKN-1, which then translocates to the nuclei of intestinal cells and induces transcription of phase 2 detoxification genes [16].
Here we report that SKN-1 is required for pathogen resistance against both Gram-negative P. aeruginosa and Gram-positive E. faecalis bacteria, consistently with an independent study [21] published after submission of this paper. We further demonstrate a Toll and Interleukin-1 Receptor domain protein (TIR-1)/PMK-1-dependent SKN-1 activation upon P. aeruginosa infection. Moreover, we show a gradual decrease of pathogen resistance and of the activation of SKN-1-dependent targets during aging, and a requirement of SKN-1 to boost immunity in response to longevity-promoting manipulations, such as reduced IIS and preconditioning H2O2 treatment. Finally, we find that hyper-activation of SKN-1 impairs pathogen resistance. Our results indicate an intricate regulation of innate immunity by SKN-1 and links pathogenic stress signaling to the xenobiotic stress response.
Results
SKN-1 is required for bacterial pathogen resistance in C. elegans
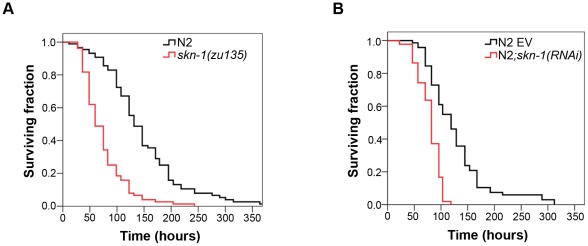
To study the role of SKN-1 in C. elegans immunity, we examined the pathogen resistance of animals in the absence of SKN-1. skn-1(zu135) allele is considered to be a genetic null mutation as it creates a premature stop codon that affects all SKN-1 isoforms [15]. skn-1(zu135) mutant worms were first exposed to the Gram-negative Pseudomonas aeruginosa (PA14) strain. As skn-1(zu135) mutants are sterile, we eliminated the difference between them and wild-type N2 strain arising from the ‘bag of worms’ phenotype, a major contributor to killing. To this end, germline development was inhibited by silencing cdc-25.1, required for embryonic mitosis and meiosis. cdc-25.1(RNAi) animals exhibit extended survival on pathogenic bacteria, as reported previously [21], [22]. In these conditions, we observed an increased susceptibility of skn-1(zu135) mutants to PA14 (Figures 1A and S1A, Tables S1A and S1E). This result was confirmed by using skn-1(RNAi) (Figures 1B and S1B, Table S1A and S1E). Furthermore, when animals were exposed to the Gram-positive Enterococcus faecalis SdB262 strain, both skn-1(zu135) and skn-1(RNAi) exhibited significantly decreased survival, though in this case the absence of SKN-1 exerted a more modest effect (Figures S1C and S1D, Table S1E). These results suggest a requirement of SKN-1 for the efficient immune response against two distinct bacterial pathogens. In subsequent experiments, we focused on further defining the role of SKN-1 in the antibacterial response against P. aeruginosa.
Figure 1. SKN-1 is required for bacterial pathogen resistance.
(A, B) Increased susceptibility to Pseudomonas aeruginosa PA14 occurs in both skn-1(zu135) mutant (p<0.0001) and skn-1(RNAi) nematodes (p<0.0001). Killing assays were performed with at least 90 young adult animals in each condition. EV: empty vector RNAi.
P. aeruginosa infection triggers SKN-1 activation
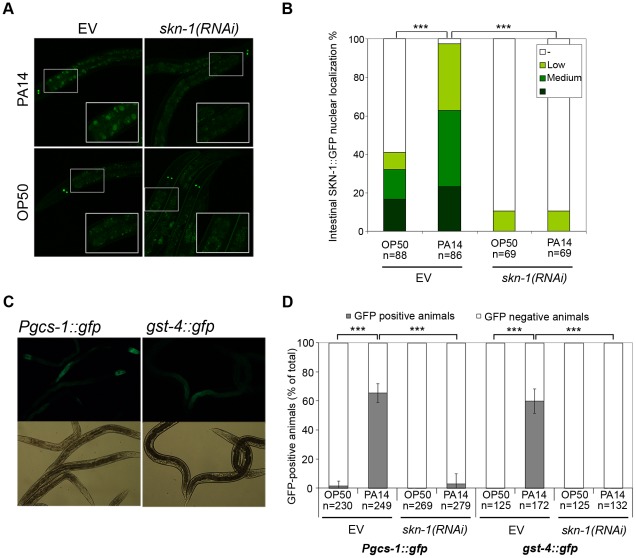
To investigate if SKN-1 nuclear translocation occurs upon PA14 exposure, we incubated skn-1::gfp L3 larvae on P. aeruginosa lawn for 5 hours. We found a massive accumulation of SKN-1::GFP in intestinal nuclei of infected larvae, compared to control animals fed by the non-pathogenic OP50 Escherichia coli strain (Figures 2A and 2B). The specificity of this response was demonstrated by a complete inhibition using a skn-1-specific double-stranded RNA. To reveal a SKN-1-dependent transcriptional activation upon PA14 infection, we examined the Pgcs-1::gfp and gst-4::gfp reporter strains. While gcs-1 is regulated exclusively by SKN-1, gst-4 is under the mutual control of both DAF-16 and SKN-1 [15]. We observed an effective intestinal induction of fluorescence to comparable extent in both strains in response to a 24 h-exposure of PA14 (Figures 2C and 2D). Both the gcs-1 promoter activation and the GST-4 expression were significantly suppressed by feeding worms with skn-1(RNAi), indicating the specific requirement of SKN-1 to elicit these responses. Thus, PA14 infection induces nuclear translocation of SKN-1 and transcriptional activation of its targets.
Figure 2. P. aeruginosa infection activates SKN-1.
(A) Representative epifluorescence image demonstrating the translocation of SKN-1::GFP in the Is007[SKN-1::GFP] strain to intestinal nuclei in L3 larvae, fed by the empty vector or skn-1 dsRNA, upon a 5-hour exposure to P. aeruginosa PA14. Note that the intestinal tissue displays autofluorescence, and in the ASI neurons SKN-1::GFP is not silenced by skn-1 RNAi treatment. (B) Quantification of SKN-1 nuclear translocation from data shown on panel (A). SKN-1::GFP-positive nuclei were counted in the intestine of 78 animals. “Low” refers to animals in which SKN-1::GFP was detected in less than 5 intestinal nuclei, while “high” indicates that SKN-1::GFP signal was present in more than 15 intestinal nuclei. (C) Representative epifluorescence microscopic image showing intestinal expression of Pgcs-1::GFP and GST-4::GFP in L3 larvae upon a 24-hour PA14 exposure. Images of control animals incubated on OP50 bacteria are shown in Figure S2. (D) Quantification of reporter expression demonstrating the SKN-1-dependence of the response. Data were obtained from panel (C) completed with the data of skn-1(RNAi) animals. Microscopic images are representatives of 3 independent experiments. EV: empty vector RNAi.
The TIR-1/PMK-1 pathway controls SKN-1 activation upon P. aeruginosa infection
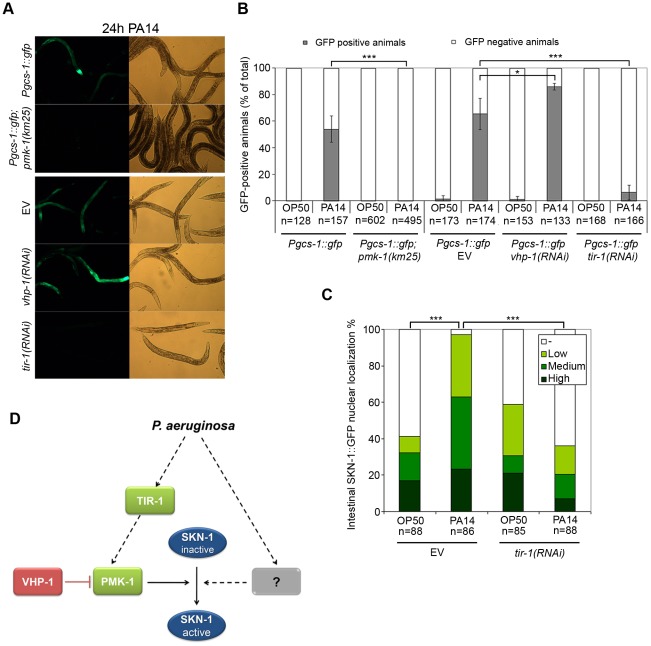
The p38 MAPK ortholog PMK-1 has a fundamental role in C. elegans innate immunity [11]. To investigate whether PMK-1 regulates SKN-1 in response to bacterial exposure, we monitored the activity of a Pgcs-1::GFP reporter in a wild-type and a pmk-1(km25) mutant genetic background (Figures 3A and 3B). We found that silencing pmk-1 entirely prevented the SKN-1-dependent activation of gcs-1 in response to PA14 infection. In physiological settings, PMK-1 is inactivated by the dual specificity MAPK phosphatase VHP-1 [23]. Suppression of VHP-1 resulted in increased PMK-1 phosphorylation and resistance to PA14 [23]. However, vhp-1(RNAi) significantly increased Pgcs-1::GFP activation upon PA14, but not upon OP50 exposure, suggesting that PMK-1 is an indispensable permissive factor for SKN-1 activation by infection.
Figure 3. The pathogen response-specific TIR-1 and p38 MAPK PMK-1 are required for SKN-1 activation upon P. aeruginosa infection.
(A) Representative epifluorescence microscopic images showing the expression of Pgcs-1::GFP in pmk-1(km25) mutants as well as in the p38 MAPK phosphatase vhp-1(RNAi), and the Toll/IL-1 resistance (TIR) domain protein tir-1(RNAi) animals in response to P. aeruginosa infection. L3 larvae were exposed to PA14 for 24 hours. Microscopic images are representatives from 3 independent experiments. (B) Quantification of reporter expression from data shown on panel (A) completed with data of control animals fed by OP50 for 24 h. (C) Quantification of SKN-1 nuclear translocation in tir-1(RNAi) L3 larvae upon 5 h PA14 exposure. Representative epifluorescence images of tir(RNAi) L3 larvae are shown in Figure S3. Please note that data in Figure 2B and 3C were derived from the same set of experiments. (D) Suggested model of SKN-1 activation during P. aeruginosa infection. Upon exposure to PA14, the TIR-1/PMK-1 pathway is indispensable but insufficient to elicit SKN-1 transactivation. We propose a second, unknown factor/pathway that is required to activate SKN-1. Whether the two pathways act in parallel or consecutively is unclear. Solid arrows indicate a direct, while dashed arrows indicate an indirect/unknown connection. EV: empty vector RNAi.
TIR-1 is a conserved Toll/IL-1 resistance (TIR) domain protein known to activate p38 MAPK signaling independently of the Toll-like receptor ortholog tol-1 during PA14 infection [24], [25]. Depletion of TIR-1 by RNAi prevented Pgcs-1::GFP fluorescence upon PA14 infection (Figures 3A and 3B). A similar inhibition in Pgcs-1::GFP expression was also observed in tir-1(qd4) mutant animals (data not shown). Moreover, silencing tir-1 prevented the nuclear translocation of SKN-1 induced by PA14 infection, but did not affect its baseline expression levels (Figures 3C and S3.). Altogether, these results suggest that the TIR-1/PMK-1 pathway is necessary to attain activation of SKN-1 by PA14 exposure (Figure 3D).
Involvement of SKN-1 in immunosenescence
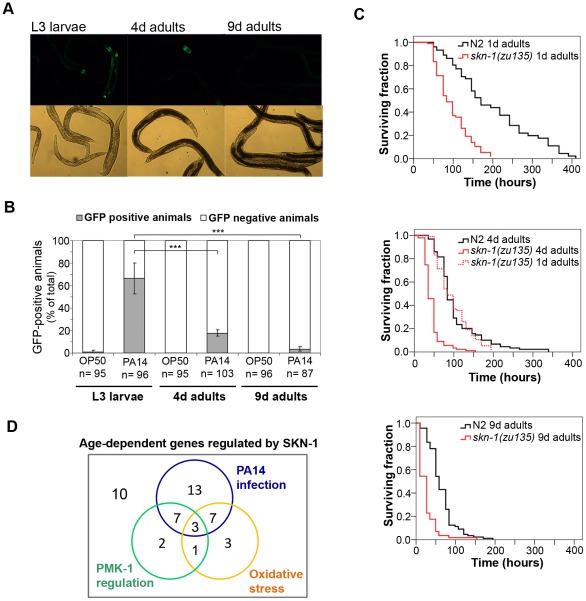
Immune function declines with age, leading to compromised immune responses to infections in the elderly. Accordingly, aged nematodes exhibit increased susceptibility to infection by various pathogens, including P. aeruginosa [26]–[28]. As SKN-1 is required for both longevity and for pathogen resistance, we asked if chronological aging affected SKN-1-dependent target gene expression in nematodes exposed to pathogenic stress. To this end we examined the promoter induction of gcs-1 by PA14 in L3 stage larvae, 4-day and in 9-day old adult worms, respectively (Figures 4A and 4B). We observed a massive age-dependent decrease in the expression of Pgcs-1::GFP reporter after 24 h of PA14 infection. To investigate, how SKN-1 activity is involved in immunosenescence, we examined the survival of 1, 4 and 9 day-old adult N2 and skn-1(zu135) mutant nematodes exposed to PA14. We observed that pathogen resistance in wild-type animals already declined at day 4 as previously described by Laws et al. [26]. Consistent with a premature decline of self defense in the absence of SKN-1 activity, 4 d adult N2 worms showed similar survival on PA14 to 1 d adult skn-1(zu135) animals (p = 0.1429). Furthermore, we found that skn-1(zu135) mutant animals exhibited increased susceptibility to PA14, compared to N2 at all ages (p>0.0001) (Figure 4C and Table S1B), indicating that SKN-1 function is also required to survive infection beyond day 9.
Figure 4. Involvement of SKN-1 in immunosenescence.
(A) Representative epifluorescence images showing the decreased induction of the gcs-1 promoter. L3 larvae, 4 d/9 d adult Pgcs-1::gfp worms were exposed to PA14 for 24 h. (B) Quantification of the epifluorescence images of panel (A). Epifluorescence images are representatives of two independent experiments. EV: empty vector RNAi. (C) Pathogen resistance of young adult (1 day-old), 4 day-old and 9 day-old adult N2 and skn-1(zu135) mutant animals. skn-1(zu135) mutant worms exhibited significantly increased susceptibility to PA14 compared to N2 wild-type animals at all ages (p<0.0001). 1 day-old adult skn-1(zu135) worms show similar pathogen resistance to 4 day-old N2 worms (p = 0.1429) (middle graph). Killing assays were performed with 3 parallel plates in each condition in 2 independent trials. (D) Venn diagram showing the distribution of age-regulated SKN-1 target genes. Data were analyzed by finding the overlaps between micro-array databases containing the genes down-regulated at least 10-fold in 15 d adult compared to 6 d adult wild-type animals [27] and SKN-1 dependent genes under non-stress [29] or oxidative stress conditions [30] using expression data from Wormbase [31]. Please note that the majority of genes belong to those regulated by PA14 infection. 10 of 46 genes could be assigned to none of the groups. For the detailed gene list please refer to Table S2.
To address the potential involvement of SKN-1-dependent gene expression in immunosenescence, we performed a bioinformatics analysis using the microarray data of Youngman et al. [27]. From the 379 genes exhibiting the most significant down-regulation during aging (>10 fold down-regulation at d15 vs. d6) we identified 46 SKN-1-regulated genes (based on skn-1(RNAi) screens [29], [30]) (Figure 4D, Table S2). Next, we examined the regulation of these genes with respect to oxidative stress, PA14 and PMK-1 dependent regulation using Wormbase expression data [31]. Strikingly, SKN-1-regulated genes subject to PA14-dependent regulation were over-represented compared to those regulated by either oxidative stress or PMK-1, respectively. These results confirm a progressive age-dependent compromise in pathogen resistance and imply that a decline in SKN-1 function contributes to immunosenescence.
Reduced IIS and oxidative preconditioning require SKN-1 for enhanced pathogen resistance
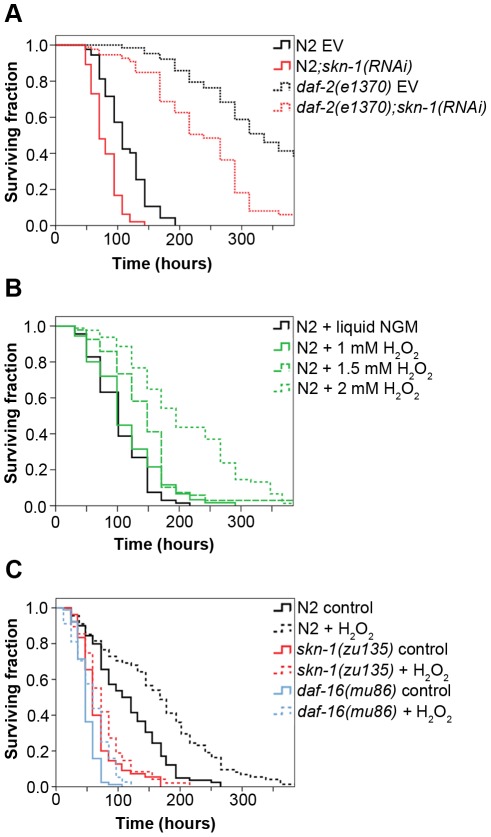
Loss-of-function mutations in the insulin/IGF-1 receptor gene, daf-2 enhance stress resistance and extend lifespan, and both processes require DAF-16 and SKN-1 activity [15]. As reduced IIS increases pathogen resistance [6], we investigated the contribution of SKN-1 to pathogen resistance in daf-2(e1370) mutant animals. In accordance with previously published data [6], daf-2(e1370) mutants exhibited robustly increased pathogen resistance against PA14 (Figure 5A and Table S1C). However silencing skn-1 by RNAi largely increased their susceptibility to PA14. These data suggest that SKN-1 is required for reduced IIS to bring about enhanced pathogen resistance against P. aeruginosa.
Figure 5. Reduced IIS and oxidative preconditioning require SKN-1 for enhanced pathogen resistance.
(A) daf-2(e1370) mutant nematodes exhibited increased resistance to P. aeruginosa, compared to that of wild-type N2 worms (p<0.0001). skn-1(RNAi) treatment of daf-2(e1370) animals increased the susceptibility to P. aeruginosa infection (p<0.0001). Killing assays were performed with at least 90 young 1-day old adult animals in each condition. (B) H2O2 pretreatment increased survival on PA14 in a concentration-dependent manner. Survival curves of N2 wild-type worms treated with various concentrations of H2O2 in liquid NGM: 1 mM (p = 0.425), 1.5 mM (p<0.0001) and 2 mM (p<0.0001) 12 h prior to the killing assay are shown. Killing assay was performed with 90 3-day old adult animals in each condition. (C) Oxidative preconditioning-induced pathogen resistance was impaired in the absence of SKN-1 or DAF-16. Increase in survival was less pronounced in either skn-1(zu135) (p = 0.0156) or daf-16(mu86) mutant (p = 0.0304), than in wild-type animals (p<0.0001). Survival curves of the same genetic background were compared in the absence and presence of H2O2. Data were combined from at least two experiments with 89 animals in average for each group. EV: empty vector RNAi.
Exposure to mild oxidative stress induces tolerance to a lethal challenge, cross-tolerance to other stresses and extends lifespan [32]. To address the impact of oxidative preconditioning on pathogen resistance, nematodes were pretreated with various concentrations of H2O2, and then exposed to PA14 infection. H2O2 preconditioning induced resistance against PA14 in a concentration-dependent manner, reaching a 2-fold increase in survival by 2 mM H2O2, compared to untreated controls (Figure 5B and Table S1D). Intriguingly, the same treatment on skn-1(zu135) mutant nematodes not only exhibited a decreased pathogen resistance, but had a strongly suppressed reaction to H2O2 (Figure 5C and Table S1D). We also found that the mutation of another major oxidative stress response regulator, DAF-16 (daf-16(mu86)), shows an even shorter basal survival, compared to skn-1(zu135), and poorly responded to H2O2 (Figure 5C and Table S1D). Thus, oxidative preconditioning requires both SKN-1 and DAF-16 for enhanced pathogen resistance against P. aeruginosa.
Excessive activation of SKN-1 by wdr-23(RNAi) impairs pathogen resistance
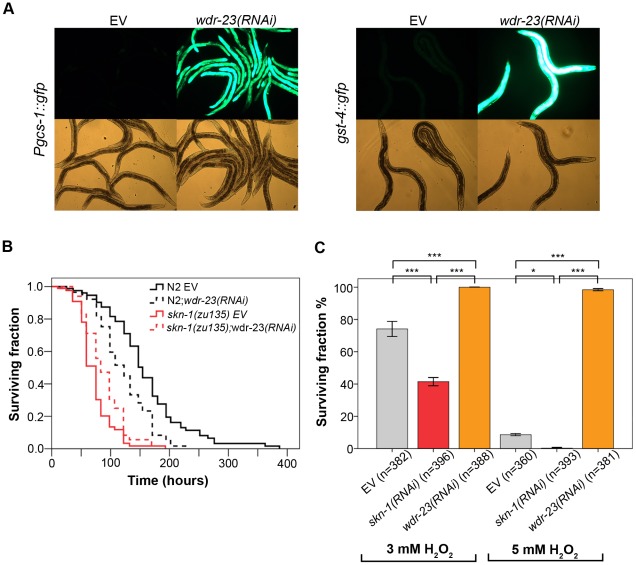
Finally, we investigated whether increased activation of SKN-1 was able to promote pathogen resistance. Stabilization of SKN-1 by RNAi against wdr-23 has been shown to induce constitutive SKN-1 activation, resistance to oxidative stress and longevity [19]. Feeding worms with wdr-23(RNAi) indeed resulted in an unexpectedly robust increase in the expression of Pgcs-1::GFP and GST-4::GFP (Figure 6A) compared to the PA14-induced expression (Figure 2C). To our surprise, wdr-23(RNAi), compared to empty vector feeding greatly reduced pathogen resistance to PA14 (Figure 6B and Table S1A). wdr-23(RNAi) did not impair survival in a skn-1(zu135) mutant background excluding a SKN-1-independent impact of WDR-23 on pathogen resistance. Interestingly, the compromised reactivity of worms to wdr-23(RNAi) was confined to pathogenic stress. Determination of oxidative tolerance revealed that wdr-23(RNAi) animals exhibited increased survival, whereas skn-1(RNAi) nematodes displayed decreased survival, compared to control worms, when exposed to 3 mM or 5 mM H2O2, respectively (Figure 6C). Our results suggest that an excessive post-translational stabilization of SKN-1 induces oxidative stress resistance but impairs resistance to bacterial infection.
Figure 6. Excessive activation of SKN-1 by wdr-23(RNAi) impairs pathogen resistance.
(A) Robust up-regulation of Pgcs-1::GFP and GST-4::GFP in 1 d adult wdr-23(RNAi) worms. (B) Pathogen resistance of wdr-23(RNAi)-fed N2 and skn-1(zu135) mutant worms. N2;wdr-23(RNAi) exhibited increased susceptibility to P. aeruginosa infection (p<0.0001). skn-1(zu135) mutant nematodes fed by wdr-23(RNAi) showed no significant difference in survival on PA14 (p = 0.1992). Killing assay was performed with at least 90 1-day old adult animals in each condition. Please note that data in Figures 1A and 6B were derived from the same set of experiments. (C) wdr-23(RNAi) treatment increased (p<0.0001, both at 3 mM and 5 mM H2O2, respectively), while skn-1 RNAi treatment decreased oxidative tolerance to H2O2 (p<0.0001 at 3 mM H2O, p<0.05 at 5 mM H2O2). Worms were treated with 3 mM or 5 mM H2O2 for 1 hour, and 24 h after challenge survival was scored. Data were combined from three experiments with 120 animals in average for each group. EV: empty vector RNAi.
Discussion
Our present study identified SKN-1 as a novel regulator of pathogen resistance against both Gram-negative P. aeruginosa and Gram-positive E. faecalis bacteria (Figures 1 and S1). We demonstrated a TIR-1/PMK-1-dependent SKN-1 activation upon P. aeruginosa infection (Figures 2 and 3). Moreover, we showed an early onset of immunosenescence with a parallel decline in SKN-1-dependent transcriptional activation (Figure 4) and a requirement of SKN-1 to efficient immunity in response to reduced IIS or preconditioning H2O2 treatment (Figure 5). Finally, we found that excessive activation of SKN-1 by blocking its turnover impaired pathogen resistance (Figure 6).
The xenobiotic stress response provides a conserved defense mechanism against oxidative and electrophilic stress via the induction of phase 2 detoxification enzymes [33]. Nrf2 and its nematode ortholog, SKN-1, are transcription factors important in oxidative and xenobiotic stress response [34], [35]. Previously, several studies demonstrated the importance of Nrf2 in innate immunity in mammals [36]–[38]. For example, Nrf2−/− mice exhibit increased susceptibility to bacterial infection and bacterial lipopolysaccharide (LPS)-induced inflammation [39]. Similarly, our present study demonstrates that SKN-1 deficiency in C. elegans impairs resistance to infection (Figures 1 and S1). During the revision of our manuscript an independent paper from the Garsin lab appeared, which obtained similar results [21]. A previous study found no significant impairment of pathogen resistance by skn-1(zu135) and skn-1(zu67) mutations in the wildtype background [40]. A possible reason of this discrepancy might be the use of cdc-25.1(RNAi) by the Garsin lab and our study, suggesting that selective bagging in wild-type vs. sterile skn-1 mutants might have masked the pathogen resistance decrease induced by loss of skn-1 in the previous investigation. Consistently with this note, the use of skn-1(RNAi) from the L1 stage, which did not induce sterility [21], confirmed the decrease in pathogen resistance. A similar finding was also reported as an earlier unpublished result of Evans et al. [41].
We observed a nuclear translocation and transcriptional activation of SKN-1 in the intestine, consistent with the primary site of infection (Figures 2 and 3). Furthermore, we could not detect any apparent change in SKN-1 intensity or nuclear localization in ASI neurons upon PA14 exposure (Figure 2). This finding is in agreement with previous reports on constitutive SKN-1B activity and a lack of interaction between SKN-1B and WDR-23 in ASI neurons, respectively [19], [34]. Together, our data imply an active role of the intestinal SKN-1C isoform and does not allow a conclusion regarding the involvement of the ASI neuronal SKN-1B in the inducible antibacterial response. However, a continuous transcriptional output of SKN-1B and/or a different mode of regulation of SKN-1B in response to infection cannot be excluded. Hence, a tissue-specific analysis of SKN-1 function may give a clue whether SKN-1 isoforms co-operate in immunity.
Our findings confirm those of van der Hoeven et al. [21] on the critical role of the p38 MAPK pathway in SKN-1 activation (Figure 3). However, the inability of vhp-1(RNAi) to activate SKN-1 on OP50 suggests that there should be additional, unidentified signals that govern SKN-1 activation in response to PA14 infection, which will certainly prompt additional studies. We showed an absolute requirement of TIR-1 for SKN-1 nuclear translocation and for gcs-1 promoter induction upon PA14 exposure, while the Garsin lab reported no to minimal involvement of TIR-1 in gst-4 and gcs-1 induction in response to E. faecalis [21]. Whether the difference between our observations beyond differences in assays and dosage/treatment by tir-1(RNAi) may be due to a differential pathogen sensing of P. aeruginosa and E. faecalis is an exciting possibility to explore. TIR-1 and PMK-1 are related to the mammalian SARM and p38 MAPK proteins, respectively [42], [43]. Although the existence of an orthologous pathway in mammals remains elusive, these findings indicate that the SKN-1-mediated response is an ancient component of innate immunity.
Immunosenescence, the age-dependent decline of immune response, is a critical problem impeding healthy ageing [44]. C. elegans provides a useful tool to investigate elements of innate immunity contributing to immunosenescence [45]. A recent systematic study reported an age-dependent progressive increase in susceptibility to PA14, detectable at day 6 of adulthood [27]. Our data on a similar age-related decline in survival, with a 45% decrease in pathogen resistance at day 4 (Figure 4C) establishes an earlier, dramatic onset of immunosenescence. Moreover, the loss of SKN-1 function phenocopies the decreased resistance of d4 worms already at day 1, and continues to negatively affect survival at day 9 (Figure 4C). A parallel strong decline in gcs-1 transactivation on day 4 and the widespread down-regulation of SKN-1 targets, including PA14-regulated genes, between day 6 and 15 of adulthood (Figure 4D, Table S2) are consistent with this observation and indicate SKN-1 as a key player in immunosenescence.
Youngman and colleagues found an involvement of PMK-1 in a decline of the innate immune response, and hypothesized intestinal deterioration as a primary event in immunosenescence [27]. Of note, the dependence of SKN-1 activation on PMK-1 ([21] and our study), the high number of age-dependent SKN-1 targets among PMK-1 targets (13 of 26; Figure 4D, Table S2 and [27]) and the impact of SKN-1 on intestinal homeostasis [30] suggest a dynamic, probably mutual interaction between SKN-1 and PMK-1 in immunosenescence. We propose that SKN-1-dependent stress responses collapse early in adulthood, which manifest in a vicious circle of decreasing intestinal homeostasis, progressive immunosenescence and increasing pathogenic load in C. elegans. This hypothesis is consistent both with the short lifespan of worms in natural conditions and with the allocation of resources to maintain the soma until the production of fit progeny (the “disposable soma theory” [46]).
Genetic or environmental interventions that operate via stress-responsive mechanisms extend both lifespan and pathogen resistance [6], [15], [32], [47], [48]. Our results on H2O2-induced pathogen resistance (Figure 5B), together with previous analogous heat-shock experiments [7] suggest that mild stresses acting early in adulthood confer resistance against pathogenic stress. Furthermore, the demonstration of the requirement of SKN-1 and DAF-16 in the enhanced pathogen resistance of both H2O2-preconditioned and daf-2(e1370) mutant nematodes suggests a dynamic cross-talk of these stress-responsive transcription networks tipping the balance between responses to nutrient availability, oxidative and pathogen stress. Though our data do not allow a clear conclusion, the recent prediction of a DAF-16-dependent regulation of SKN-1 [49] is in line with the proposed functional interaction between SKN-1 and DAF-16 and is a subject of future interesting studies.
Evidence on mammals indicates a defensive role of Nrf2 against inflammation-induced tissue damage [36]–[39]. An analogous nematode model raises the question, whether SKN-1 affects immunity independently of its impact on aging. Indeed, longevity, stress resistance and pathogen resistance are intimately linked in short-lived C. elegans. However, a greater reduction of survival in skn-1(zu135) mutants on PA14 than on non-pathogenic OP50 (51% vs. 19% compared to N2, Figures 1 and S4, Tables S1 and S3) suggests a stronger impact of SKN-1 on pathogen resistance than on longevity. The pathogen-induced activation of SKN-1 and the large number of PA14-regulated SKN-1-targets including immune-related CUB-like domain proteins (Figure 4D, Table S2) [29] support SKN-1's active involvement in the pathogen response. Finally, it has previously been shown that knock-down of skn-1 in the daf-2(e1370) mutant selectively suppresses stress resistance but not lifespan [15]. Thus, our findings demonstrating a SKN-1-dependent increase of pathogen resistance by this allele (Figure 5A), suggest an immune-specific effect of SKN-1. Studies investigating SKN-1-dependent responses on OP50 vs. pathogens would help reveal the downstream mediators of SKN-1 and to determine the immune-specific and other branches of SKN-1 action.
SKN-1 activation by wdr-23(RNAi) impairs pathogen resistance, a result in contrast with those of the Garsin lab [21]. The reason may lie in the use of cdc-25.1(RNAi) by us, or in the different dosage/duration of RNAi treatment in the two experimental protocols. Nevertheless, our findings on the adverse effects of excessive SKN-1 activity are consistent with those reporting that loss of WDR-23 activity slows growth via SKN-1 [19], expression of SKN-1 from high-copy arrays is toxic [15], and that SKN-1 mediates increased susceptibility to PA14 in the absence of BLI-3 [21]. Combining the two wdr-23(RNAi) results ([21] and our study) clearly shows that this type of activation can dissociate immunity from oxidative stress resistance. As a potential mechanism, excessive SKN-1 activation may remodel the transcriptional response in favor of anti-oxidative defense and/or may repress pathogen-specific defenses. Indeed, differential SKN-1 transcriptional outputs were demonstrated [29]. It is logical to assume that negative and positive inputs regulating SKN-1 allow fine-tuning of stress resistance, growth and immunity. An analogous deterioration of pathogen resistance by the excessive activation of DAF-16 [10] underscores the necessity of tight control of stress responses to avoid deleterious consequences during infection.
Taken together, we propose that an optimal enhancement of SKN-1 activity in proper time-frame may enhance immune responses and delay immunosenescence without compromising longevity. In recent years, C. elegans has become a versatile model not only for studying innate immunity, host-pathogen interactions, but for testing pharmacological interventions in drug discovery [50]. The results presented herein may prompt studies on drugs targeting SKN-1/Nrf2 to modulate the innate immune response. In conclusion, our findings indicate an intricate regulation of innate immunity by SKN-1, and link pathogenic stress signaling to xenobiotic and oxidative stress responses.
Materials and Methods
C. elegans strains and maintenance
Nematodes were maintained and propagated on E. coli OP50 as described by Brenner [51] at 20°C. The following C. elegans strains were obtained from the Caenorhabditis Genetics Center and were used in this study: N2, EU31 skn-1(zu67)IV/nT1[unc-?(n754) let-?](IV;V), KU25 pmk-1(km25)IV., ZD101 tir-1(qd4)III. Further strains were used: LD001 Is007 [skn-1::gfp], CF1038 daf-16(mu86)I., CB1370 daf-2(e1370)III. (Tibor Vellai, Eötvös Loránd University, Budapest, Hungary), LD1171 Is003 [Pgcs-1::gfp] (T. Keith Blackwell, Harvard Medical School, Boston MA, USA) and MJCU017 kIs17[gst-4::gfp, pDP#MM016B]X. (Johji Miwa, Chubu University, Kasugai, Japan). Nematodes were treated with cdc-25.1(RNAi) to avoid the bacterial infection induced ‘bag of worms’ phenotype in all experiment.
Crossing and genotyping by PCR
Pgcs-1::gfp;pmk-1(km25) and Pgcs-1::gfp;tir-1(qd4) strains were created by mating male pmk-1(km25) or tir-1(qd4), respectively, with LD1171 Is003 [Pgcs-1::gfp] hermaphrodites. Transgenic rol progeny was isolated with the correct genotype as scored by PCR. PCR primers were obtained from Sigma. The primers pmk-1-OF (5′-GGATACGGAAGAAGAGCCAATG-3′) and pmk-1-OR (5′-CAACAGTCTGCGTGTAATGC-3′) were used to detect the pmk-1(km25) deletion allele. The wild-type pmk-1 allele amplified a 1195-bp fragment compared to a 882-bp fragment from pmk-1(km25) allele. Homozygous pmk-1(km25) mutants were identified by PCR using primers pmk-1-IF (5′-TCCTATAAGTTGCCATGACCTCAG-3′) and pmk-1-IR (5′-CCCGAGCGAGTACATTCAGC-3′) from inside the deletion region. Wild-type animals generated a 469-bp fragment, while the homozygous pmk-1(km25) allele did not produce any fragment. The primers tir-1-OF (5′-TGGGTAAATGAGGAAGAGAGAGAG-3′) and tir-1-OR (5′-TCGGTTGACGAGTCGAATTTGG-3′) were used to detect the tir-1(qd4) deletion allele. The wild-type tir-1 allele amplified a 1368-bp fragment compared to a 228-bp fragment from tir-1(qd4) allele. Homozygous tir-1(qd4) mutants were identified by PCR using primers tir-1-OF and tir-1-IR (5′-CACAAGAACGTGCAACATCG-3′) from inside the deletion region. Wild-type animals generated a 327-bp fragment, while the homozygous tir-1(qd4) allele did not produce any fragment.
RNA interference (RNAi)
The HT115(DE3) E. coli bacteria producing dsRNA against cdc-25.1 (Andy Golden NIDDK/NIH, Bethesda MD, USA), skn-1 (T. Keith Blackwell, Harvard Medical School, Boston MA, USA), wdr-23 (Keith P. Choe, University of Florida, Gainesville FL, USA), vhp-1 and tir-1 (Source BioScience Geneservice, Cambridge, United Kingdom) were used in our study. RNAi feeding E. coli clones were grown overnight in LB medium containing 100 µg/ml ampicillin. RNAi treatment was performed as described by Shapira et al. [22]. Worms were grown on RNAi bacteria from hatching till young adult stage. If several RNAi constructs were used in one condition, ON cultures of the feeding bacteria strains were mixed equally. Empty vector containing HT115(DE3) bacteria (EV) was used as a control in all cases.
Preparation of pathogenic bacteria
Different human opportunistic bacteria, such as Pseudomonas aeruginosa and Enterococcus faecalis [28], [52] are ubiquitously used as pathogen models. Gram-negative Pseudomonas aeruginosa PA14 (David W. Wareham, Queen Mary University of London, London, UK) and Gram-positive Enterococcus faecalis SdB262 (Jonathan J. Ewbank, Centre d'Immunologie de Marseille-Luminy, Marseille, France) bacteria were maintained and prepared for experiments as described by Powell and Ausubel [53]. BHI agar was supplemented with 100 µg/ml rifampicin for E. faecalis killing assay.
Killing assay
Killing assays were performed with young adult animals at 25°C on slow killing plates (P. aeruginosa) or rifampicin BHI plates (E. faecalis), or otherwise as it was noted in the figure legend. Dead worms were scored every 12 hours till complete extinction of the population. Viability was determined by assaying for movement in response to gentle prodding. Worms died on the wall of the Petri dish or crawled into the gel were censored. 30 animals per condition were tested with 3 parallel plates in at least two independent trials, except that an H2O2-concentration dependence of preconditioned pathogen resistance was established in one trial. For studying pathogen resistance of daf-2 mutants, nematodes were grown at 15°C. For oxidative preconditioning 2-day old adult animals were treated with 0 (control), 1 mM, 1.5 mM and 2 mM H2O2 (Sigma) in liquid NGM for 2 hours at 20°C. Before the killing assay worms were transferred to OP50 seeded NGM plates for a 12-hour recovery period. To test the effect of aging on pathogen resistance, worms were maintained on OP50-seeded NGM plates before the challenge. To avoid ‘bag of worms’ phenotype, animals were fed by cdc-25.1(RNAi).
Fluorescence microscopy
Nematodes were treated as indicated in the figure legends (Figures 2, 3, 4 and 6). After treatments at least 40 worms per condition were placed on a 2% agarose pad, and immobilized by adding 40 mM levamisole in M9 buffer. Images were taken by a Leica DMI6000B epifluorescence microscope with a DFC480 camera. Epifluorescent microscopic images are representatives of at least 3 experiments. To analyze Pgcs-1::GFP and GST-4::GFP expression upon PA14 infection, two groups of animals were determined depending on the detected GFP level in the intestine: GFP positive and GFP negative animals. To study the nuclear localization of SKN-1 in response to PA14 infection minimum 15 skn-1::gfp worms per condition were analyzed in at least 3 independent trials. Images were captured by a Zeiss LSM510 confocal laser scanning microscope equipped with a 40×/1.3 oil immersion objective (Plan-Neofluar, Zeiss).
Analysis of SKN-1 dependent targets amongst genes down-regulated by aging
A list of 379 genes exhibiting the most significant age-dependent decline in their expression (>10-fold at d6 vs. d15) was acquired from [27]. Data were analyzed by finding the overlaps between genes subject to SKN-1 dependent genes under non-stress [29] or oxidative stress conditions [30]. Then the expression of the identified genes was analyzed based on Wormbase data, focusing on PA14-, oxidative stress- or PMK-1-dependent regulation [31].
Oxidative stress tolerance
Young, 1-day old adult worms were incubated in liquid NGM for 1 hour at 20°C with 3 mM and 5 mM H2O2 (Sigma). After oxidative challenge animals were transferred to OP50 seeded NGM plates and viability was tested 24 hours later. 35 animals per plate were examined in each condition with 3 parallel plates in 3 independent trials.
Statistical analysis
Data were analyzed by using the SPSS software 15.0 (SPSS Inc., Chicago, IL, USA). Survival curves were compared by Kaplan-Meyer log-rank test. To compare the means of survival (oxidative tolerance assay) or the GFP expression of the Pgcs-1::gfp, gst-4::gfp, skn-1::gfp strains variables were analyzed by one-way ANOVA test. Results are expressed as mean ± standard deviation (SD). Statistical significance was indicated as follows: * p<0.05, ** p<0.001, *** p<0.0001.
Accession numbers
C. elegans proteins/genes: Q17941, Q9XTG7, Q2MGF0, Q17450, O61213, P54145, Q9U3Q6, O02215, G5EC10, Q18198, Q9XUH3, Q9XUF9, O44552, Q27487, Q18938, O17725, O16849, Q968Y9, Q9XVB4, Q9XVA9, Q19223, O62146, G5EGH6, Q8MNR8, Q19774, O02357, Q09321, Q9UAQ9, P91316, Q20770, Q20840, G5EC22, P90893, Q20968, Q21009, Q20117, Q9U2Q9, Q21355, Q9XW45, Q21381, Q09991, Q94269, Q94271, P34528, Q17446, Q2PJ68, P34707, P41977, O02364, Q9XUC0, Q86DA5, Q10038, P90794, Q8WRF1, Q9U309, G5EFR9, Q9GR66, G5ECJ8, Q86S61, O76725, Q9N4X8, Q9NAB1, Q23498, Q23564
Supporting Information
SKN-1 is required for pathogen resistance against both P. aeruginosa and E. faecalis .
(DOC)
PA14-induced activation of Pgcs-1 ::GFP and GST-4::GFP expression.
(DOC)
Suppression of PA14-induced SKN-1 nuclear localization by tir-1(RNAi) .
(DOC)
Lifespan of N2 and skn-1(zu135) mutant worms.
(DOC)
Statistical analysis of killing assays.
(DOC)
List of SKN-1-dependent genes down-regulated by aging.
(DOC)
Statistical analysis of lifespan assays.
(DOC)
Acknowledgments
We thank T. Keith Blackwell, Johji Miwa, Tibor Vellai and the Caenorhabditis Genetics Center for C. elegans strains, T. Keith Blackwell, Andy Golden, Keith P. Choe for RNAi strains and Jonathan J. Ewbank and David W. Wareham for Pseudomonas aeruginosa PA14 and Enterococcus faecalis SdB262 strains, respectively, and Wormbase for collecting and providing data on C. elegans. We are grateful to Attila Mócsai and Gergő Szanda (Semmelweis University) for help in fluorescence/confocal microscopy, Beatrix Gilányi for technical help, Jennifer Tullet and members of the Sőti Group for discussions, Tibor Vellai and Tamás Korcsmáros for critical reading of the manuscript and to the Editor and the anonymous Reviewers for their helpful comments.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the EU (FP6-518230, FP7-200970, TÁMOP-4.2.2/B-10/1-2010-0013), a joint grant of the Hungarian Science Foundation and Norway Grants (NNF-78794), the Hungarian Science Foundation (OTKA K69105 and OTKA-K83314). C.S. is a Bolyai Research Scholar of the Hungarian Academy of Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bolm M, Jansen WT, Schnabel R, Chhatwal GS. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect Immun. 2004;72:1192–1194. doi: 10.1128/IAI.72.2.1192-1194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen WT, Bolm M, Balling R, Chhatwal GS, Schnabel R. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect Immun. 2002;70:5202–5207. doi: 10.1128/IAI.70.9.5202-5207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 4.Chavez V, Mohri-Shiomi A, Garsin DA. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun. 2009;77:4983–4989. doi: 10.1128/IAI.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohri-Shiomi A, Garsin DA. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem. 2008;283:194–201. doi: 10.1074/jbc.M707956200. [DOI] [PubMed] [Google Scholar]
- 6.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 7.Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh V, Aballay A. Regulation of DAF-16-mediated Innate Immunity in Caenorhabditis elegans. J Biol Chem. 2009;284:35580–35587. doi: 10.1074/jbc.M109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 12.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 13.Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213:237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 18.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 21.Hoeven R, McCallum KC, Cruz MR, Garsin DA. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011;7:e1002453. doi: 10.1371/journal.ppat.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DH, Liberati NT, Mizuno T, Inoue H, Hisamoto N, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci U S A. 2004;101:10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 26.Laws TR, Harding SV, Smith MP, Atkins TP, Titball RW. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol Lett. 2004;234:281–287. doi: 10.1016/j.femsle.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Youngman MJ, Rogers ZN, Kim DH. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002082. doi: 10.1371/journal.pgen.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B109–114. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- 33.Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv Enzyme Regul. 2003;43:121–134. doi: 10.1016/s0065-2571(02)00038-9. [DOI] [PubMed] [Google Scholar]
- 34.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, Cha YN, Surh YJ. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res. 2010;690:12–23. doi: 10.1016/j.mrfmmm.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Nagai N, Thimmulappa RK, Cano M, Fujihara M, Izumi-Nagai K, et al. Nrf2 is a critical modulator of the innate immune response in a model of uveitis. Free Radic Biol Med. 2009;47:300–306. doi: 10.1016/j.freeradbiomed.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey CJ, Thimmulappa RK, Sethi S, Kong X, Yarmus L, et al. Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci Transl Med. 2011;3:78ra32. doi: 10.1126/scitranslmed.3002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans EA, Kawli T, Tan MW. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 2008;4:e1000175. doi: 10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 43.Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, et al. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 44.Aw D, Silva AB, Palmer DB. Immunosenescence: emerging challenges for an ageing population. Immunology. 2007;120:435–446. doi: 10.1111/j.1365-2567.2007.02555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alper S. Model systems to the rescue: The relationship between aging and innate immunity. Commun Integr Biol. 2010;3:409–414. doi: 10.4161/cib.3.5.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirkwood TB. Understanding ageing from an evolutionary perspective. J Intern Med. 2008;263:117–127. doi: 10.1111/j.1365-2796.2007.01901.x. [DOI] [PubMed] [Google Scholar]
- 47.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 48.Singh V, Aballay A. Heat shock and genetic activation of HSF-1 enhance immunity to bacteria. Cell Cycle. 2006;5:2443–2446. doi: 10.4161/cc.5.21.3434. [DOI] [PubMed] [Google Scholar]
- 49.Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, et al. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewbank JJ, Zugasti O. C. elegans: model host and tool for antimicrobial drug discovery. Dis Model Mech. 2011;4:300–304. doi: 10.1242/dmm.006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Meth Mol Biol. 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SKN-1 is required for pathogen resistance against both P. aeruginosa and E. faecalis .
(DOC)
PA14-induced activation of Pgcs-1 ::GFP and GST-4::GFP expression.
(DOC)
Suppression of PA14-induced SKN-1 nuclear localization by tir-1(RNAi) .
(DOC)
Lifespan of N2 and skn-1(zu135) mutant worms.
(DOC)
Statistical analysis of killing assays.
(DOC)
List of SKN-1-dependent genes down-regulated by aging.
(DOC)
Statistical analysis of lifespan assays.
(DOC)