Abstract
Catechol-O-methyltransferase (COMT) activity has been reported to be higher in African Americans (AA) than Caucasians (Cau). COMT converts 2- and 4-hydroxy (OH) estrogens to 2- and 4-methoxyestrogens, respectively, and can increase estrogenic milieu locally in tissues. To assess whether the increased incidence of preterm birth (PTB) among AA women is associated with single-nucleotide polymorphism (SNP) in the COMT gene, we examined variations in maternal and fetal COMT genes and their association with pregnancy outcomes (term vs preterm pregnancies) using 4 functional SNPs: rs4633, rs4680, rs4818, and rs6269 in both AA and Cau. We analyzed samples from 267 AA women (191 term and 76 preterm pregnancies) and 339 Cau (194 term and 145 preterm pregnancies) in this study. The results showed a significant difference (P < .05) in allele and genotype frequencies between term and preterm AA and Cau women in 3 SNPs in both maternal and fetal DNA. The analysis revealed that in AA fetal COMT genes, SNP rs4818 is associated with PTB at the allele (C; P < .001), genotype (C/C; P < .01), and 2- (P < .03) and 3 (P < .04)-window haplotype levels. Multidimensionality reduction analysis also showed a significant (P < .01) association between rs4818 and PTB. In conclusion, our study demonstrated that a synonymous polymorphism, rs4818 in the fetal COMT gene, is associated with PTB in AA.
Keywords: preterm birth, African Americans, Caucasians, single-nucleotide polymorphism, catechol-O-methyltransferase
Introduction
Preterm birth ([PTB] <37 weeks of gestation) accounts for 70% of perinatal mortality and 50% of morbidity and is a major clinical dilemma in modern obstetrics.1 In the United States, approximately 12.7% of all births are preterm,2 costing more than $26 billion/year. The incidence of PTB among African American (AA) women is significantly higher than that of other ethnic groups.3 The literature indicates that most socioeconomic parameters and pathophysiological pathways do not explain the ethnic disparity in PTB.4 Novel data from our laboratory have shown that catechol-O-methyltransferase (COMT), which mediates the methylation of hypoestrogenic 2-hydroxyestrogen to hyperestrogenic 2-methoxyestrogen, is high among AA compared with Caucasians (Cau).5 Because estrogens play an important role in labor and delivery, we hypothesize that the genetic variants of the COMT gene that regulate estrogenic activity have a role in the increased incidence of PTB in AA.
COMT is an intracellular enzyme that plays an important role in metabolizing catecholestrogens. COMT expression has been reported in many tissues, including fetal membranes.6–8 Gene expression9 of COMT is controlled by 2 distinct promoters located in Exon 3 of Chromosome 22. The distal 5′ promoter (P2) regulates the synthesis of both membrane-bound (MB-COMT) and soluble COMT (S-COMT) proteins, 10–12 while the P1 promoter regulates the synthesis of S-COMT. In vertebrates, COMT appears mostly in the soluble form, and only a minor fraction is in the particulate form (as MB-COMT).13 COMT converts 2- and 4-hydroxy (OH) estrogens to 2- and 4-methoxy estrogens, respectively. 2-Hydroxyestrogens have a low affinity to the estrogen receptor and therefore have little or no estrogenic effect.14–16 2-Methoxyestrone is one of the most abundant estrogen metabolites in human plasma17 and has a very high affinity for the sex hormone-binding protein.18 2-Methoxyestradiol has been reported to inhibit breast cancer by inhibiting tubulin polymerization.19 It was reported that COMT knockout mice exhibited preeclampsia-like symptoms,20 suggesting that COMT plays an important role during pregnancy. However, the role of COMT in human pregnancy and delivery is not clear.
Variants in the COMT genotype (Val/Val, Val/Met, and Met/Met) have been reported to be associated with various physiological abnormalities. It was recently reported that the mean birth weight of infants may be related to their high active COMT Val/Val genotype.21 Studies conducted in our laboratory to determine the frequency of COMT polymorphisms among ethnically diverse women revealed a significantly higher prevalence of the COMT Val/Val genotype in AA (47%) versus Cau (19%) or Hispanic women (30%).5 In addition, we observed a significant association between the high active Val/Val variation of the COMT gene and a higher prevalence of uterine leiomyoma (fibroids), an estrogen-dependent disorder, in AA women. Therefore, we speculate that functional single-nucleotide polymorphisms (SNPs) contribute to higher COMT activity and higher estrogen milieu in AA, resulting in their higher PTB rate. The objective of this study is to test the association between known functional COMT SNPs and PTB in AA and Cau, using both maternal and fetal DNA samples.
Materials and Methods
Participants
The AA and Cau women of non-Hispanic origin included in the study were recruited at Centennial Women’s Hospital in Nashville, TN, between September 2003 and December 2005. Race was identified by a self-report questionnaire that traced the ancestry of respondents back to 3 generations, starting from the fetus.22 Individuals who had more than 1 ethnic ancestry were excluded from the study. Participants included in the study were between the ages of 18 and 40 years and had singleton live births. Gestational age was determined by last menstrual period and corroborated by ultrasound dating. The participants were recruited at the time of active labor, defined as the presence of regular uterine contractions (2 contractions/10 minutes) with documented cervical changes, followed by delivery. Cases consisted of women who delivered preterm (prior to 370/7 weeks gestation). Participants with multiple gestations, preeclampsia, preterm premature rupture of membranes, placenta previa, fetal anomalies, gestational diabetes, poly- and oligohydramnios, and other complications, such as surgeries during pregnancy, were excluded. The controls were women who had term deliveries (≥370/7 weeks of gestation) with no medical or obstetrical complications during pregnancy. We used noncontiguous gestational ages to define the cases and controls to minimize phenotypic overlap. The study was approved by the TriStar Nashville Institutional Review Board (IRB) at Centennial Medical Center and the Meharry Medical College IRB, Nashville, TN. All participants provided informed consent.
Sample Collection
The samples for the study consisted of 267 AA women (191 controls and 76 cases) and 339 Cau women (194 controls and 145 cases). DNA was extracted from maternal (606) and cord blood (568) samples collected from both controls and cases using the Autopure automated system (Gentra Systems, Minneapolis, Minnesota). DNA samples included Cau (maternal: 145 cases and 194 controls; fetal: 140 cases and 179 controls) and AA (maternal: 76 cases and 191 controls; fetal: 66 cases and 183 controls).
Genotyping and Genetic Analysis
The SNPs selected for our investigation included the commonly used Val/Met sequence variant (SNP; rs4680) and an SNP (rs4633) within exon 3 of the MB-COMT transcript or exon 1 of the S-COMT transcript that defines a synonymous C-T (His/His) change. To have better coverage of the 27-kb genomic region, SNP rs4818, a synonymous C-G (Leu/Leu), and rs6269 from promoter region were also analyzed based on their ability to tag surrounding variants from the HapMap (http://www.hapmap.org). The criterion for Cau tag SNPs was a minor allele frequency (MAF) of 0.07; the criterion for AA was an MAF of 0.20. For both, we used a linkage disequilibrium (LD) cutoff of r2 = .8. The rs numbers of SNPs from the National Institutes of Health (NIH) database, chromosome, and base pair position on chromosome, are shown in Table 1. Genotyping was completed using the Illumina GoldenGate genotyping system (Illumina, San Diego, California).
Table 1.
Single-Nucleotide Polymorphisms (SNPs): ID Numbers (RS Numbera) and Their Location in COMT Gene.
| SNP | Gene | Chromosome | Locationb | Variation | Amino Acid |
|---|---|---|---|---|---|
| RS6269 | COMT | 22 | 18,329,952 | A/G | Promoter region |
| RS4633 | COMT | 22 | 18,330,235 | C/T | His62/His |
| RS4818 | COMT | 22 | 18,331,207 | C/G | Leu136/Leu |
| RS4680 | COMT | 22 | 18,331,271 | A/G | Val158/Met |
Abbreviation: COMT, catechol-O-methyltransferase.
a Nackley et al.32
b Location is from ENSEMBL using Build 36.
Statistical Analysis
The Shapiro–Wilk test of normality was performed for gestational age, gestational and birth weight, Appearance, Pulse, Grimace, Activity, Respiration (APGAR) score at 1 and 5 minutes, and maternal age. All measurements deviated significantly from normality, with the exception of maternal age; as a result, the Mann–Whitney 2-sample rank sum test was used to test for statistical differences between medians of cases and controls. Student t tests were used to test for statistical differences between case and control means for maternal age.
Genetic Analysis
Single-locus allele frequency differences within the Cau and AA cases and controls were analyzed using the software Tools for Population Genetic Analysis (TFPGA), Version 1.3, available at http://www.marksgeneticsoftware.net/index.html.23 Differences between the cases and controls in genotype distributions at single loci were assessed with the Metropolis algorithm to obtain P values, as implemented in the program R X C.24 Single-locus Hardy–Weinberg equilibrium (HWE) analyses were also performed using TFPGA software. Statistical significance for the above was determined using Fisher exact tests.
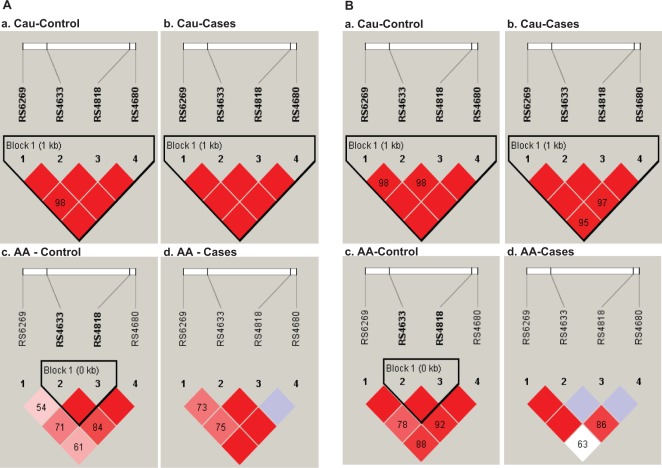
Haplotype frequencies from the genotype data and differences in case–control haplotype distributions were tested using the FastEH+ program, which uses the expectation maximization (EM) algorithm.25 Significant deviations were detected in random haplotypes using empirically derived P values with this software. Haplotype blocks were assigned using D′ confidence interval algorithm.26 The number below the lines between blocks is the multiallelic D′ between blocks (Figure 1). This gives a measure of LD and indicates the D′ statistic for 2 consecutive SNPs. D′ = 1 indicates complete LD and decreases as the value of D′ decreases. Regions with high LD (D′ = 1 and log of the odds [LOD] > 2) as well as SNPs in the strong LD blocks are shown in red, with the intensity decreasing with decreased D value. Regions with weak LD (0.21 < D′ < 1 and LOD > 2) are shown in blue, and regions with no LD and LOD scores (LOD < 2) are shown in white. Pairwise measures of D′ and r 2 were performed with HaploView software to assess LD.
Figure 1.
HaploView-generated LD map of 4 SNPs within the COMT gene. Haplotype blocks are assigned using the D′ confidence interval algorithm created by Gabriel et al26. D' = 1 indicates complete LD and decreases as the value of D′ decreases. Regions with high LD (D′ = 1 and LOD > 2) as well as SNPs in the strong LD blocks are shown in red. Decrease in intensity suggests decreased D value and LD. Regions with weak LD (0.21 < D′ < 1 and LOD > 2) are shown in blue. Regions with no LD (D′ < 1 and LOD < 2) are shown in white. A, Linkage disequilibrium (LD) maps in maternal samples: (a) Cau: control*; (b) Cau: cases**; (c) AA: control; (d) AA: cases. B, Linkage disequilibrium (LD) map in fetal samples: (a) Cau: control; (b) Cau: cases; (c) AA: control; (d) AA: cases. SNP, single-nucleotide polymorphism; COMT, catechol-O-methyltransferase; LOD, log of the odds; Cau, Caucasians; AA, African Americans; *term delivery; **preterm delivery.
Multilocus Analysis
Multilocus analysis was performed using the multifactor dimensionality reduction (MDR) method.27,28 Multifactor dimensionality reduction collapses all the genetic data into 2 categories, high and low risk, by comparing all single-locus and all multilocus combinations and then categorizing each genotype into either high risk or low risk based on the ratio of cases to controls that have that genotype. Multifactor dimensionality reduction ultimately selects 1 genetic model, either single- or multilocus, which most successfully predicts phenotype or disease status. The prediction error of the model is estimated using 10-fold cross-validation (CV). Simply put, the data are divided into 10 equal parts, and 9 of the 10 data are used to generate the model. The model is then tested on the remaining 1 of the 10 data. This method is performed for each of the 9 of 10 and 1 of 10 combinations. The 10-fold CV is repeated 10 times to ensure that the results are not due to chance divisions of the data. The average number of times that the same best model comes up is given as the CV consistency and is represented as a continuous value from 1 to 10. Both the CV consistency and the prediction error minimization are used to choose the single best model. Statistical significance is determined empirically by permuting the case and control labels 1000 times. We eliminated the problem of multiple testing by generating the P values with permutations.
Multifactor dimensionality reduction is best suited for equal number of cases and controls. Because of the excess of controls in our sample, both undersampling of the control group and oversampling of the case group were implemented with the MDR open source software, available at www.epistasis.org. These methods have been shown to be effective in determining the best genetic model using MDR.29,30 Results for the 2 resampling methods were compared for consistency. Odds ratios, sensitivity, and specificity were calculated using the multilocus model determined from the MDR analysis, as implemented in the software.
Results
As expected, the cases and controls differed in gestational age, birth weight, and APGAR scores in both ethnic groups (Table 2). There were no significant differences in maternal age and socioeconomic factors (education, annual income, and smoking) between the cases and controls (data not shown).
Table 2.
Maternal Characteristics and Pregnancy Outcomes in Women That Delivered Preterm (Cases) and Term (Controls)
| Characteristics | Caucasians | African Americans | ||||
|---|---|---|---|---|---|---|
| Controls | Cases | 2-Tailed P Value | Controls | Cases | 2-Tailed P Value | |
| Gestational age, weeks, mean (SD) | 38.9 (1.0) | 32.9 (3.1) | <.001 | 39.0 (1.0) | 32.3 (3.9) | <.001 |
| Gestational weight, g | 3446 (2100–4661) | 2150 (370–3790) | <.001 | 3190 (1952–4517) | 2240 (462–3782) | <.001 |
| Birth weight, g, mean (SD) | 3407 (427) | 2026 (719) | <.001 | 3226 (436) | 2094 (642) | <.001 |
| APGAR 1 | 8 (4–9) | 8 (1–9) | <.001 | 8 (3–9) | 8 (1–9) | <.001 |
| APGAR 5 | 9 (7–10) | 9 (1–9) | <.001 | 9 (7–10) | 9 (6–10) | <.001 |
Abbreviation: SD, standard deviation; APGAR 1 and 5, Appearance, Pulse, Grimace, Activity, Respiration at first and five minutes respectively.
Hardy Weinberg equilibrium (HWE) Analysis
All SNPs in the maternal and fetal samples obtained from both the Cau and AA cases and controls were used in HWE (Table 3). The data were analyzed using PowerMarker software,31 and statistical significance for these analyses was determined using Fisher exact test.
Table 3.
Comparison For Hardy–Weinberg Equilibrium in COMT Gene in Caucasian and African American Women That Delivered Preterm (Cases) and Term (Controls)a
| Caucasians | African Americans | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | MA | Cases | Controls | Cases | Controls | ||||
| n | MAF | n | MAF | n | MAF | n | MAF | ||
| Maternal COMT gene | |||||||||
| RS6269 | G | 68 | 0.456 | 188 | 0.414 | 41 | 0.317 | 223 | 0.346 |
| RS4633 | T | 68 | 0.455 | 188 | 0.489 | 41 | 0.375 | 223 | 0.346 |
| RS4818 | G | 68 | 0.463 | 188 | 0.411 | 41 | 0.171 | 223 | 0.220 |
| RS4680 | A | 68 | 0.455 | 188 | 0.487 | 41 | 0.293 | 223 | 0.287 |
| Fetal COMT gene | |||||||||
| RS6269 | G | 70 | 0.343 | 196 | 0.408 | 35 | 0.286 | 219 | 0.393 |
| RS4633 | C | 70 | 0.443 | 197 | 0.500 | 35 | 0.371 | 217 | 0.300 |
| RS4818 | G | 70 | 0.314 | 193 | 0.391 | 35 | 0.086 | 221 | 0.251 |
| RS4680 | G | 70 | 0.450 | 195 | 0.490 | 35 | 0.357 | 217 | 0.288 |
Abbreviations: COMT, catechol-O-methyltransferase; SNP, single-nucleotide polymorphism; MA, minor allele; MAF, minor allele frequency.
aSignificance by Fisher exact test.
Allele and Genotype Frequencies
A total of 16 comparisons were made (4 SNPs and 4 different groups: maternal controls and cases as well as fetal controls and cases) between races for both allele and genotype frequencies (Table 4). Significant single SNP differences were observed for allele frequencies in maternal controls between races at rs6269 (A; P < .05) and rs4633 (C; P = .001), and rs4818 (C; P < .001) and rs4680 (A; P = .001). Similarly, allele frequencies in the maternal cases were significantly different at SNPs rs4818 (P < .001), rs6269 (P = .03), rs4633 (P = .001), and rs4680 (P = .001). Allele frequencies at SNP rs6269 in the fetal controls and cases did not show any difference; however, the frequencies at the remaining 3 SNPs were significantly different (P ≤ .001).
Table 4.
Allele and Genotype Frequency Differences Between Caucasian and African Americansa
| P Value | |||||
|---|---|---|---|---|---|
| SNP | Allele/Genotype | Maternal | Fetal | ||
| Cases | Control | Cases | Control | ||
| Allele | |||||
| RS6269 | A | .03 | .04 | .34 | .65 |
| RS4633 | C | .25 | .001 | .01 | <.001 |
| RS4818 | C | <.001 | <.001 | <.001 | <.001 |
| RS4680 | A | .02 | .001 | .005 | <.001 |
| Genotype | |||||
| RS6269 | A/A | .06 | .14 | .57 | .11 |
| RS4633 | C/C | .06 | <.001 | .03 | .001 |
| RS4818 | C/C | < .001 | .001 | .012 | <.001 |
| RS4680 | A/A | .04 | .001 | .052 | .001 |
Abbreviation: SNP, single-nucleotide polymorphism.
a P values are derived using PowerMarker software (http://statgen.ncsu.edu/powermarker/).
Genotype frequencies in the maternal controls showed significant (P ≤ .001) differences at rs4818(C/C), rs4633 (C/C), and rs4680 (A/A) but not at rs6269 (A/A). In the maternal cases, genotype frequencies showed differences at rs4818 (C/C; P < .001) and rs4680 (A/A; P < .04) but not at rs4633(C/C) or rs6269 (A/A). In the fetal samples, genotype frequencies showed significant differences in both the controls and cases at rs4818, rs4633, and rs4680 but not at site rs6269 (Table 4).
Association Analysis of Individual SNPs
Allele association
Single-locus analysis of SNP rs4818 in AA fetal COMT DNA revealed a significant association (P = .001) between allele C and PTB, with odds ratio (OR) 3.58 (95% confidence interval [CI]: 1.51-8.49; Table 5). However, no such association was seen between rs4818 and PTB in fetal COMT DNA obtained from Cau. None of the alleles in the other 3 SNPs (rs6269, rs4633, and rs4680) showed any association with PTB either in Cau or in AA fetal DNA. None of the SNPs analyzed in the maternal DNA of either the Cau or AA women showed any association with PTB (Table 5).
Table 5.
Allele Association of SNPs Within COMT Gene With Preterm Birth in Caucasians and African Americans
| SNP | Allele | Maternal | P Value | Fetal | P Valuea |
|---|---|---|---|---|---|
| Odds Ratio (95% CI) | Odds Ratio (95% CI) | ||||
| Caucasians | |||||
| RS6269 | A | 0.84 (0.57,1.25) | .36 | 0.76 (0.51,1.13) | .17 |
| G | |||||
| RS4633 | C | 1.15 (0.77,1.70) | .45 | 0.79 (0.54,1.17) | .28 |
| T | |||||
| RS4818 | C | 0.81 (0.54,1.21) | .29 | 1.40 (0.93,2.12) | .11 |
| G | |||||
| RS4680 | A | 0.88 (0.59,1.31) | .53 | 1.27 (0.86,1.88) | .18 |
| G | |||||
| African Americans | |||||
| RS6269 | A | 1.14 (0.69,1.89) | .69 | 1.62 (0.93,2.81) | .10 |
| G | |||||
| RS4633 | C | 0.88 (0.54,1.45) | .52 | 0.72 (0.43,1.23) | .27 |
| T | |||||
| RS4818 | C | 1.37 (0.74,2.53) | .38 | 3.58 (1.51,8.49) | <.001b |
| G | |||||
| RS4680 | A | 1.03 (0.61,1.73) | .89 | 1.37 (0.80,2.34) | .21 |
| G | |||||
Abbreviations: COMT, catechol-O-methyltransferase; SNP, single-nulceotide polymorphism; CI, confidence interval.
a P value obtained by Fisher exact test.
b Single-nucleotide polymorphism NP rs4818 shows a significant association with preterm birth in African Americans.
Genotype association
Genotype analysis showed a strong association (P = .01) between the C/C genotype of SNP rs4818 and PTB (Table 6) in AA fetal COMT DNA. There was no association between the other SNPs and PTB in fetal DNA. None of the COMT SNPs studied showed associations with PTB in AA maternal DNA. In Cau, we did not see association between any of these SNPs and PTB either in maternal or in fetal DNA.
Table 6.
Genotype Association of SNPs Within COMT Gene With Preterm Birth in Caucasians and African Americansa
| SNP | Genotype | Maternal | Frequency | ||||
|---|---|---|---|---|---|---|---|
| Cases | Controls | P Value | Cases | Controls | P Value | ||
| Caucasians | |||||||
| RS6269 | A/A | 0.26 | 0.36 | .29 | 0.44 | 0.33 | .22 |
| RS4633 | C/C | 0.28 | 0.27 | .57 | 0.21 | 0.23 | .27 |
| RS4818 | C/C | 0.25 | 0.35 | .32 | 0.49 | 0.34 | .08 |
| RS4680 | A/A | 0.19 | 0.25 | .65 | 0.33 | 0.23 | .29 |
| African Americans | |||||||
| RS6269 | A/A | 0.48 | 0.42 | .72 | 0.48 | 0.39 | .16 |
| RS4633 | C/C | 0.47 | 0.46 | .50 | 0.37 | 0.49 | .39 |
| RS4818 | C/C | 0.73 | 0.60 | .08 | 0.83 | 0.57 | .01a |
| RS4680 | A/A | 0.12 | 0.08 | .65 | 0.14 | 0.08 | .40 |
Abbreviations: COMT, catechol-O-methyltransferase; SNP, single-nucleotide polymorphism.
a P values from Fisher exact test.
b Significant association with preterm birth in African Americans.
Haplotype Analysis
Haplotype-based association analysis was performed for all 3 possible SNP combinations within COMT, using PowerMarker statistical software. Separate haplotype analyses were performed on the maternal and the fetal DNA samples obtained from Cau and AA to study the descent pattern of a combination of these alleles at different sites on a single chromosome. In 2-marker sliding window haplotype analysis, rs4633-rs4818 and rs4818-rs4680 in the fetal DNA of AA showed a significant association with PTB (P = .03; Table 7). No significant haplotype associations were found in AA maternal samples or Cau (maternal and fetal) samples (Table 7). In 3-marker sliding window haplotype analysis, haplotype rs4680-rs4818-rs4633 in fetal DNA showed a significant (P = .04) association with PTB (Table 7). However, in 4-marker sliding window haplotype analysis, no association was observed with PTB (Table 7).
Table 7.
Association of COMT Haplotype With Preterm Birth in African Americans
| SNPs | Maternal | Fetal | ||||
|---|---|---|---|---|---|---|
| Haplotype | Frequency | P Value | Haplotype | Frequency | P Value | |
| Two-marker sliding window haplotype analysisa | ||||||
| RS6269-RS4633 | G-T | 0.051 | .65 | G-T | 0.37 | .42 |
| RS4633-RS4818 | T-C | 0.35 | .75 | T-C | 0.30 | <.03 |
| RS4818-RS4680 | G-G | 0.21 | .79 | C-A | 0.29 | <.03 |
| Three-marker sliding window haplotype analysis | ||||||
| RS6269-RS4633-RS4818 | A-T-C | 0.30 | .63 | A-T-C | 0.30 | .10 |
| RS4680-RS4818-RS4633 | T-C-G | 0.08 | .23 | C-C-G | 0.44 | <.04 |
| Four-marker sliding window haplotype analysis | ||||||
| RS6269-RS4633-RS4818-RS4680 | G-C-G-G | 0.17 | .41 | A-T-C-A | 0.28 | .14 |
Abbreviations: COMT, catechol-O-methyltransferase; SNP, single-nucleotide polymorphism.
a PowerMarker and HaploView statistical softwares were used.
Linkage disequilibrium Characterization
Analysis showed that all 4 SNPs (rs4633, rs4680, rs4818, and rs6269) had D′ values of 1.0 and LOD > 2 in Cau maternal (Figure 1A; Panels a and b) and fetal samples (Figure 1B; Panels a and b) in both the controls and cases (shown in red). African American (AA) maternal (Figure 1A; Panel c) and fetal control (Figure 1B; Panel c) samples showed a single haplotype block of 1.0 kb, encompassing rs4818 and rs4633 with 0.21 < D′ < 1 and LOD > 2. In the AA cases, no haplotype block was observed and the D′ values (LOD < 2) were low (maternal: Figure 1A, Panel d; fetal: Figure 1B, Panel d).
Multifactor Dimensionality Reduction Analysis
Multilocus analysis performed in fetal DNA obtained from AA showed a significant association (Table 8) between rs4818 and PTB (P = .01). The test showed a CV consistency of 10/10. However, we did not see any significant association between SNPs and PTB, either in AA maternal samples or in Cau maternal or fetal samples.
Table 8.
Multifactor Dimensionality Reduction Analysis in African American Fetal Samples
| Model | Permutation P Value | CV Consistency |
|---|---|---|
| RS4818 | .01 | 10/10 |
Abbreviation: CV, cross-validation.
Discussion
To explain the high COMT activity in AA compared with Cau, we examined genetic variations in maternal and fetal DNA and their association with pregnancy outcome (case vs control). In this study, we demonstrated significant differences in the frequency of SNPs in both the maternal and the fetal DNA of cases and controls and the between-race differences in SNPs’ association with PTB. We investigated 4 SNPs (rs4633, rs4680, rs4818, and rs6269) in the COMT gene, based on the prior knowledge that these SNPs affect COMT enzyme activity.32 Because SNPs rs4633, rs4680, and rs4818 showed significant differences in allele and genotype frequencies in the fetal samples of cases and controls, we considered them for association analysis. Our results showed that SNP rs4818 in the fetal COMT gene is strongly associated with PTB in AA. However, in Cau, no such association was found with PTB in fetal or maternal DNA. This study demonstrated that a synonymous polymorphism, rs4818 in the COMT gene, is associated with PTB in AA fetal DNA at the allele and genotype levels.
Our study revealed that rs4818 in the COMT gene is associated with PTB in AA fetal DNA at the allele and genotype levels. Allele or genotype frequency differences in rs6269 and rs4633 were not significantly different in the maternal cases and controls. Therefore, we did not expect any association with PTB at the allele and genotype levels for the maternal data. The COMT30 is reported to create an 18-fold difference in expressed enzyme activity by modifying the secondary structure of messenger RNA (mRNA), which affects the mRNA stability.32 This is the first study in which mutation from C to G, a synonymous polymorphism (Leu/Leu) within the COMT gene that does not alter the structure of the COMT protein but impacts expression, is linked to a specific reproductive disorder, such as PTB. However, SNP rs4818 did not show any significant association with PTB in maternal DNA in either race or in Cau fetal DNA.
After the onset of spontaneous labor, there is an increase in the E2/P4 ratio in amniotic fluid, probably because of alterations in the steroidogenic activity of the fetal membranes.33 The elevated COMT expression observed in the amnion of laboring participants and the inhibition of prostaglandin E2 (PGE2) by a specific COMT inhibitor in the amnion explants8 suggests that COMT secreted by fetal tissues increases the local estrogenic milieu and plays a role in labor and delivery. Results from our study suggest that SNP rs4818 in the fetal COMT gene is associated with PTB in AA and may explain the increased COMT activity in the AA cases. Data analysis revealed that the C allele at rs4818 in fetal COMT DNA showed a significant association with PTB. We also showed that this association was seen at genotype C/C and even at the haplotype (2- and 3-marker sliding window), suggesting that rs4818 in fetal COMT DNA is strongly associated with PTB. A study conducted by Roussos et al34 suggests that a synonymous polymorphism within the COMT gene rs4818 accounts for greater variation in COMT activity compared with the functional Val158Met polymorphism. Their study revealed that variants in rs4818 impart strong and differential effects on prefrontal cortex functions. Elevated COMT expression in the amnion of laboring women and PGE2 inhibition in amnion explant cultures treated with COMT-specific inhibitor in our laboratory suggest that COMT secreted by fetal membranes, particularly the amnion, regulates the onset of labor in term pregnancy.8 Thus, polymorphism in the COMT gene in fetal DNA can at least partially explain the higher incidence of PTB in AA.
The haplotype analysis documented the strong influence of rs4818 in AA fetal DNA and its association with PTB. Because the allele and genotype frequencies of rs6269 were not significantly different in the fetal DNA of the controls and cases, we did not expect any involvement of this SNP in PTB. The pairwise LD values for the 4 SNPs within the COMT gene were high in Cau, indicating a high degree of LD; these values were not different between the controls and cases for both maternal and fetal samples. As expected, in AA maternal and fetal control samples, the LD was not as dominant as it was in Cau. However, a single haplotype block of 1.0 kb, encompassing rs4818 and rs4633, was seen in the maternal and fetal controls, In AA maternal cases, LD values were lower than the controls and in the fetal cases where SNP rs4818 showed association with PTB no LD (white block) was evident. However, more data are required to confirm the findings. A CV consistency value of 10/10 and a P value of .01 after multilocus analysis (MDR) for rs4818 in fetal COMT DNA suggest the single strongest effect of rs4818, as no interactions were observed with any of the other SNPs studied. These results suggest that the SNP at rs4818 is strongly associated with PTB in AA.
In summary, our results provide evidence of the role of fetal COMT gene variant rs4818 in the incidence of PTB in AA. However, no such association was observed with either AA maternal samples or Cau maternal or fetal samples. Further investigations with a larger population are needed to determine whether COMT activity differs in early, mid, late, and recurrent PTBs. Additional studies are needed to understand the physiological role of COMT in the incidence of PTB.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The authors disclosed receipt of the following financial support for the, authorship, and/or publication of this article: Regional Centers for Minority Institution 5G12 RR003032-27.
References
- 1. Hamilton BE, Martin JA, Ventura SJ. Births: preliminary data for 2005. Natl Vital Stat Rep. 2006;55(11):1–18 [PubMed] [Google Scholar]
- 2. Martin JA, Hamilton BE, Sutton PD, Ventura SJ, Menacker F, Kirmeyer S. Births: final data for 2004. Natl Vital Stat Rep. 2006;55(1):1–101 [PubMed] [Google Scholar]
- 3. Creasy RK. Preterm birth prevention: where are we? Am. J Obstet Gynecol. 1993;168(4):1223–1230 [DOI] [PubMed] [Google Scholar]
- 4. Goldenberg RL, Cliver SP, Mulvihill FX, et al. Medical, psychosocial, and behavioral risk factors do not explain the increased risk for low birth weight among black women. Am J Obstet Gynecol. 1996;175(5):1317–1324 [DOI] [PubMed] [Google Scholar]
- 5. Al-Hendy A, Salama SA. Catechol-O-methyltransferase polymorphism is associated with increased uterine leiomyoma risk in different ethnic groups. J Soc Gynecol Investig. 2006;13(2):136–144 [DOI] [PubMed] [Google Scholar]
- 6. Ding Y-S, Gatley SJ, Fowler JS, Chen R, Volkow ND. Mapping catechol-O-methyltransferase in vivo: initial studies with [18F] Ro41-0960. Life Sci. 1996;58(3):195–208 [DOI] [PubMed] [Google Scholar]
- 7. Wentz MJ, Shi SQ, Shi L, et al. Treatment with an inhibitor of catechol-O-methyltransferase activity reduces preterm birth and impedes cervical resistance to stretch in pregnant rats. Reproduction. 2007;134(6):831–839 [DOI] [PubMed] [Google Scholar]
- 8. Harirah H, Thota C, Wentz MJ, Zaman W, Al-Hendy A. Elevated expression of catechol-O-methyltransferase is associated with labor and increased prostaglandin E(2) production by human fetal membranes. Am J Obstet Gynecol. 2009;201(5):496.e1-496.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salminen M, Lundström K, Tilgmann C, Savolainen R, Kalkkinen N, Ulmanen I. Molecular cloning and characterization of rat liver catechol-O-methyltransferase. Gene. 1990;93(2):241–247 [DOI] [PubMed] [Google Scholar]
- 10. Tenhunen J, Ulmanen I. Production of rat soluble and membrane-bound catechol-O-methyltransferase forms from bifunctional mRNAs. Biochem J. 1993;296(pt 3):595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tenhunen J, Salminen M, Jalanko A, Ukkonen S, Ulmanen I. Structure of the rat catechol-O-methyltransferase gene: separate promoters are used to produce messenger RNAs for soluble and membrane-bound forms of the enzyme. DNA Cell Biol. 1993;12(3):253–263 [DOI] [PubMed] [Google Scholar]
- 12. Tenhunen J, Salminen M, Lundström K, Kiviluoto T, Savolainen R, Ulmanen I. Genomic organization of the human catechol O-methyltransferase gene and its expression from two distinct promoters. Eur J Biochem. 1994;223(3):104 9-1059. [DOI] [PubMed] [Google Scholar]
- 13. Guldberg HC, Marsden CA. Catechol-O-methyltransferase: pharmacological aspects and physiological role. Pharmacol Rev. 1975;27(2):135–206 [PubMed] [Google Scholar]
- 14. MacLusky NJ, Riskalla M, Krey L, Parvizi N, Niftolin F. Anovulation in female rats induced by neonatal administration of the catechol estrogens, 2-hydroxy-estradiol and 4-hydroxy-estradiol. Neuroendocrinology. 1983;37(5):321–327 [DOI] [PubMed] [Google Scholar]
- 15. Van Aswegen CH, Purdy RH, Wittliff JL. Binding of 2-hydroxyestradiol and 4-hydroxyestridiol to estrogen receptor human breast cancers. J Steroid Biochem. 1989;32(4):485–492 [DOI] [PubMed] [Google Scholar]
- 16. Feigelson HS, Henderson BE. Estrogens and breast cancer. Carcinogenesis. 1996;17(11):2279–2284 [DOI] [PubMed] [Google Scholar]
- 17. Kraychy S, Gallagher TF. 2-Methoxyestrone, a new metabolite of estradiol-17 β in man. J Biol Chem. 1957;229(1):519–526 [PubMed] [Google Scholar]
- 18. Dunn JF. Transport of estrogens in human plasma. In: Merriem GR, Lipsett MB, eds. Catechol Estrogens. New York, NY: Raven Press; 1983:167–176 [Google Scholar]
- 19. Klauber N, Parangi S, Flynn E, Hamel E, D’Amato RJ. Inhibition of angiogenesis and breast cancer in mice by microtubule inhibitors 2-methoxyestradiol and taxol. Cancer Res. 1997;57:81–86 [PubMed] [Google Scholar]
- 20. Kanasaki K, Palmsten K, Sugimoto H, et al. Deficiency in catechol-O-methyltransferase and 2-methoxyoestradiol is associated with pre-eclampsia. Nature. 2008;453(7198):1117–1121 [DOI] [PubMed] [Google Scholar]
- 21. Thapar A, Langley K, Fowler T, et al. Catechol-O-methyltransferase gene variant and birth weight predict early onset antisocial behavior in children with attention deficit/hyperactivity disorder. Arch Gen Psychiatry. 2005;62(11):1275–1278 [DOI] [PubMed] [Google Scholar]
- 22. Menon R, Velez DR, Thorsen P, et al. Ethnic differences in key candidate genes for spontaneous preterm birth: TNF-alpha and its receptors. Hum Hered. 2006;62(2):107–118 [DOI] [PubMed] [Google Scholar]
- 23. Zaykin DV, Westfall PH, Young SS, et al. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53(2):79–91 [DOI] [PubMed] [Google Scholar]
- 24. Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49(6):1280–1283 [DOI] [PubMed] [Google Scholar]
- 25. Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–157 [DOI] [PubMed] [Google Scholar]
- 26. Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229 [DOI] [PubMed] [Google Scholar]
- 27. Weiss GM, Provost F. Learning when training data are costly: the effect of class distribution on tree induction. J Artif Intell Res. 2003;19(1):315–354 [Google Scholar]
- 28. Japkowicz N, Stephen S. The class imbalance problem: a systematic study. Intell Data Anal. 2002;6(5):429–449 [Google Scholar]
- 29. Fleiss JL. Statistical Methods for Rates and Proportions. New York: Wiley; 1981 [Google Scholar]
- 30. Diatchenko L, Slade GD, Nackley AG, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14 ( 1 ):135–143 [DOI] [PubMed] [Google Scholar]
- 31. Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129 [DOI] [PubMed] [Google Scholar]
- 32. Nackley AG, Shabalina SA, Tchivileva IE, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314(5807):1930–1933 [DOI] [PubMed] [Google Scholar]
- 33. Keelan JA, Coleman M. The molecular mechanisms of term and preterm labor: recent progress and clinical implications. Clin Obstet Gynecol. 1997;40(3):460–478 [DOI] [PubMed] [Google Scholar]
- 34. Roussos P, Giakoumaki SG, Pavlakis S, Bitsios P. Planning, decision making and the COMT rs4818 polymorphism in healthy males. Neuropsychologia. 2008;46(2):757–763 [DOI] [PubMed] [Google Scholar]