Abstract
Maternal undernutrition results in offspring nephron number reduction and hypertension that are hypothesized to begin as compensatory changes in fetal gene expression during gestation. To evaluate mechanisms of dysregulated nephrogenesis, pregnant Sprague Dawley rats were 50% food restricted from embryonic day (E) 10 to E20. At E20, fetal male kidneys were examined by microarray analysis. A total of 476 differentially expressed transcripts were detected including those regulating development and differentiation, mitosis and cell cycle, chromatin assembly, and steroid hormone regulation. Differentially regulated genes were detected in MAPK/ERK, Wnt, and Notch signaling pathways. Validation of the microarray results was performed for the Notch signaling pathway, an important pathway in nephron formation. Protein expression of Notch pathway factors by Western blotting showed significantly decreased Notch2 and downstream effector Hey1 protein expression, while Ctbp1 co-repressor was increased. These data together show that maternal undernutrition results in developmental disruption in fetal nephrogenesis gene expression signaling.
Keywords: kidney, transcriptional profiling, gene expression, intrauterine growth, nephron number
Introduction
As first proposed by Barker,1,2 gestational programming is a process in which stressors during a critical point in gestation impact fetal development resulting in life-long consequences. Poor maternal nutrition is a known stressor resulting in low–birth-weight offspring at increased risk of developing metabolic syndrome (obesity, type II diabetes, hypertension, cardiovascular disease) in humans and animal models.3–5 The kidney is a key at-risk organ for gestational programming due to preferential shunting of nutrients to more critical organs such as the brain.6 Growth restricted offspring of undernourished mothers demonstrate significantly reduced nephron numbers with nephron deficits ranging from 13% to 41% in animal models.7–9 This decrease in nephron number is associated with hypertension and reduced renal function.10–12 In humans, epidemiological studies have shown associations of low nephron numbers with adult hypertension, and of low-birth-weight with chronic kidney disease.13–17 The mechanisms for nephropenia in the maternal undernourished model currently are unknown but may include effects on gene expression regulating ureteric branching or nephron maturation, nephron anatomy, ion transport, apoptosis, glucocorticoid responsiveness, and epigenetic modulation.11,18–22
Fetal nephrogenesis is regulated by spatial and temporal transcription/growth factors, which are potential candidates for mediation of gestational programming. Critical signaling pathways and events such as ureteric bud branching, apoptosis, cell survival, epithelial to mesenchymal transformation, and vascularization occur during kidney development and may be disrupted by maternal undernutrition (MUN). We have used an established rat model of MUN-induced intrauterine growth restriction (IUGR) in which the dam is 50% food restricted during the second half of gestation. This model results in MUN offspring with significantly decreased glomerular number at 3 weeks and blood pressure elevation at 12 weeks of age.23 At embryonic day 20 (E20), MUN fetal kidneys demonstrate downregulation of Gdnf and its downstream ureteric bud branching factors Wnt4 and Wnt11, as well as dysregulation of Wt1 and Pax2, factors that regulate the mitogenic activity of Gdnf.20 To more fully evaluate mechanisms of disrupted development in the MUN offspring late fetal kidney, we subjected MUN E20 kidneys to DNA microarray analysis and categorized known genes into ontological groups and signaling pathways. Among the signaling pathways showing dysregulation, Notch signaling was prominent and is well-known to be involved in development of kidney nephrons.24,25 Therefore, we validated the differential regulation of MUN on this important signaling pathway.
Materials and Methods
Maternal Rat Diets
Studies were approved by the Animal Care and Use Committee of the Los Angeles Biomedical Research Institute at Harbor–University of California, Los Angeles, and were in accordance with the American Association for Accreditation of Laboratory Care and National Institutes of Health guidelines. A model of 50% undernutrition (MUN) pregnant rat dams was used. First-time pregnant Sprague Dawley rats (Charles River Laboratories, Hollister, California) were housed in a facility with constant temperature and humidity and a controlled 12:12-hour light/dark cycle. At 10 days of gestation, rats were provided either an ad libitum diet of standard laboratory chow (Lab Diet 5001, Brentwood, MO; protein, 23%; fat, 4.5%; metabolizable energy, 3030 kcal/kg) or a 50% diet (MUN) determined by quantification of normal intake in the ad libitum-fed rats. The respective diets were given from 10 days of pregnancy until gestational day 20 (E20).
Fetal kidney removal
The dams (4 control (C) and 4 MUN) were anesthetized with isoflurane and E20 fetuses quickly removed and placed on ice cold dishes. Litter sizes ranged from 12 to 16. Fetuses from dams with less than 12 or more than 16 fetuses were not used in order to avoid any possible effects of over or undernutrition related to litter size. Fetuses were dissected, and intact kidneys and a portion of liver were removed and flash frozen in liquid nitrogen. Each kidney used represented a fetus from a specific dam; therefore, 4 control and 4 MUN kidneys represents 4 individual control dams compared to 4 individual MUN dams. At the time of tissue harvesting, fetal sex was assessed based on the presence or absence of visible testes or uterine horns. To confirm fetal sex, polymerized chain reaction (PCR) analysis was performed to identify the presence or absence of the SRY Y chromosome gene. Frozen liver from each fetus (20 mg) was homogenized and DNA extracted using the DNazol kit (Invitrogen, Carlsbad, California). Polymerized chain reaction conditions were identical to a previously published report.26
RNA Isolation, cDNA Synthesis, Hybridization, and Gene Chip Analysis
Male kidneys were shipped on dry ice to NeuroInDx Inc, Signal Hill, California, for processing RNA quality assessment, cRNA synthesis, and determination of gene expression data for each RNA sample using the Agilent microarray workstation and accompanying feature extraction software. Total RNA was isolated from tissue using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. One kidney from each male fetus was homogenized in 1 mL Trizol Reagent. Male kidneys were chosen versus females or unsexed kidneys based on previous reports that male kidneys are generally more susceptible to the effects of IUGR than female kidneys. Final RNA was resuspended in 100 µL DEPC-treated water and stored at –80°C until use.8–13 RNA quality was checked using an Agilent Bioanalyser 2100 system and demonstrated excellent quality based on chromatographs and A260/280 ratios. Complementary RNA was synthesized and biotin-labeled using the Agilent Direct-Label cDNA Synthesis Kit according to the manufacturer's instructions. Total RNA was used for first and second strand cDNA synthesis followed by an in vitro transcription to amplify cy3 and cy5 labeled cRNA. The cRNA was quality-checked and then hybridized to the rat Agilent Arrays. Following hybridization, washing, and scanning, the resulting data were globally normalized and genes demonstrating a significant difference of P < .01 by paired Student t-test were further evaluated for fold expression using Genesifter software (Geospiza, Seattle, WA).
Ontology and pathway analysis
Genes that were upregulated or downregulated by 1.5-fold or greater were considered significantly differentially expressed. These genes then were assigned to specific ontological groups using Database for Annotation, Visualization, and Integrated Discovery (DAVID)27,28 and to specific pathways using Kyoto Encyclopedia of Genes and Genome (KEGG) pathways.29
Western Blot
The primary and secondary antibodies were NOTCH2 (Santa Cruz SC-5545), primary 1:1000, secondary 1:5000; CTBP1 (Santa Cruz SC-5963) primary 1:1000, secondary 1:10,000; RPBJK (Santa Cruz SC-55019), primary 1:1000, secondary 1:5,000; SKIP (Santa Cruz SC-30139), primary 1:1000, secondary 1:10,000; MAML1 (Millipore AB15546), primary 1:1000, secondary 1:10,000; HEY1 (Santa Cruz SC-28746), primary 1:1000, secondary 1:5,000; and B-actin (Sigma A5441): primary 1:10,000, secondary 1:10,000. Secondary horseradish peroxidase conjugated antibodies were anti-rabbit (Bio-Rad 170-6515) and anti-mouse (Bio-Rad 170-6516). All commercial antibodies were optimized for binding specificity and bands depicted have the expected molecular weights.
Protein extraction
Protein was extracted in radioimmuno precipitation assay (RIPA) buffer that contained protease inhibitors (HALT cocktail, Pierce) by homogenization of frozen kidneys on ice. Supernatants were clarified by 20-minute micro centrifugation at 12 000g. Supernatant protein concentration was determined by bicinchoninic acid (BCA) solution (PIERCE, Rockford, Illinois). Supernatants were frozen at −80°C until use.
Western blot
Protein expression was analyzed as previously described and each blot was performed at least twice.30 A total of 5 to 6 samples were run for C and MUN. Equal amounts of protein (25 μg) were mixed with Criterion sodium dodecyl sulfate sample buffer (Bio-Rad, Hercules, California), boiled for 3 minutes, and separated on a Criterion 4% to 12% Bis-Tris denaturing gel. The separated proteins were transferred electrophoretically to a nitrocellulose membrane (Bio-Rad Laboratories) for 2 hours at 120 volts, 4°C. Nonspecific antibody binding was blocked by incubation for 1 hour at RT with 5% nonfat dry milk in Tris-buffered saline solution containing 0.1% Triton X-100 (TBST; Bio-Rad). The membrane was incubated with the appropriate primary antibody in 5% milk in TBST overnight, washed 3 times for 10 minutes each with TBST + 0.1% Triton X-100 at room temperature. Blots were incubated for 1 hour with anti-rabbit or anti-mouse secondary antibodies as appropriate, followed by washing as before and a final TBS only wash, each 10 minutes. Chemi-Glow Chemiluminescent Substrate (Pierce) was used to detect the targeted protein. The band density on the X-ray film was optically scanned and quantitated using BioRad software. Blots were stripped with Restore stripping buffer (Pierce), reprobed and normalized to the reference protein (β-actin), and presented as fold change relative to the control level.
Statistical analysis
Microarray data were analyzed using paired Student t-test and differences were considered significant at P < .01. Differences in protein expression were analyzed by unpaired Student t-test with significance of P < .05. Means were transformed into fold changes with MUN values relative to control value. Control fold change was represented as 1.0. SEM was calculated for each mean and expressed as a proportional value to the fold change.
Results
DNA Microarray Expression Results
Four control and four MUN male fetal kidneys, each from a separate dam, were processed for RNA extraction and examined by pairwise comparison of gene expression by microarray analysis. Of the approximately 41 000 oligonucleotides representing unique rat mRNA transcripts on the Agilent microarray, 3547 transcripts were both detectable and differed statistically between C and MUN (P < .01). At the chosen significance threshold of 1.5 fold, 434 transcripts were increased and 45 transcripts were decreased by MUN in the E20 kidney, indicating that the majority of differentially expressed genes were induced rather than repressed by MUN. Ratios of differentiation ranged from 10.3-fold increased to 2.2-fold decreased. All differentially expressed targets are listed in the supplemental materials.
Functional Analysis of Microarray Data
The gene identifiers for the 479 differentially expressed transcripts were submitted for ontological classification using DAVID (6th version).27,28 Of these 479 gene identifiers, 122 could be identified and categorized into the Gene Ontology functional category of biological process. Classification of the genes overrepresented in the ontology category of biological process revealed 20 categories having between 9 and 2 members (Table 1 ). The biological process groups overlap to a large extent but several unique groups of genes were prominently represented.
Table 1.
Ontological Summary of Gene Expression by Biological Functional Categorya
| Biological Process | No. per Group | Accession Number | Symbol | Name | Fold Change |
|---|---|---|---|---|---|
| GO:0048666~neuron development/differentiation | |||||
| 9 | NM_203409 | Ncam2 | Neural cell adhesion molecule 2 | 6.1 | |
| XM_001072877 (BQ196383) | Tiam1 | T-cell lymphoma invasion and metastasis 1 | 8.1 | ||
| NM_013191 | S100b | S100 protein, beta polypeptide, neural | 6.8 | ||
| AF073379 | Grin3a | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 9.5 | ||
| NM_001012150 (AA997325) | Enah | Enabled homolog | –1.7 | ||
| NM_019621 | Dlg4 | Discs, large homolog 4 (Drosophila) | 5.8 | ||
| NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | ||
| NM_001001508 (BF557101) | Lppr4 | Lipid phosphate phosphatase-related protein type 4 | 4.7 | ||
| NM_001029902 | Ptprk | Protein tyrosine phosphatase, receptor type, K, extracellular region | 2.2 | ||
| GO:0008610~lipid biosynthetic process | |||||
| 8 | NM_138884 | Akr1d1 | Aldo-keto reductase family 1, member D1 | 7.9 | |
| NM_017274 (BI288209) | Gpam | Glycerol-3-phosphate acyltransferase, mitochondrial | 3.1 | ||
| NM_078622 (BF548541) | Pcyt1a | Phosphate cytidylyltransferase 1, choline, alpha isoform | 1.7 | ||
| NM_001011933 (AA997788) | Far1 | Fatty acyl CoA reductase 1 | 5.2 | ||
| NM_001024370 (AW434228) | Pigy | Phosphatidylinositol glycan anchor biosynthesis, class Y | 6.8 | ||
| NM_012942 | Cyp7a1 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | 6.5 | ||
| NM_022626 | Phka1 | Phosphorylase kinase alpha 1 | 8.5 | ||
| NM_017232 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | –1.4 | ||
| GO:0006396~RNA processing | |||||
| 9 | XM_236501 (CF109785) | SR140 | U2-associated SR140 protein | 7.4 | |
| NM_053662 (AA998342) | Ccnl1 | Cyclin L1 | 7.3 | ||
| XM_233805 (AA901271) | Hnrpll | Heterogeneous nuclear ribonucleoprotein L-like | 7.4 | ||
| XM_001058589 (AA926313) | Nova1 | Neuro-oncological ventral antigen 1 | 8.2 | ||
| NM_001115021 (AI172068) | Qk | Quaking | –1.8 | ||
| NM_001008293 (AA849810) | Tfb2m | Transcription factor B2, mitochondrial | 5.8 | ||
| NM_001025738 (AI058614) | Fusip1 | FUS interacting protein (serine-arginine rich) 1 | 6.7 | ||
| NM_001106585 (AA924863) | Lsm8 | LSM8 homolog, U6 small nuclear RNA associated (S. cerevisiae) | –1.6 | ||
| NM_001107156 (AI556918) | Sart3 | Squamous cell carcinoma antigen recognized by T-cells 3 | 7.3 | ||
| GO:0032990~cell morphogenesis | |||||
| 7 | NM_203409 | Ncam2 | Neural cell adhesion molecule 2 | 6.1 | |
| NM_013191 | S100b | S100 protein, beta polypeptide, neural | 6.1 | ||
| AF073379 | Grin3a | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 9.5 | ||
| NM_001012150 (AA997325) | Enah | Enabled homolog | –1.7 | ||
| NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | ||
| NM_001001508 (BF557101) | Lppr4 | Lipid phosphate phosphatase-related protein 4 | 4.7 | ||
| NM_001029902 | Ptprk | Protein tyrosine phosphatase, receptor type, K, extracellular region | 2.2 | ||
| GO:0009792~embryonic development ending in birth | |||||
| 5 | NM_001012150 (AA997325) | Enah | Enabled homolog | –1.7 | |
| NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | ||
| NM_001100700 (BF396545) | Sfrp2 | Secreted frizzled-related protein 2 | 8.9 | ||
| NM_012502 | Ar | Androgen receptor | 8.6 | ||
| NM_013172 | Myf6 | Myogenic factor 6 | 9.0 | ||
| GO:0007067~mitosis | |||||
| 6 | NM_021689 | Ereg | Epiregulin | 3.6 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_031583 (AI501054) | Smc3 | Structural maintenace of chromosomes 3 | 7.8 | ||
| NM_022256 | Btc | Betacellulin | 8.5 | ||
| XM_001080369 (AA875348) | Ska2 | Spindle and kinetochore associated complex subunit 2 | 5.4 | ||
| NM_001108008 (BE104462) | Eml4 | Echinoderm microtubule associated protein like 4 | 6.2 | ||
| GO:0043009~chordate embryonic development | |||||
| 6 | NM_001012150 (AA997325) | Enah | Enabled homolog | 0.58 | |
| NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | ||
| NM_001100700 (BF396545) | Sfrp2 | Secreted frizzled-related protein 2 | 8.9 | ||
| NM_024355 (BF398114) | Axin2 | Axin2 | 10.2 | ||
| NM_012502 | Ar | Androgen receptor | 8.6 | ||
| NM_013172 | Myf6 | Myogenic factor 6 | 9.0 | ||
| GO:0000087~M phase of mitotic cell cycle | |||||
| 6 | NM_021689 | Ereg | Epiregulin | 3.6 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_031583 (AI501054) | Smc3 | Structural maintenace of chromosomes 3 | 7.8 | ||
| XM_001080369 (AA875348) | Ska2 | Spindle and kinetochore associated complex subunit 2 | 5.4 | ||
| NM_001108008 (BE104462) | Eml4 | Echinoderm microtubule associated protein like 4 | 6.2 | ||
| NM_022256 | Btc | Betacellulin | 8.5 | ||
| GO:0021700~developmental maturation | |||||
| 5 | NM_019621 | Dlg4 | Discs, large homolog 4 (Drosophila) | 5.8 | |
| NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | ||
| NM_021689 | Ereg | Epiregulin | 3.6 | ||
| NM_017232 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 8.6 | ||
| NM_001107954 (AI029408) | Reck | Reversion-inducing-cysteine-rich protein with kazal motifs | 5.3 | ||
| GO:0006333~chromatin assembly or disassembly | |||||
| 4 | XM_001063352 (BE107091) | Chd4 | Chromodomain helicase DNA binding protein 4 | 5.8 | |
| AI145002 | Nap1l3 | Nucleosome assembly protein 1-like 3 | 10.3 | ||
| NM_001107470 (BE104039) | Shprh | SNF2 histone linker PHD RING helicase | 9.4 | ||
| NM_001014145 | Cdyl | Chromodomain protein, Y chromosome-like | 5.9 | ||
| GO:0008654~phospholipid biosynthetic process | |||||
| 3 | NM_017274 (BI288209) | Gpam | Glycerol-3-phosphate acyltransferase, mitochondrial | 3.2 | |
| NM_001024370 (AW434228) | Pigy | Phosphatidylinositol glycan anchor biosynthesis, class Y | 6.8 | ||
| NM_022626 | Pcyt1a | Phosphate cytidylyltransferase 1, choline, alpha isoform | 1.7 | ||
| GO:0046394~carboxylic acid biosynthetic process | |||||
| 4 | NM_138884 | Akr1d1 | Aldo-keto reductase family 1, member D1 | 7.9 | |
| NM_012942 | Cyp7a1 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | 6.5 | ||
| NM_017232 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 8.6 | ||
| NM_138884 | Akr1d1 | Aldo-keto reductase family 1, member D1 | 7.9 | ||
| GO:0045787~positive regulation of progression through cell cycle | |||||
| 4 | NM_031131 (BF420705) | Tgfb | Transforming growth factor, beta 2 | 7.3 | |
| NM_021689 | Ereg | Epiregulin | 3.6 | ||
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_022256 | Btc | Betacellulin | 8.5 | ||
| GO:0035282~segmentation | |||||
| 3 | NM_001100700 (BF396545) | Sfrp2 | Secreted frizzled-related protein 2 | 8.9 | |
| NM_024355 (BF398114) | Axin2 | Axin2 | 10.2 | ||
| NM_013172 | Myf6 | Myogenic factor 6 | 9.0 | ||
| GO:0016127~sterol catabolic process | |||||
| 2 | NM_138884 | Akr1d1 | Aldo-keto reductase family 1, member D1 | 7.9 | |
| NM_012942 | Cyp7a1 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | 6.5 | ||
| GO:0046474~glycerophospholipid biosynthetic process | |||||
| 3 | NM_078622 (BF548541) | Pcyt1a | Phosphate cytidylyltransferase 1, choline, alpha isoform | 1.7 | |
| NM_001024370 (AW434228) | Pigy | Phosphatidylinositol glycan anchor biosynthesis, class Y | 6.8 | ||
| NM_022626 | Phka1 | Phosphorylase kinase alpha 1 | 8.5 | ||
| GO:0045840~positive regulation of mitosis | |||||
| 3 | NM_021689 | Ereg | Epiregulin | 3.6 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_022256 | Btc | Betacellulin | 8.5 | ||
| GO:0001756~somitogenesis | |||||
| 3 | NM_001100700 (BF396545) | Sfrp2 | Secreted frizzled-related protein 2 | 8.9 | |
| NM_024355 (BF398114) | Axin2 | Axin2 | 10.2 | ||
| NM_013172 | Myf6 | Myogenic factor 6 | 9.0 | ||
| GO:0006706~steroid catabolic process | |||||
| 2 | NM_012942 | Cyp7a1 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | 6.5 | |
| NM_138884 | Akr1d1 | Aldo-keto reductase family 1, member D1 | 7.9 | ||
| GO:0007176~regulation of epidermal growth factor receptor activity | |||||
| 2 | NM_021689 | Ereg | Epiregulin | 3.6 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
a All genes differentially expressed by 1.5-fold or greater were submitted to analysis by Database for Annotation, Visualization, and Integrated Discovery (DAVID). The 122 identified genes were sorted into biological functional categories. Some genes are represented in more than 1 category. Genbank accession numbers represent the most current identifiers. Accession numbers in parenthesis are taken from the Agilent microarray identifier. Downregulation of genes is represented as a negative-fold change.
Gene Ontology Functional Categories
Development and differentiation involved genes were the most predominant. Strongly upregulated genes included the cell adhesion molecule Neural cell adhesion molecule 2 (Ncam2), T-cell lymphoma invasion and metastasis 1 (Tiam1), transforming growth factor β2 (TGFβ2), secreted frizzle-related protein (Sfrp2), and Axin2. There was downregulation of enabled homolog (Enah).
Mitosis and cell cycle regulation dysregulated genes were also prominent. Upregulated genes included epiregulin (Ereg), an epidermal growth factor (EGF)-family ligand, and TGFα and Betacellulin (Btc), both of which bind the fetal epidermal growth factor (EGF) receptor.
Chromatin assembly genes often were augmented including chromodomain helicase DNA binding 4 (Chd4), nucleosome assembly protein 1-like 3 (Nap1/3), SNF histone linker PHD ring helicase (Shprh), and chromodomain protein, Y chromosome-like (Cdyl). These genes have not been identified previously in the kidney.
Steroid hormone regulation genes were strongly increased and included androgen receptor (AR), Aldo-keto reductase D1 (Akr1d1, also known as 5-beta reductase), glycerol-3-phosphate acyltransferase expressed in the mitochondria (Gpam), phosphate cytidylyltransferase 1 alpha (Pcyt1a), phosphorylase kinase alpha 1, also known as Pcyt1b (Phka1), and prostaglandin-endoperoxide synthase 2, also known as Cox2 (Ptgs2), which catalyzes conversion of arachidonic acid products to prostaglandin.
There are other interesting dysregulated genes covering a variety of biological processes. S100b (S100 protein, beta polypeptide), a calgranulin family member, was upregulated by MUN. Ion-regulation may be affected as Dlg4 (discs, large homolog 4, also known as Psd95 [post-synaptic density protein 95]) and Grin3A were both strongly upregulated. Finally, a large number of RNA processing genes were increased and decreased. Most notable were augmented mitochondrial transcription factor B2 (Tfb2m), increased Fus interacting protein 1 (Fusip1, also called Sfrs13a), downregulated LSM8(Lsm8) homolog, and upregulated squamous cell carcinoma antigen recognized by T-cells 3 (Sart3).
KEGG Pathways
We next used DAVID to assign genes to known KEGG pathways. A total of 47 pathways were reported and of those we selected pathways demonstrating at least 3 genes each with > 1.5 fold change in expression detected in a given pathway for further classification. This resulted in 15 pathways impacted by MUN (Table 2 ). Of the 15 KEGG pathways identified, we previously detected 2 which were dysregulated in MUN kidneys, the MAPK/ERK kinase and Wnt signaling pathways.20,23
Table 2.
Ontological Summary of Gene Expression by KEGG Pathway Category a
| Ontological Categorization | No. Per Group | Accession Number | Symbol | Name | Fold Change |
|---|---|---|---|---|---|
| Adipocytokine signaling pathway | |||||
| 3 | NM_013091 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.6 | |
| NM_012675 | Tnf | Tumor necrosis factor superfamily, member 2 | 1.6 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| Antigen processing and presentation | |||||
| 3 | NM_001008831 | RT1-Ba | Butyrophilin-like 2 (mch class ii associated) | 2.7 | |
| NM_001008885 | Slc39a7 | rt1 class I, locus ke4 | 1.5 | ||
| NM_013156 | Ctsl1 | Cathepsin L1 | 7.0 | ||
| Apoptosis | |||||
| 4 | NM_012908 | FasL | Tumor necrosis factor (ligand) superfamily, member 6 | 7.0 | |
| NM_013091 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.6 | ||
| NM_012675 | Tnf | Tumor necrosis factor superfamily, member 2 | 1.6 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| Calcium signaling pathway | |||||
| 4 | NM_052799 | Nos1 | Nitric oxide synthase 1, neuronal | 1.6 | |
| NM_001004279 | Ppid | Peptidylprolyl isomerase d | 1.6 | ||
| NM_078619 | Slc8a2 | Solute carrier family 8 (sodium/calcium exchanger), member 2 | 9.4 | ||
| NM_022626 | Phka1 | Phosphorylase kinase alpha 1 | 8.5 | ||
| Cell adhesion molecules (CAMs) | |||||
| 4 | NM_203409 | Ncam2 | Neural cell adhesion molecule 2 | 6.1 | |
| NM_001008831 | RT1-Ba | Butyrophilin-like 2 (mch class ii associated) | 2.7 | ||
| NM_013082 | Sdc2 | Syndecan 2 | 10.2 | ||
| NM_001008885 | Slc39a7 | rt1 class I, locus ke4 | 1.5 | ||
| Cytokine–cytokine receptor interaction | |||||
| 4 | NM_012908 | FasL | Tumor necrosis factor (ligand) superfamily, member 6 | 7.0 | |
| NM_013091 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.6 | ||
| XM_231873 (BQ194449) | Il12rb2 | Interleukin 12 receptor, beta 2 | –1.6 | ||
| NM_012675 | Tnf | Tumor necrosis factor superfamily, member 2 | 1.6 | ||
| ECM–receptor interaction | |||||
| 3 | NM_001108118 (AI385193) | Itga5 | Integrin alpha 5 (fibronectin receptor alpha) | 1.5 | |
| NM_013082 | Sdc2 | Syndecan 2 | 10.2 | ||
| XM_342325 | Col11a1 | Procollagen, type xi, alpha 1 | 4.5 | ||
| ErbB signaling pathway | |||||
| 4 | NM_022256 | Btc | Betacellulin | 8.5 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| NM_021689 | Ereg | Epiregulin | 3.6 | ||
| Fc epsilon RI signaling pathway | |||||
| 3 | NM_134366 | Rac | Ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1) | 1.4 | |
| NM_012675 | Tnf | Tumor necrosis factor superfamily, member 2 | 1.6 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| Focal adhesion | |||||
| 4 | NM_001108118 (AI385193) | Itga5 | Integrin alpha 5 (fibronectin receptor alpha) | 1.5 | |
| NM_131914 | Cav2 | Caveolin 2 | 5.7 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| XM_342325 | Col11a1 | Procollagen, type xi, alpha 1 | 4.5 | ||
| MAPK signaling pathway | |||||
| 6 | NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | |
| NM_012908 | FasL | Tumor necrosis factor (ligand) superfamily, member 6 | 7.0 | ||
| NM_013091 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.6 | ||
| NM_012675 | Tnf | Tumor necrosis factor superfamily, member 2 | 1.6 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| NM_022223 (AF348523) | Fgf14 | Fibroblast growth factor 14 | 5.7 | ||
| Notch signaling pathway | |||||
| 4 | NM_019201 | Ctbp1 | C-terminal binding protein 1 | 6.8 | |
| NM_001109279 (AA944168) | Skip | Similar to nuclear protein skip (ski-Interacting protein Snw1) | –1.8 | ||
| NM_001106997 (BQ195789) | Maml1 | Mastermind like 1 | –1.8 | ||
| NM_024358 | Notch2 | Notch2 | –1.8 | ||
| Regulation of actin cytoskeleton | |||||
| 3 | NM_001108118 (AI385193) | Itga5 | Integrin alpha 5 (fibronectin receptor alpha) | 1.5 | |
| XM_001072877 (BQ196383) | Tiam1 | T-cell lymphoma invasion and metastasis 1 | 8.1 | ||
| NM_022223 (AF348523) | Fgf14 | Fibroblast growth factor 14 | 5.7 | ||
| Renal cell carcinoma | |||||
| 3 | NM_031131 (BF420705) | Tgfb2 | Transforming growth factor, beta 2 | 7.3 | |
| NM_012671 (BG670310) | Tgfa | Transforming growth factor alpha | 6.0 | ||
| NM_017093 | Akt2 | Thymoma viral proto-oncogene 2 | 1.5 | ||
| Wnt signaling pathway | |||||
| 3 | NM_001100700 (BF396545) | Sfrp2 | Secreted frizzled-related protein 2 | 8.9 | |
| NM_019201 | Ctbp1 | C-terminal binding protein 1 | 7.0 | ||
| NM_024355 (BF398114) | Axin2 | Axin2 | 10.2 | ||
a All genes differentially expressed by 1.5-fold or greater were submitted to analysis by Database for Annotation, Visualization, and Integrated Discovery (DAVID). The 122 identified genes were sorted into KEGG pathways. Only pathways with 3 or more identified genes are listed. Some genes are represented in more than 1 category. Genbank accession numbers represent the most current identifiers. Accession numbers in parenthesis are taken from the Agilent microarray identifier. Downregulation of genes is represented as a negative-fold change.
MAPK/ERK signaling pathway genes identified were all upregulated and included TGFβ2, Fas ligand (FasL), tumor necrosis factor receptor (Tnfrsf1a), tumor necrosis factor alpha (TNFα), Akt2 (Thymoma viral proto-oncogene 2), and fibroblast growth factor 14 (FGF14). Inflammatory signaling genes that were moderately upregulated include Rac (ras-related c3 botulinium toxin substrate 1). It is regulated by the GTP exchange factor t-cell lymphoma cell invasion and metastasis 1 (Tiam1),31 which was strongly induced.
Wnt signaling genes induced by MUN included secreted frizzled-related protein 2 (Sfrp2), a secreted glycoprotein molecule that structurally resembles cell surface frizzled receptors but lacks the transmembrane domain,32 and Axin2 and Ctbp1, both of which were highly upregulated by MUN.
Apoptosis genes were upregulated including FasL, TNFα1, the TNFα1 receptor, and Akt2. The induction of possibly Fas-mediated apoptotic activity suggests increased apoptosis/inflammation in the MUN kidney, possibly localized to mesangial cells.33
Calcium signaling demonstrated strong upregulation of Slc8a2 mRNA (solute carrier family 8 member 2, also called Ncx2), a sodium-calcium exchanger involved in calcium resabsorption and phosphorylase kinase α 1 (Phka1), a calcium-responsive subunit involved in the repression of glycogen metabolism. Two moderately induced calcium-regulated genes detected were Nitric oxide synthase 1, neuronal (Nos1) and peptidyprolyl isomerase d (Ppid, also known as cyclophilin D).
Cell-cell interaction and extra-cellular matrix signaling KEGG pathways also appear to be dysregulated by MUN, including cell adhesion molecules (CAMs), extracellular membrane (ECM) receptor interactions, and focal adhesion. The most strongly upregulated genes included neural cell adhesion molecule 2 (Ncam2), syndecan 2 (Sdc2, also referred to as heparan sulfate proteoglycan, HSG), and caveolin 2 (Cav2). Integrin, fibronectin receptor alpha 5 (Itga5 mRNA) and Col11a1 were both moderately overexpressed.
Immunity- and defense-related mRNAs were upregulated by MUN. These included T-cell-mediated immunity and MHC class II antigen processing members Rrt1-Ba (butyrophilin-like 2) and cathepsin L1 (Ctsl1), and MCH class I member Slc39a7 (rt1 class I, locus ke4). Cathepsin L has additional functions as a lysosomal protease.34,35
The pleiotropic ErbB (Epidermal growth factor) signaling pathway was upregulated by MUN. Key MUN-augmented genes detected were Btc, Ereg, and TGFα, all EGF receptor ligands. Betacellulin was one of the most highly upregulated MUN mRNAs at over 8-fold induction.
Notch signaling pathway members were dysregulated, which appears to be one of the most intriguing effects of MUN. Ctbp1, a co-repressor of recombination signal binding protein for immunoglobulin kappa J region (Rbpj, also known as Csl), had augmented mRNA expression. Two coactivators of Rbpj showed reduced mRNA expression, specifically Skip and Maml1 (Table 2). Additionally, Notch2 showed reduced mRNA expression. These data strongly suggested that the Notch pathway is affected by MUN and we sought to validate this result by measuring protein expression of Notch signaling components.
Western Blot Validation of Notch Signal Pathway MUN Expression
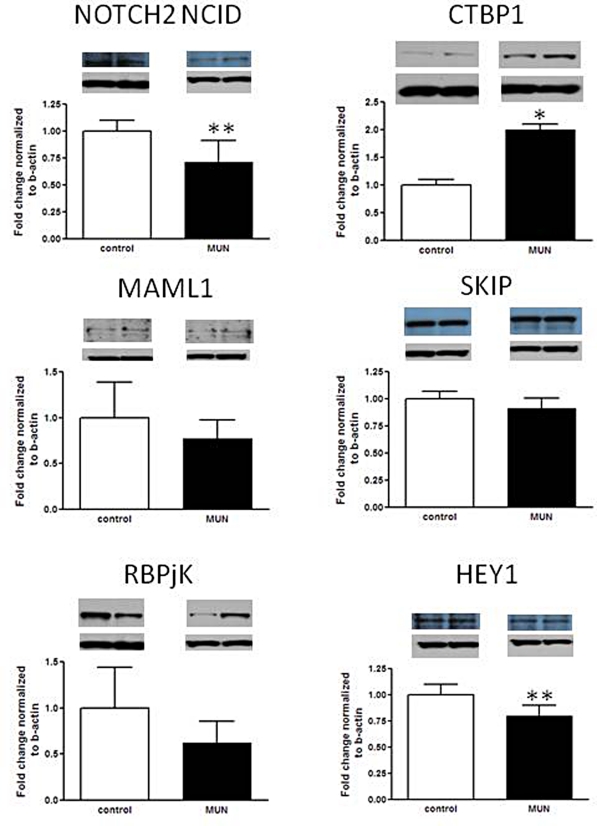
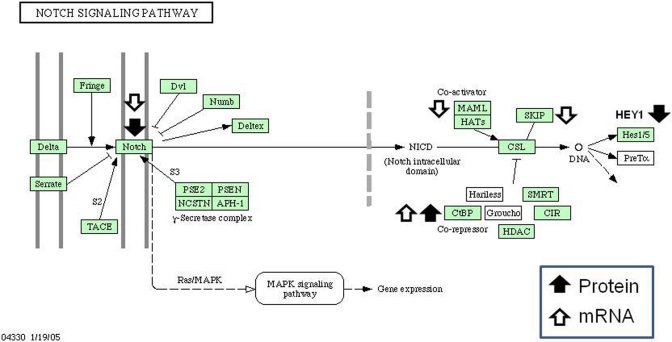
Based on the microarray data analysis, 4 Notch signaling pathway genes demonstrated altered expression. The Notch signaling pathway is involved in developmental patterning in the kidney and is critical for the development of glomeruli,36–38 suggesting that alteration of this pathway may contribute to IUGR programmed nephropenia. Protein expression of the 4 genes identified by the microarray (Notch2, Ctbp1, Maml1, Skip) were measured as well as other members of the Notch-signaling pathway including the transcription factor Rpbjk (recombination signal binding protein for immunoglobulin kappa J region; also called Csl), and a Notch pathway regulated gene hairy/enhancer-of-split related with YRPW motif 1 (Hey1; Figure 1 ). In concordance with the microarray results, Notch2 (-1.4-fold; P = .001) and Ctbp1 (2.0-fold; P = .04) were downregulated and upregulated, as expected. Maml showed a trend downward but was not statistically significant (-1.3 fold, P = .09) and Skip protein expression was unchanged (-1.1-fold, P = .34). The signaling transcription factor Rbpjk appeared reduced, but not significantly (-1.6-fold, P = .10) whereas the downstream gene Hey1 was downregulated (-1.2-fold, P = .001). These data overall point to a reduction in Notch pathway protein expression and are diagrammed in Figure 2 .
Figure 1.
Protein expression of Notch signaling pathway factors as determined by Western blotting. Five to six Control versus MUN kidney lysates; each protein probed with a specific primary antibody. β-actin represents a housekeeping protein that is not affected by maternal undernutrition (MUN) and was used for normalization of protein loading. Representative bands for the target protein and b-actin are depicted. *P ≤ .05, ** P ≤ .01.
Figure 2.
Differential expression of mRNA and proteins encoding factors in the Notch signaling pathway. Arrow direction indicates up or downregulation detected by DNA microarray (open arrows) or by Western blotting (filled arrows). Diagram is a modification of the Kyoto Encyclopedia of Genes and Genome (KEGG) based Notch pathway.
Discussion
In this study, we demonstrated specific changes in gene expression in fetal E20 kidneys from MUN dams. Statistically significant changes in gene expression greater than 1.5-fold were evaluated and organized into ontological biological processes and KEGG pathways. Ontological categorization revealed that MUN prominently affects development and differentiation genes, which are involved in kidney morphogenesis. These encompass Tiam1 and TGFβ2 involved in mesenchymal to epithelial transformation,39,40 while Sfrp2 and Axin2 modulate Wnt4 expression and signaling, required for pretubular aggregate formation.41,42 Mitosis and cell-cycle genes participating in renal development were dysregulated. Ereg is a target of WT1 and involved in branching morphogenesis.43 TGFα and Btc bind the fetal EGF receptor stimulating cell proliferation and may be involved in renal dysplasia.44–46 Epigenetics is thought to play a role in IUGR regulation and the upregulation of chromatin assembly genes may be an important mechanism for fetal programming.47
Additional genes involved in hormonal control, stress, and RNA processing were detected. Steroid hormone regulation genes were strongly increased and included AR, which is expressed in nascent tubules and may increase responsiveness to testosterone and eventually increase hypertension.48 Additionally increased were Gpam, which regulates triacylglycerol levels in response to nutrient stress in the kidney,49 phosphate cytidylyltransferase 1 alpha (Pcyt1a) involved in the synthesis of phospholipids known as plasmalogens, which are a significant component in kidney membranes,50 and Ptgs2, which is hormonally regulated during nephrogenesis.51 A large number of RNA processing genes were dysregulated suggesting modification of RNA posttranscriptional processing by MUN. Increased genes include Tfb2m, which may increase mitochondrial transcription52 and Fusip1, which binds to preprocessed RNA and represses splicing under stress conditions.53 Genes with decreased expression were Lsm8 involved in ribosomal RNA maturation which causes decreased ribosome content,54 and Sart3, which has been implicated in the regulation of mRNA splicing.55,56
Kidney organ development regulation appears to be strongly affected by gestational programming with the overall pattern suggesting a fairly broad effect of MUN on nephrogenesis. Of the 15 KEGG pathways identified, we previously reported 2 that were dysregulated in MUN kidneys, the MAPK/ERK kinase and Wnt signaling pathways.20,23 In this study, we identified additional factors in these pathways. Fas ligand and the TNFs previously have been detected in kidney associated with conditions such as diabetes and inflammation57,58 or chronic kidney disease.59,60 Several of the MAPK/ERK signaling pathway factors, namely Fas ligand, TNFα, TNFα receptor, and Akt2, are critical for apoptotic signaling by both the extrinsic and intrinsic pathways.61,62 Increased gene expression of these factors suggests an increase in apoptosis of the programmed kidney, possibly by inflammatory signaling. Other upregulated inflammatory signaling genes include Rac, which regulates cell−cell adhesion and migration in renal epithelial cells63 and is regulated by the GTP exchange factor Tiam1,31 which was strongly induced by MUN. Cathepsin L, an upregulated immunity and defense gene, has additional functions as a lysosomal protease, which can be produced by podocytes during inflammatory responses, again suggestive of an MUN-elicited inflammatory response.34,35 Interestingly, relatively few mRNAs appeared to be downregulated by MUN. One downregulated factor was interleukin 12 receptor β2 (Il12rb2), a cytokine receptor that interacts with Jak, and leads to phosphorylation of multiple signal pathways including the MAPK/ERK signaling pathway.64 Due to the anti-inflammatory effects of IL-12,65 it is possible that reduction of this receptor’s expression contributes to an increased inflammatory response.
Alterations in genes participating in Wnt signaling were not unexpected in MUN offspring kidneys based on our previous findings,20 Sfrp2 is a secreted glycoprotein molecule that can inhibit Wnt binding to its receptor.66 This family of proteins has been implicated in organ development and cell fate.67 C-terminal binding protein 1 (Ctpb1) is a corepressor mediating beta-catenin/TCF transcriptional activities.68 Additionally, Axin2 and Ctbp1, both of which were highly upregulated by MUN, repress the Wnt pathway at separate points.42 The pleiotropic ErbB signaling pathway genes were augmented in MUN kidneys; this pathway impacts cell proliferation, survival, differentiation, and angiogenesis. Excessive synthesis of pathway members results in abnormal growth factor responses. We identified increased mRNA for EGF receptor ligands, and these are known to be critical nephrogenic morphogens.46 Of these, epiregulin stimulates proliferation and migration of renal proximal tubular cells in cell culture69 and TGFα has been implicated in the development of the metanephros.70
Finally, Notch signaling pathway members were dysregulated, one of the most intriguing effects of MUN. Ctbp1, a co-repressor of Rbpj (Csl), the transcription factor mediating Notch ligand activity, was upregulated in mRNA expression while coactivators were reduced, suggesting inhibition of Notch signaling. Additionally, Notch2, a key Notch receptor involved in nephrogenesis, showed reduced mRNA expression. The Notch signaling pathway is highly conserved across species regulating cell proliferation, differentiation, fate, and death. Notch signaling molecules are expressed throughout nephron development with the Notch2 receptor identified in pretubular aggregates, comma and S bodies, immature glomerular epithelial cells, and tubular and collecting duct cells.71 The Notch2 ligand, Jagged 1 (Jag1), is expressed in a similar profile.71,72 While Notch1 and 2 are found in developing mammalian kidneys, Notch2 activity is essential for patterning of the proximal regions of the nephron development.36–38 Disruptions of Notch signaling is associated with developmental syndromes,72–77 in humans and animal models. For example, mutations or absence of Jag1 and Notch2 cause Alagille syndrome, an autosomal dominant disorder affecting several organs, including the kidney.37,71,78–81
There are several developmental stages during which decrements in Notch2 may impact nephron formation. Maternal undernutrition downregulates Notch2 and a downstream effector, Hey1. Reduction in Notch2 activity results in abnormal glomerulogenesis and small kidneys, while Notch2-deficient kidneys also have abnormalities of distal and proximal tubular development.72 Early in metanephric development, the Notch signaling pathway interacts with Gdnf and Ret to participate in primary ureteric budding and subsequent branching,38,75 and subsequent interactions with Gdnf and Maml impact further branching.42 Hey1 might play a role in determining the precise timing of renal vesicle transformation to comma-shaped bodies since it is expressed abundantly in the S-shaped body, from precursor cells of the loop of Henle to podocytes.72 Our previous study of E20 MUN kidneys demonstrated dysregulation of Wnt4, Wnt11, Bmp4, Bmp7, and Fgf2, some of which may cross-signal with the Notch pathway.20,82–84 Notch signaling pathway co-activators and co-repressors also are shared with other pathways, i.e. Ctbp1 and Wnt signaling, and their dysregulation may have effects on gene expression, affecting kidney development beyond the Notch pathway.36,71,82,85
In summary, MUN leads to >1.5-fold altered expression of 479 mRNA transcripts in the E20 MUN kidney, suggesting that gestational programming has a large impact on nephrogenesis. There are a number of signaling pathways disrupted by MUN, some expected and some novel. Not all dysregulated processes will be identified by microarray analysis; nevertheless, this type of analysis affords a means to survey developmental disruption in the fetal kidney due to MUN. Microarray analysis identified the Notch pathway as one with significant alterations, and we further analyzed this pathway to validate the microarray data. We found reductions in Notch2, co-activators and a downstream target as well as upregulation of a co-repressor. Dysregulation of the Notch signaling pathway, which is critical for determination of cell fate in the developing kidney, is likely to be an important target of gestational programming leading to nephron reduction.
Acknowledgments
We would like to thank Linda Day, Stacy Behare, Gloria Chang, and Lindsay Chun for their technical assistance. We would also like to thank Antonia Enriquez and Diane Park for their assistance with manuscript preparation.
Footnotes
The authors declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
The authors disclosed receipt of the following financial support for the research and/or authorship of this article: Supported by NIH R03HD060782 to TRM.
References
- 1. Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1(8489):1077–1081 [DOI] [PubMed] [Google Scholar]
- 2. Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261(5):412–417 [DOI] [PubMed] [Google Scholar]
- 3. Sullivan EL, Grove KL. Metabolic imprinting in obesity. Forum Nutr. 2010;63:186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ross MG, Beall MH. Adult sequelae of intrauterine growth restriction. Semin Perinatol. 2008;32(3):213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rhodes P, Craigon J, Gray C, Rhind SM, Loughna PT, Gardner DS. Adult-onset obesity reveals prenatal programming of glucose-insulin sensitivity in male sheep nutrient restricted during late gestation. PLoS ONE. 2009;4(10):e7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simmons RA, Gounis AS, Bangalore SA, Ogata ES. Intrauterine growth retardation: fetal glucose transport is diminished in lung but spared in brain. Pediatr Res. 1992;31(1):59–63 [DOI] [PubMed] [Google Scholar]
- 7. Langley-Evans SC, Welham SJ, Jackson AA. Fetal exposure to a maternal low protein diet impairs nephrogenesis and promotes hypertension in the rat. Life Sci. 1999;64(11):965–974 [DOI] [PubMed] [Google Scholar]
- 8. Brennan KA, Kaufman S, Reynolds SW, et al. Differential effects of maternal nutrient restriction through pregnancy on kidney development and later blood pressure control in the resulting offspring. Am J Physiol Regul Integr Comp Physiol. 2008;295(1):R197–R205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langley-Evans SC, Langley-Evans AJ, Marchand MC. Nutritional programming of blood pressure and renal morphology. Arch Physiol Biochem. 2003;111(1):8–16 [DOI] [PubMed] [Google Scholar]
- 10. Schreuder MF, Nauta J. Prenatal programming of nephron number and blood pressure. Kidney Int. 2007;72(3):265–268 [DOI] [PubMed] [Google Scholar]
- 11. Woods LL. Maternal nutrition and predisposition to later kidney disease. Curr Drug Targets. 2007;8(8):906–913 [DOI] [PubMed] [Google Scholar]
- 12. Almeida JR, Mandarim-de-Lacerda CA. Maternal gestational protein-calorie restriction decreases the number of glomeruli and causes glomerular hypertrophy in adult hypertensive rats. Am J Obstet Gynecol. 2005;192(3):945–951 [DOI] [PubMed] [Google Scholar]
- 13. Gross ML, Amann K, Ritz E. Nephron number and renal risk in hypertension and diabetes. J Am Soc Nephrol. 2005;16(suppl 1):S27–S29 [DOI] [PubMed] [Google Scholar]
- 14. Painter RC, Roseboom TJ, van Montfrans GA, et al. Microalbuminuria in adults after prenatal exposure to the Dutch famine. J Am Soc Nephrol. 2005;16(1):189–194 [DOI] [PubMed] [Google Scholar]
- 15. Dotsch J, Plank C, Amann K, Ingelfinger J. The implications of fetal programming of glomerular number and renal function. J Mol Med. 2009;87(9):841–848 [DOI] [PubMed] [Google Scholar]
- 16. Abitbol CL, Ingelfinger JR. Nephron mass and cardiovascular and renal disease risks. Semin Nephrol. 2009;29(4):445–454 [DOI] [PubMed] [Google Scholar]
- 17. Baum M. Role of the kidney in the prenatal and early postnatal programming of hypertension. Am J Physiol Renal Physiol. 2010;298(2):F235–F247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. do Carmo Pinho Franco M, Nigro D, Fortes ZB, et al. Intrauterine undernutrition--renal and vascular origin of hypertension. Cardiovasc Res. 2003;60(2):228–234 [DOI] [PubMed] [Google Scholar]
- 19. Fowden AL, Giussani DA, Forhead AJ. Intrauterine programming of physiological systems: causes and consequences. Physiology (Bethesda). 2006;21:29–37 [DOI] [PubMed] [Google Scholar]
- 20. Abdel-Hakeem AK, Henry TQ, Magee TR, et al. Mechanisms of impaired nephrogenesis with fetal growth restriction: altered renal transcription and growth factor expression. Am J Obstet Gynecol. 2008;199(3):252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox LA, Nijland MJ, Gilbert JS, et al. Effect of 30 per cent maternal nutrient restriction from 0.16 to 0.5 gestation on fetal baboon kidney gene expression. J Physiol. 2006;572(pt 1):67–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Symonds ME, Stephenson T, Gardner DS, Budge H. Tissue specific adaptations to nutrient supply: more than just epigenetics?. Adv Exp Med Biol. 2009;646:113–118 [DOI] [PubMed] [Google Scholar]
- 23. Henry TQ, Mansano RZ, Nast CC, et al. GDNF and MAPK-ERK pathway signaling is reduced during nephrogenesis following maternal under-nutrition. J Dev Origins Health Dis. 2010;1:67–74 [DOI] [PubMed] [Google Scholar]
- 24. Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28(3):339–363 [DOI] [PubMed] [Google Scholar]
- 25. Carlson ME, O'Connor MS, Hsu M, Conboy IM. Notch signaling pathway and tissue engineering. Front Biosci. 2007;12:5143–5156 [DOI] [PubMed] [Google Scholar]
- 26. McClive PJ, Sinclair AH. Rapid DNA extraction and PCR-sexing of mouse embryos. Mol Reprod Dev. 2001;60(2):225–226 [DOI] [PubMed] [Google Scholar]
- 27. Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57 [DOI] [PubMed] [Google Scholar]
- 28. Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 29. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(database issue):D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi GY, Tosh DN, Garg A, Mansano R, Ross MG, Desai M. Gender-specific programmed hepatic lipid dysregulation in intrauterine growth-restricted offspring. Am J Obstet Gynecol. 2007;196(5):477. [DOI] [PubMed] [Google Scholar]
- 31. Buchsbaum RJ, Connolly BA, Feig LA. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol Cell Biol. 2002;22(12):4073–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brophy PD, Lang KM, Dressler GR. The secreted frizzled related protein 2 (SFRP2) gene is a target of the Pax2 transcription factor. J Biol Chem. 2003;278(52):52401–52405 [DOI] [PubMed] [Google Scholar]
- 33. Wornle M, Schmid H, Merkle M, Banas B. Effects of chemokines on proliferation and apoptosis of human mesangial cells. BMC Nephrol. 2004;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mason RW, Gal S, Gottesman MM. The identification of the major excreted protein (MEP) from a transformed mouse fibroblast cell line as a catalytically active precursor form of cathepsin L. Biochem J. 1987;248(2):449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Asanuma K, Shirato I, Ishidoh K, Kominami E, Tomino Y. Selective modulation of the secretion of proteinases and their inhibitors by growth factors in cultured differentiated podocytes. Kidney Int. 2002;62(3):822–831 [DOI] [PubMed] [Google Scholar]
- 36. Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng HT, Kim M, Valerius MT, et al. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134(4):801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng HT, Kopan R. The role of Notch signaling in specification of podocyte and proximal tubules within the developing mouse kidney. Kidney Int. 2005;68(5):1951–1952 [DOI] [PubMed] [Google Scholar]
- 39. Malliri A, van ES, Huveneers S, Collard JG. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J Biol Chem. 2004;279(29):30092–30098 [DOI] [PubMed] [Google Scholar]
- 40. Sims-Lucas S, Caruana G, Dowling J, Kett MM, Bertram JF. Augmented and accelerated nephrogenesis in TGF-beta2 heterozygous mutant mice. Pediatr Res. 2008;63(6):607–612 [DOI] [PubMed] [Google Scholar]
- 41. Lescher B, Haenig B, Kispert A. sFRP-2 is a target of the Wnt-4 signaling pathway in the developing metanephric kidney. Dev Dyn. 1998;213(4):440–451 [DOI] [PubMed] [Google Scholar]
- 42. Valenta T, Lukas J, Korinek V. HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31(9):2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim HS, Kim MS, Hancock AL, et al. Identification of novel Wilms' tumor suppressor gene target genes implicated in kidney development. J Biol Chem. 2007;282(22):16278–16287 [DOI] [PubMed] [Google Scholar]
- 44. Solari V, Shima H, Puri P. Increased expression of EGFR and TGF-alpha in segmental renal dysplasia in duplex kidney. Pediatr Surg Int. 2004;20(4):243–247 [DOI] [PubMed] [Google Scholar]
- 45. Rogers SA, Ryan G, Hammerman MR. Metanephric transforming growth factor-alpha is required for renal organogenesis in vitro. Am J Physiol. 1992;262(4 pt 2):F533–F539 [DOI] [PubMed] [Google Scholar]
- 46. Sakurai H, Tsukamoto T, Kjelsberg CA, Cantley LG, Nigam SK. EGF receptor ligands are a large fraction of in vitro branching morphogens secreted by embryonic kidney. Am J Physiol. 1997;273(3 pt 2):F463–F472 [DOI] [PubMed] [Google Scholar]
- 47. Joss-Moore LA, Lane RH. The developmental origins of adult disease. Curr Opin Pediatr. 2009;21(2):230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baserga M, Bares AL, Hale MA, et al. Uteroplacental insufficiency affects kidney VEGF expression in a model of IUGR with compensatory glomerular hypertrophy and hypertension. Early Hum Dev. 2009;85(6):361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewin TM, Granger DA, Kim JH, Coleman RA. Regulation of mitochondrial sn-glycerol-3-phosphate acyltransferase activity: response to feeding status is unique in various rat tissues and is discordant with protein expression. Arch Biochem Biophys. 2001;396(1):119–127 [DOI] [PubMed] [Google Scholar]
- 50. Poloumienko A, Cote A, Quee AT, Zhu L, Bakovic M. Genomic organization and differential splicing of the mouse and human Pcyt2 genes. Gene. 2004;325:145–155 [DOI] [PubMed] [Google Scholar]
- 51. Carey LC, Valego NK, Chen K, Rose JC. Thyroid hormone regulates renocortical COX-2 and PGE2 expression in the late gestation fetal sheep. Reprod Sci. 2008;15(6):598–603 [DOI] [PubMed] [Google Scholar]
- 52. Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35(12):4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427(6974):553–558 [DOI] [PubMed] [Google Scholar]
- 54. Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D. Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J Biol Chem. 2003;278(4):2147–2156 [DOI] [PubMed] [Google Scholar]
- 55. Kawagoe N, Shintaku I, Yutani S, et al. Expression of the SART3 tumor rejection antigen in renal cell carcinoma. J Urol. 2000;164(6):2090–2095 [PubMed] [Google Scholar]
- 56. Medenbach J, Schreiner S, Liu S, Luhrmann R, Bindereif A. Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol Cell Biol. 2004;24(17):7392–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kelly DJ, Stein-Oakley A, Zhang Y, et al. Fas-induced apoptosis is a feature of progressive diabetic nephropathy in transgenic (mRen-2)27 rats: attenuation with renin-angiotensin blockade. Nephrology (Carlton). 2004;9(1):7–13 [DOI] [PubMed] [Google Scholar]
- 58. Fornoni A, Ijaz A, Tejada T, Lenz O. Role of inflammation in diabetic nephropathy. Curr Diabetes Rev. 2008;4(1):10–17 [DOI] [PubMed] [Google Scholar]
- 59. Vielhauer V, Mayadas TN. Functions of TNF and its receptors in renal disease: distinct roles in inflammatory tissue injury and immune regulation. Semin Nephrol. 2007;27(3):286–308 [DOI] [PubMed] [Google Scholar]
- 60. Scherberich JE. Proinflammatory blood monocytes: main effector and target cells in systemic and renal disease; background and therapeutic implications. Int J Clin Pharmacol Ther. 2003;41(10):459–464 [DOI] [PubMed] [Google Scholar]
- 61. Chuang PY, He JC. Signaling in regulation of podocyte phenotypes. Nephron Physiol. 2009;111(2):9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patel VA, Lee DJ, Longacre-Antoni A, et al. Apoptotic and necrotic cells as sentinels of local tissue stress and inflammation: response pathways initiated in nearby viable cells. Autoimmunity. 2009;42(4):317–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sander EE, van DS, ten Klooster JP, et al. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143(5):1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. la SA, Gadina M, Kelsall BL. G(i)-protein-dependent inhibition of IL-12 production is mediated by activation of the phosphatidylinositol 3-kinase-protein 3 kinase B/Akt pathway and JNK. J Immunol. 2005;175(5):2994–2999 [DOI] [PubMed] [Google Scholar]
- 65. Schwertschlag US, Trepicchio WL, Dykstra KH, Keith JC, Turner KJ, Dorner AJ. Hematopoietic, immunomodulatory and epithelial effects of interleukin-11. Leukemia. 1999;13(9):1307–1315 [DOI] [PubMed] [Google Scholar]
- 66. Rattner A, Hsieh JC, Smallwood PM, et al. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA. 1997;94(7):2859–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811–820 [DOI] [PubMed] [Google Scholar]
- 68. Zhang Z, Deb A, Zhang Z, et al. Secreted frizzled related protein 2 protects cells from apoptosis by blocking the effect of canonical Wnt3a. J Mol Cell Cardiol. 2009;46(3):370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhuang S, Yan Y, Daubert RA, Schnellmann RG. Epiregulin promotes proliferation and migration of renal proximal tubular cells. Am J Physiol Renal Physiol. 2007;293(1):F219–F226 [DOI] [PubMed] [Google Scholar]
- 70. Carev D, Saraga M, Saraga-Babic M. Expression of intermediate filaments, EGF and TGF-alpha in early human kidney development. J Mol Histol. 2008;39(2):227–235 [DOI] [PubMed] [Google Scholar]
- 71. McCright B, Gao X, Shen L, et al. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128(4):491–502 [DOI] [PubMed] [Google Scholar]
- 72. Chen L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol. 2005;288(5):F939–F952 [DOI] [PubMed] [Google Scholar]
- 73. Leimeister C, Schumacher N, Gessler M. Expression of Notch pathway genes in the embryonic mouse metanephros suggests a role in proximal tubule development. Gene Expr Patterns. 2003;3(5):595–598 [DOI] [PubMed] [Google Scholar]
- 74. Piscione TD, Wu MY, Quaggin SE. Expression of Hairy/Enhancer of Split genes, Hes1 and Hes5, during murine nephron morphogenesis. Gene Expr Patterns. 2004;4(6):707–711 [DOI] [PubMed] [Google Scholar]
- 75. Cheng HT, Miner JH, Lin M, Tansey MG, Roth K, Kopan R. Gamma-secretase activity is dispensable for mesenchyme-to-epithelium transition but required for podocyte and proximal tubule formation in developing mouse kidney. Development. 2003;130(20):5031–5042 [DOI] [PubMed] [Google Scholar]
- 76. Wang P, Pereira FA, Beasley D, Zheng H. Presenilins are required for the formation of comma- and S-shaped bodies during nephrogenesis. Development. 2003;130(20):5019–5029 [DOI] [PubMed] [Google Scholar]
- 77. Reidy KJ, Rosenblum ND. Cell and molecular biology of kidney development. Semin Nephrol. 2009;29(4):321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McDaniell R, Warthen DM, Sanchez-Lara PA, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79(1):169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(spec no 1):R9–R13 [DOI] [PubMed] [Google Scholar]
- 80. Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16(3):243–251 [DOI] [PubMed] [Google Scholar]
- 81. McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129(4):1075–1082 [DOI] [PubMed] [Google Scholar]
- 82. Herpin A, Cunningham C. Cross-talk between the bone morphogenetic protein pathway and other major signaling pathways results in tightly regulated cell-specific outcomes. FEBS J. 2007;274(12):2977–2985 [DOI] [PubMed] [Google Scholar]
- 83. Naylor RW, Jones EA. Notch activates Wnt-4 signalling to control medio-lateral patterning of the pronephros. Development. 2009;136(21):3585–3595 [DOI] [PubMed] [Google Scholar]
- 84. Wahl MB, Deng C, Lewandoski M, Pourquie O. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development. 2007;134(22):4033–4041 [DOI] [PubMed] [Google Scholar]
- 85. Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126(15):3415–3424 [DOI] [PubMed] [Google Scholar]