Abstract
Differences in gene expression and imprinting have been reported, comparing in vivo versus in vitro generated preimplantation embryos. Furthermore, mouse studies have shown that placenta development is altered following in vitro culture. However, the molecular mechanisms underlying these findings are unknown. We therefore isolated trophectoderm (TE) and inner cell mass (ICM) cells from in vivo and in vitro fertilization (IVF) embryos and evaluated their transcriptome using microarrays. We found that the transcriptomes of in vitro produced ICM and TE cells showed remarkably few differences compared to ICM and TE cells of in vivo generated embryos. In vitro fertilization embryos showed a reduced number of TE cells compared to in vivo embryos. In addition, TE of IVF embryos showed significant downregulation of solute transporter genes and of genes involved in placenta formation (Eomesodermin, Socs3) or implantation (Hbegf). In summary, IVF and embryo culture significantly affects the transcriptome of ICM and TE cells.
Keywords: ICM, TE, gene expression, mouse embryo, microarray
Introduction
The first differentiation event in the embryo occurs at the blastocyst stage, when the formation of trophectoderm (TE) and inner cell mass (ICM) occurs.1 The ICM will give rise to the embryo proper and extraembryonic tissues, while the TE will generate the placenta.2 Many morphological and functional changes characterize this differentiative event. For example, in addition to having a more restricted developmental potential than the ICM, the TE cells are epithelial and possess tight and gap junctional connections. In addition, they are polarized and capable of pumping fluid into the blastocyst to form the blastocoel. The ICM cells, in contrast, are not polarized and totipotent. Also, de novo methylation of genes occurs at a much higher level in the ICM than in the TE.3,4
This structural and functional differentiation is reflected in and caused by changes in the transcriptional profiles of each of these 2 cell types. Gene expression profiling provides a global picture of gene expression and has provided much information on the changes in gene expression that occur during preimplantation development. Various gene clusters have been identified based on their temporal pattern of expression and further investigation has led to the discovery of new genes.3
By the 64-cell stage, the blastomeres of developing preimplantation embryos differ in their gene expression patterns.5 In vitro derivatives of ICM (embryonic stem [ES] cells) and TE (trophoblast stem [TS] cells) have significant differences in their gene expression patterns.6
Embryos subjected to artificial reproductive technologies (ART), including in vitro fertilization (IVF), in vitro culture (IVC), and intracytoplasmic sperm injection (ICSI), must undergo preimplantation development including their first lineage differentiation under artificial and presumably suboptimal conditions. Disruption of normal fetal and placental development can occur when embryos are subjected to manipulations in vitro.7 While ART is currently considered to be safe, a series of reports have documented an increase in complications, including low-birth-weight singletons born at term,8 increased birth defects,8,9 and increased epigenetic disorders such as Angelman and Beckwith-Wiedemann syndromes.10 Interestingly, ART pregnancies display a significant increase in maternal complication that can be attributed to abnormal placentation, such as placental abruption, placenta praevia, excessive bleeding at the time of delivery, premature rupture of the membranes, and preeclampsia.11
While it is possible that infertility, per se, could increase these risks,12 it is also likely that changes in adult phenotype are secondary to the IVC of preimplantation embryos.13 While it is known that IVC14–16 and IVF17 of preimplantation mouse embryos alter the global gene expression patterns of whole blastocysts, it is unknown how the transcriptomes of TE and ICM cells are specifically affected by the culture process. This is important, since both IVF and IVC yield blastocysts with significantly fewer TE cells.17 In addition, we have found that when mouse IVF blastocysts are transferred to recipient females and allowed to develop to fetal stages, both the placentas and the fetuses have alteration in their size.18
In this report, we describe for the first time, the gene expression profile of mouse ICM and TE isolated from IVF embryos. While the gene expression patterns of TE and ICM from in vivo blastocysts are very different from each other, suggesting extensive differentiation, we found relatively little difference in the gene expression patterns of TE and ICM of IVF blastocysts. This indicates a significant alteration in the primary differentiation event of IVF blastocysts.
Materials and Methods
Embryo Collection and Separation of ICM and TE
In vitro and in vivo produced embryos were isolated from super-ovulated dams as previously described.17 Briefly, CF-1 female mice were injected with 5 IU Pregnant Mare Serum Gonadotropin and 46 to 48 hours later with 5 IU human chorionic gonadotropin (hCG). For the in vitro group, oocytes were obtained the following morning from the ampullae and incubated in Whitten medium (WM) containing 15 mg/mL bovine serum albumin (BSA) and sperm obtained from the cauda epididymis of male B6D2F1/J mice for 4 hours. Fertilized oocytes were washed and cultured in WM to the expanded blastocyst stage under 5% CO2 in humidified air at 37°C. In vivo blastocysts were flushed out of the uterus 94 to 96 hours post hCG.
It is possible to select embryos based on timing (eg, 96 hours after hCG) or based on morphology (similarly appearing blastocysts). Because embryos produced in vitro reach the blastocyst stage later than the in vivo developing embryos, we selected blastocysts using morphological, rather than temporal, criteria as we have done before.14,17 In fact, IVF embryos 96 hours post hCG were, on average, at the early blastocyst stage, while the majority of their in vivo counterparts were already at the expanded blastocyst stage. Expanded blastocysts of similar morphology were therefore collected 96 hours after hCG (in vivo) and 112 hours after hCG (IVF). We have previously shown that this experimental protocols results in IVF embryos having a similar number of ICM cells compared to in vivo embryos (12.8 ± 0.4 vs 13.8 ± 0.5, NS) but a reduced number of TE cells (33.5 ± 1.2 vs 49.6 ± 1.5; P < .05).17
Inner cell mass samples were collected by immunosurgery, and TE cells were collected by manual dissection, following published protocols.19,20 Different TE cell collection methods were utilized because the RNA quality (assessed by the bioanalyzer method21) of the TE cells post immunosurgery was consistently poor.
For immunosurgery, the TE cells were lysed by incubating embryos in 20 μg/mL anti-mouse rabbit antibodies (Sigma-Aldrich Corp, St Louis, Missouri) in WM for 30 minutes followed by 30-minute incubation in 5 μg/mL of rabbit complement (Invitrogen Corp, Carlsbad, California) in WM at 37°C. The ICM samples were cleaned of destroyed TE cells by repeated pipetting using a glass pipette with an inner diameter of 30 to 40µm. The TE samples were collected by manual dissection from whole blastocysts using a glass pipette with an inner diameter of 30 to 40 µm under dissecting microscope.22 Upon collection, the ICM and TE samples were immediately transferred to cell lysis buffer provided in the PicoPure RNA isolation kit (Molecular Devices, Sunnyvale, California) and frozen at −80°C. Collection of ICM was repeated 3 times for each treatment group, in vivo and in vitro. The same was done for TE collection. All animal experiments were approved by the Institutional Animal Care and Use Committee of University of California San Francisco.
RNA extraction and amplification
Total RNA was extracted using PicoPure RNA Isolation Kit (Molecular Devices) according to the manufacturer’s instructions from 3 pools of 40 ICM samples and 3 pools of 120 TE samples per treatment.17 A larger number of TE samples were collected to gather a sufficient amount of total RNA (>1.5 ng). In column DNase treatment was performed using RNase-free DNase set (Qiagen Inc, Valencia, California) as described in the user guide of the PicoPure RNA isolation kit to remove the residual DNA. Five hundred picogram of RNA from each sample was used for reverse transcription followed by linear amplification of antisense complementary DNA (cDNA) strand, fragmentation and biotin labeling using NuGEN WT-Ovation Pico System (NuGEN Technologies Inc, San Carlos, California). This is a robust method of amplification,23 especially well suited for experiments involving small amounts of starting material. Briefly, RNA is converted into cDNA with a unique DNA/RNA heteroduplex at one end. A linear isothermal DNA amplification process was conducted using DNA/RNA chimeric primer, DNA polymerase, and RNase H in a homogeneous isothermal assay that provides highly efficient amplification of DNA sequences. The amplified cDNA strands were subjected to an enzymatic fragmentation and the fragmented product was then labeled with biotin using NuGENs FL-Ovation cDNA Biotin V2 Kit (NuGEN Technologies Inc). RNA and cDNA mass and size distribution were determined before and after amplification and after fragmentation using the Agilent Bioanalyzer (Agilent Technologies Inc, Santa Clara, California). RNA samples with RNA integrity number 8 and above were selected for amplification. Complementary DNA yield before fragmentation and labeling was 6.6 to 7.4 µg for ICM and 5.0 to 7.8 µg for TE samples. For fragmentation, biotin labeling, and hybridization, 5 µg of amplified cDNA from each sample were used.
Microarray Preparation
Fragmented and biotin-labeled cDNA samples were submitted to the Genomic Core Facility of University of California San Francisco for GeneChip hybridization. The samples were hybridized to mouse Affymetrix 430 2.0 GeneChips that covers 39 000 transcripts or 15 288 genes. Samples were washed and stained on fluidic stations and scanned at 3 µm resolution according to the manufacturer’s instructions (GeneChip Analysis Technical Manual, www.affymetrix.com). Microarrays were repeated 3 times per treatment with amplified, fragmented, and labeled cDNA obtained from 3 different pools of ICM and TE samples.
Complementary DNA Preparation and Real-Time Polymerase Chain Reaction Analysis
ICM and TE of blastocysts were collected as described above and total RNA was isolated. Reverse transcription (RT) was accomplished by utilizing the commercially available first strand cDNA synthesis kit (iScript cDNA Synthesis Kit, Bio-Rad Laboratories, Hercules, California) following the kit manufacturer’s protocol, as we have done before.24 For each treatment group, the RT reaction was repeated 3 times with RNA from different sets of ICM and TE. To confirm the ability of this microarray analysis to resolve the differences in expression level, 6 genes that showed a statistically significant gene expression were selected.
Briefly, the real-time polymerase chain reaction (PCR) was performed using SyBr green PCR supermix (Bio-Rad Laboratories); primers for 3 ICM (Lifr, Sox2, and Fgf4),25,26 3 TE (Gata3, Cdx2, and Eomes) genes27,28 and Gapdh 24 with 0.1 embryo equivalent of cDNA of each treatment group were used. Primers were designed using Primer3 software. Triplicates were used for each real-time PCR reaction and a minus template served as control. The comparative threshold cycle method was used for quantification (Bio-Rad Laboratories).24
Microarray Analysis
Statistical analysis was carried out using the R software (http://www.r-project.org/) and the appropriate Bioconductor packages (http://www.bioconductor.org/) run under R (see below).
In order to remove all the possible sources of variation of a nonbiological origin between arrays, densitometry values between arrays were normalized using the RMA normalization function implemented in the Bioconductor affylmGUI. Statistically significant differences between groups were identified using the rank product nonparametric test implemented in the Bioconductor RankProd package. Applying a Student t test with such a limited amount of samples (3 in each experimental group) is problematic because the obtained statistical significance is not robust; in this situation the mean and the standard deviation could be easily biased by outliers; thus we have carried out a nonparametric statistical test as a rough filter to narrow down the list of most relevant genes. Moreover, the rank product approach includes a multiple hypothesis test for raw P value correction (here named pfp correction) to ascertain a false-positive rate similar to false discovery rate (FDR) correction.
Genes flagged as “present” or “marginal” in at least 1 hybridization based on raw perfect match (PM) and mismatch (MM) as determined by “mas5calls” function on the Bioconductor affy package were considered expressed genes.
Functional annotations were carried out using the Ingenuity Pathways Analysis platform (http://www.ingenuity.com/).
Results
Here, we report a detailed analysis of the transcriptome of TE and ICM cells derived from mouse blastocysts generated in vivo or in vitro. The in vitro embryos were cultured in suboptimal conditions, as a way to increase the detection rate of genes and pathways altered by the culture conditions.
Validation of Results and Identification of ICM and TE Marker Genes
To confirm the reliability and reproducibility of the microarray data, we performed multiple quality control analyses. First, when we performed in silico quality control analysis of the amplified material, we found that cDNA of 1 IVF sample (1 of the 3 samples of TE cells from IVF embryos) was suboptimal. Hence, this sample was eliminated for further analysis (data not shown).
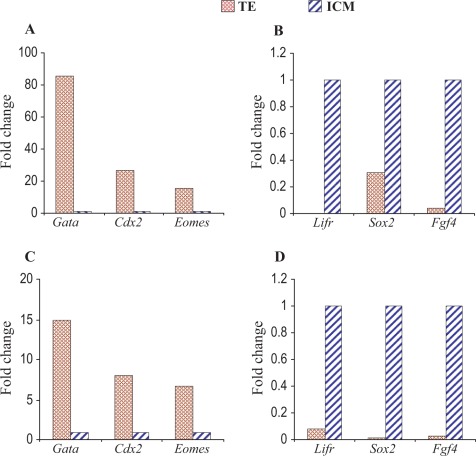
To demonstrate accurate collection and proper tissue separation, we selected several known marker genes of ICM (Lifr, Sox2, and Fgf4)25,26 and TE (Gata3, Cdx2, and Eomes)27,28 and performed real-time PCR. All the ICM marker genes showed appropriately higher expression in ICM cells, and TE marker genes showed higher expression in TE cells (Figure 1).
Figure 1.
Microarray validation by quantitative PCR in vivo samples. The PCR data confirmed the microarray results. Upper (A, B) and lower (C, D) panels show the real-time PCR and microarray data, respectively. Left panel (A, C) shows increased expression of Gata 3, Cdx2, and Eomes in TE cells. Expression of Gata 3, Cdx2, and Eomes in ICM samples was normalized to 1. Right panel (B, D) shows increased expression of Lifr, Sox2, and Fgf4 in ICM cells. Expression of Lifr, Sox2, and Fgf4 in TE samples was normalized to 1. PCR indicates polymerase chain reaction; ICM, inner cell mass; TE, trophectoderm.
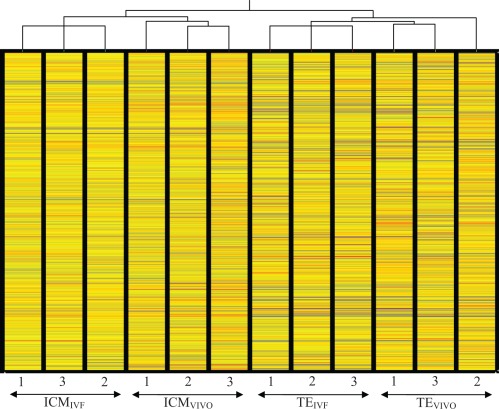
Finally, results of the unsupervised clustering (condition tree) analysis using all the entire probe set provided an excellent internal control: all the ICM samples clustered together and separately from the TE samples; in addition, the ICM and TE samples separated according to the method of fertilization (in vitro or in vivo), indicating that the collection of the samples was consistent and reliable (Figure 2).
Figure 2.
Unsupervised clustering (condition tree) of all expressed genes. Importantly the ICM samples and TE samples separated in 2 different groups, confirming an appropriate technical separation of the tissues. Furthermore, the IVF samples and in vivo samples of each cell type clustered separately, indicating an effect of the method of conception on the global pattern of gene expression of each cell types. IVF indicates in vitro fertilization.
Effect of IVF on the Differentiation Process
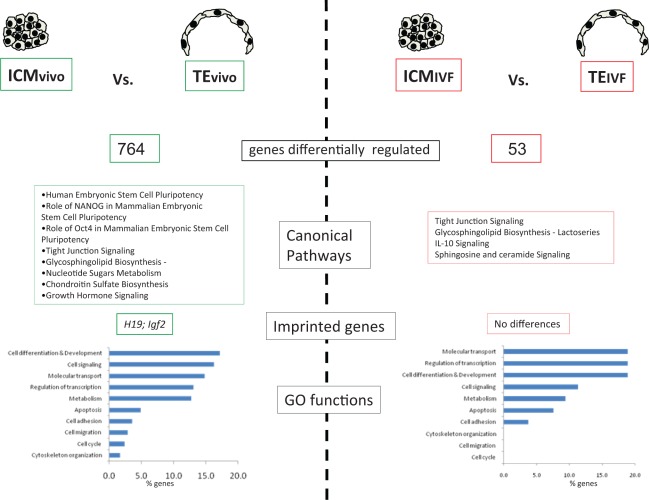
Overall, a similar but slightly reduced number of genes was expressed in TEIVF (n = 10 547) and ICMIVF (n = 11 170) of in vitro embryos compared to those of in vivo embryos (n = 10 687 and 11 302, respectively, P < .01). Strikingly, only 53 genes were differentially expressed comparing ICMIVF to TEIVF (Tables 1 and 2, Supplemental Table 1, and Figure 3), while 764 were differentially expressed between ICM and TE of in vivo embryos (Tables 1 and 2, Supplemental Table 1, and Figure 4). The small number of differentially expressed genes in the IVF embryos generally belonged to similar functional categories as those of the ICMvivo and TEvivo. For example, canonical pathway analysis revealed that genes involved in tight junction signaling and glycosphingolipid biosynthesis were altered both in vivo and in vitro generated embryos (Figure 3).
Table 2.
Up- and Downregulated Genes (and Probes) in the Different Comparisons According to Cell Type and Mode of Conceptiona
| Comparison | VIVO TE/ICM | IVFTE/ICM | ICM ivf/vivo | TEivf/vivo |
|---|---|---|---|---|
| Upregulated | 397 (883) | 45 (63) | 132 (196) | 20 (28) |
| Downregulated | 367 (829) | 8 (12) | 178 (293) | 89 (117) |
| Total | 764 (1712) | 53 (85) | 310 (489) | 109 (145) |
| Gene list | Supplemental Table 1a | Supplemental Table 1b | Supplemental Table 1c | Supplemental Table 1d |
Abbreviations: ICM, inner cell mass; TE, trophectoderm; IVF, in vitro fertilization.
a Corrected P value < .05.
Figure 3.
Difference in pathways and Gene Ontology (GO) functions between TEIVF and ICMIVF and between TEvivo and ICMvivo. The x-axis in the function graph reports the percentage of statistically different gene altered in each class out of the total genes belonging to that particular class. ICM indicates inner cell mass; TE, trophectoderm; IVF, in vitro fertilization.
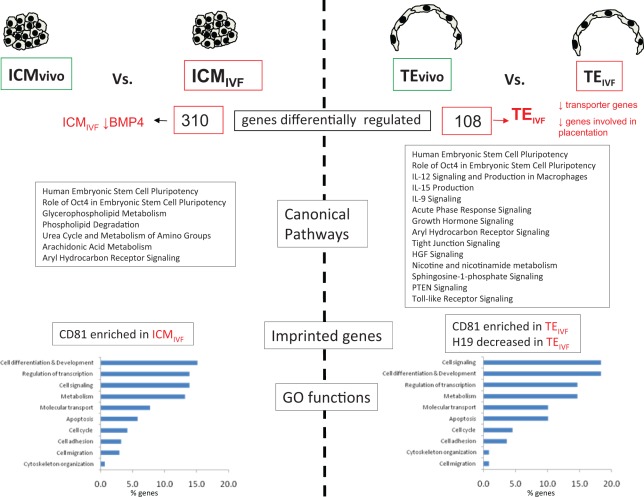
Figure 4.
Difference in pathways and Gene Ontology functions between TEvivo and TEIVF or ICMvivo and ICMIVF . The x-axis in the function graph reports the percentage of statistically different gene altered in each class out of the total genes belonging to that particular class. ICM indicates inner cell mass; TE, trophectoderm; IVF, in vitro fertilization.
Table 1.
Number and Percentage of Expressed Genes in Each Cellular Type
| Cellular Type | Expressed Genes | Percentage % |
|---|---|---|
| TEivf | 10 547 | 69.11 |
| TEvivo | 10 687 | 70.02 |
| ICMivf | 11 170 | 73.19 |
| ICMvivo | 11 302 | 74.05 |
Abbreviations: ICM, inner cell mass; TE, trophectoderm; IVF, in vitro fertilization.
We further analyzed the differences between in vivo and in vitro derived ICM and TE. Forty-nine (92%) out of 53 genes that were differentially expressed in the ICM and TE of in vitro embryos were also different between ICM and TE of in vivo embryos. Only 2 ICM genes (Socs3 and Tshz3) and 2 TE genes (Slfn10 and Morc4) were uniquely different in IVF embryo.
Comparison of the Transcriptome of ICM Obtained From in vivo and in vitro Produced Embryos
Inner cell mass of IVF or in vivo embryos differed in the expression of 310 genes (132 genes were more highly expressed in ICMIVF than in ICMvivo and 178 genes were more highly expressed in ICMvivo than in ICMIVF (Tables 1 and 2 and Supplemental Table 1).
In vitro fertilization and culture altered several fundamental processes of the ICMIVF cells (Figure 3), including specific functions like cell differentiation and development, cell signaling, metabolism, and regulation of transcriptions (more than 10% of genes belonging to these classes were different between ICMIVF and ICMvivo). In addition, Oct4 and human ES cell pluripotency pathways were altered, together with pathways involved in phospholipid metabolism. BMP4 a gene important in inducing differentiation of human ES cells into TE cells was downregulated in ICMIVF.
Comparison of the Transcriptome of TE Cells Obtained From in vivo and in vitro Produced Embryo
Trophectoderm of IVF or in vivo embryos differed in the expression of 108 genes. Compared to TEvivo, 20 genes were more highly expressed and 88 genes were less highly expressed in TEIVF (Tables 1 and 2 and Supplemental Table 1).
In TEIVF genes involved in cell cycle, apoptosis, connective tissue development, implantation, and metabolism (lipid, carbohydrate and protein) were differentially regulated compared to TEvivo (Figure 4). Several pathways were different between TE cells of IVF and in vivo embryos. Oct4 and human ES cell pluripotency pathways were altered in TEIVF compared to TEvivo, similarly to what was found in the ICM comparison. In addition, interleukin (IL), toll-like receptor, and tight junction signaling were altered.
Numerous genes involved in solute transport (Slc24a5, Slc39a2, Slc30a10, Slc43a2, Slc7a3, Slc18a2, Slc35f3, and Slc22a4) and placentation (Socs3, Eomes, and Hbegf) were downregulated in TEIVF (Supplemental Table 1).
Methylation and Imprinted Genes
Overall few imprinted genes or genes involved in the DNA methylation process were differentially expressed between ICM and TE cells. Only, the methylation gene Mbd2 was expressed at a higher level in TEvivo compared to ICMvivo, while no methylation genes were different in ICMIVF and TEIVF.
Two imprinted genes (H19 and Igf2) were found to be differentially expressed between TEvivo and ICMvivo, while none was different between TEIVF and ICMIVF (Figure 4). On the contrary, CD81 was enriched in both TE and ICM following IVC (Figure 3).
Because imprinted genes tend to cluster in specific chromosomal locations, we further investigated whether certain chromosomes or chromosomal locations showed increased misregulation of genes in IVF embryos. Analysis revealed that the misexpressed genes did not locate in preferential loci (data not shown).
Discussion
In this report, we have measured the differences in gene expression of isolated, normal mouse ICM and TE from in vivo conceived blastocysts and compared them to the ICM and TE of IVF embryos. This analysis has led us to several conclusions.
First, ICM and TE from in vivo blastocysts, in agreement with data published by Guo et al, exhibit many differences in their transcriptomes.5 In contrast, the transcriptomes of ICM and TE from IVF blastocysts showed strikingly few differences from each other, indicating that there is a muted level of differentiation between the 2 groups of cells following IVF. In particular, only 53 genes were statistically different between TEIVF and ICMIVF, while 764 were different between ICMvivo and TEvivo (Tables 1 and 2 and Supplemental Table 1). In other words, the transcriptomes of ICMIVF and TEIVF cells are more similar than the transcriptomes of ICMvivo and TEvivo cells. These results, together with our prior findings that 1912 genes are different between whole blastocysts produced in vivo and IVF blastocysts,17 offer important insights. It is possible that IVC challenges the embryo, with many (1912) genes being differentially expressed, both in the ICM and in the TE. At the same time, there is a reduced activity of other “nonstress” pathways with only 53 genes being different between ICM and TE. In contrast, in vivo embryos have a more varied gene expression pattern.
Supporting these findings, the imprinted genes H19 and Igf2 were not differentially expressed in TEIVF and ICMIVF, while both genes were differentially expressed between TEvivo and ICMvivo (both increased in TE cells). The lack of increase of Igf2 in TEIVF is potentially significant, as deletion of the placental specific Igf2 gene leads to reduced growth of the placenta, followed several days later by fetal growth restriction.29 It is well known that imprinting errors can occur following culture in vitro,30 and indeed an increase in imprinting disorders has been suggested following human ART.31 The increased expression of the imprinted gene CD81 noted in both TEIVF and ICMIVF compared to TEvivo and ICMvivo is notable but of unclear significance. CD81 is a tetraspanin involved in reorganizing the cell membrane and stabilizing signaling molecules in response to different molecules such as integrins and Major histocompatibility complex (MHC) class II.32
A second significant finding is that messenger RNA (mRNA) of numerous transporter genes is altered in TEIVF. Our prior work has shown that transporter genes were increasingly misregulated in whole blastocysts cultured in WM—a suboptimal medium compared to Potassium Simplex Optimized Medium (KSOM) with amino acids—an optimal medium.14 These current experiments showed that the majority of transporter genes are downregulated in TEIVF (Supplemental Table 1). For example, the zinc transporter Slc39a2 is transiently expressed in trophoblast giant cells,33 the first definitive cell type to differentiate after fertilization. After implantation, giant cells will form part of the parietal yolk sac. Downregulation of Slc39a2 suggests a delay in the differentiation of trophoblast giant cells or an impairment of their functioning.
Third, the in vitro process seems to alter genes involved in implantation and placentation. Overall, the comparison of TEIVF and TEvivo suggests increased stress following IVF; there is a dysregulation of acute phase response signaling, inflammation (IL-9, IL-12, and IL-15 pathways) and apoptosis (sphingosine signaling). This early activation of stress-related pathways may have effects on later placentation. Indeed we have found that placentas of mice produced by IVF have altered fetal/placenta ratio compared to in vivo produced animals, after controlling for developmental stage.18 Among genes decreased in TEIVF compared to TEvivo, the downregulation of Eomesodermin (Eomes decreased 2.3-fold) and heparin-binding Epidermal growth factor (EGF)-like growth factor (Hbegf decreased 2.9-fold) is particularly significant. Eomes is a T-box gene essential for mesoderm formation; mouse embryos lacking Eomes arrest at the blastocyst stage because the trophoectoderm does not differentiate into trophoblast.34 Hbegf is expressed both in trophoblast cells throughout gestation and in the endometrium, and it has been involved in implantation.35 Of the genes uniquely different between TEIVF and ICMIVF, important is the downregulation of Suppressor of Cytokine signaling 3, (Socs3, downregulated 7.9 fold in TEIVF) as this gene is involved in placentation.36 Mice with a deletion of Socs3 die at midgestation due to placental insufficiency, and Socs3 null placentae have increased numbers of mature trophoblast giant cells, disruption of the labyrinthine layer, and a decrease in the spongiotrophoblast layer.36
BMP4 is another key signaling molecule downregulated in ICMIVF compared to ICMvivo. BMP4 is known to be expressed in ICM37 and when added to culture media induce differentiation of human ES cells into TE cells.38 It is tempting to postulate that the reduced BMP4 present in ICMIVF is an additional factor responsible for the reduced number of TE cells noted in IVF embryos.17
One interesting observation by Pampfer is that ICM cells are more sensitive to embryotoxic agents than TE and are predisposed to apoptosis.39 This author proposes that this is due to the distinct microenvironments (internal blastocoel fluid for the ICM vs external uterine fluid for TE) these cells are exposed to during development. Our data analysis suggests a more complex interpretation. Supporting the Pampfer statement is the finding that more genes (310 vs 108) were dysregulated in ICM compared to TE following IVF. On the other hand, we have shown that IVF embryos have reduced TE cell number17; in addition, there are more apoptosis genes differentially expressed between TEIVF and TEvivo (10%) than ICMIVF and ICMvivo (5%); finally the fold changes for ICMIVF over ICMvivo are all relatively small (∼ 4-fold) in comparison to the differences observed in TEIVF over TEvivo (∼ 8-fold). Taken together, these findings suggest that both cell populations are affected by the IVC but in different fashions.
Our results show similarity and also some notable differences with a similar study performed using human embryos.19 Overall, similar developmental pathways are represented in ICM and TE of human and mouse embryos. However, Adjaye et al19 identified 78 genes differentially expressed in ICM and TE within 1 of the blastocysts and none of these genes match the 53 genes that are statistically different in ICM and TE cells of IVF mouse embryos. Instead, 2 of the TE markers (Camk1 and Fabp3) were found to correspond to differentially expressed genes in in vivo mouse embryos. This difference may be either due to species-related differences or due to differences in protocol used. The Adjaye study is significantly different because these authors compared gene expression difference in ICM and TE in IVF human blastocysts, while we compared ICM and TE differences in IVF and in vivo mouse blastocysts. In addition, their work was conducted using a total of only 2 embryos, because of the obvious limited availability of embryos.
The data presented should be interpreted with a few caveats. In the current study, we used suboptimal culture conditions (WM) to amplify the effects of IVC and, as in a pharmacologic experiment, increase the detection rate of genes and pathways affected by the in vitro differentiation process. This strategy, though it does not allow a direct comparison with the culture media used in current human IVF clinics, increases the discovery rate of abnormal gene expression and more clearly point to pathways that might be altered after culture in vitro.
Inner cell mass cells were separated by immunosurgery,19 while collection of TE cells was performed by manual dissection. The reason to use this approach was that the quality of RNA obtained from TE cells after immunosurgery19 was consistently poor based on bioanalyzer traces. In particular, the lysate of TE cells showed a significant RNA degradation and was not suitable for amplification and microarray analysis. By contrast, the manual separation method resulted in RNA of excellent quality. A potential limitation of this method of isolation is that not all TE cells are collected, but only the TE cells not in contact with the ICM, the cells that later will become mural TE cells. Mural TE gives rise to trophoblastic giant cells, while polar TE, formed by TE in direct contact with the ICM, results in the ectoplacental cone and extraembryonic ectoderm.40
Finally, it is important to note that results provided in this manuscript might be specific for the strain of mice tested (CF1 X B6D2F1/J) and could be partially different in other mice strains41 or different species. In addition, data using microarray might be incomplete compared to the information obtained using Whole Transcriptome Shotgun Sequencing.42 In fact, RNA sequencing methods provide a wider range of information, allowing detection of posttranscriptional mutations, identification of noncoding RNAs, and recognition of allelic variants. However, next generation sequencing approaches can be 10- to 100-fold more expansive than microarray and can suffer from technical bias related to ligation steps and complex computational analysis42
In summary, these data offer a comprehensive and robust analysis of gene expression in ICM and TE of mouse embryos and provide fundamental resources for understanding how culture conditions affect differentiation and how gene expression is involved in early tissue differentiation. Importantly, we found that the differential gene expression between TE and ICM cells is blunted by IVF and culture. Analysis of the data provided can be useful to identify genes and transcription factors critical for early cell differentiation during normal development as well. Future work must examine the mechanisms by which this altered gene expression pattern leads to fewer TE in cultured embryos, suboptimal placentation, and possibly defects within the embryo and fetus.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by NICHD/NIH through cooperative agreement 1U54HD055764 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. JAH was funded by the José Castillejo’s scholarship from the Spanish Government.
References
- 1. Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol. 2008;322(1):133–144 [DOI] [PubMed] [Google Scholar]
- 2. Dyce J, George M, Goodall H, Fleming TP. Do trophectoderm and inner cell mass cells in the mouse blastocyst maintain discrete lineages? Development. 1987;100(4):685–698 [DOI] [PubMed] [Google Scholar]
- 3. Ko MS. Expression profiling of the mouse early embryo: reflections and perspectives. Dev Dyn. 2006;235(9):2437–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Senner CE, Hemberger M. Regulation of early trophoblast differentiation - lessons from the mouse. Placenta. 2010;31(11):944–950 [DOI] [PubMed] [Google Scholar]
- 5. Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–685 [DOI] [PubMed] [Google Scholar]
- 6. Tanaka TS, Kunath T, Kimber WL, et al. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12(12):1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farin CE, Farmer WT, Farin PW. Pregnancy recognition and abnormal offspring syndrome in cattle. Reprod Fertil Dev. 2010;22(1):75–87 [DOI] [PubMed] [Google Scholar]
- 8. Schieve LA, Meikle SF, Ferre C, et al. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346(10):731–737 [DOI] [PubMed] [Google Scholar]
- 9. Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346(10) 725–730 [DOI] [PubMed] [Google Scholar]
- 10. DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson RA, Gibson KA, Wu YW, Croughan MS. Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstet Gynecol. 2004;103(3):551–563 [DOI] [PubMed] [Google Scholar]
- 12. Kovalevsky G, Rinaudo P, Coutifaris C. Do assisted reproductive technologies cause adverse fetal outcomes? Fertil Steril. 2003;79(6):1270–1272 [DOI] [PubMed] [Google Scholar]
- 13. Rinaudo PF, Lamb J. Fetal origins of perinatal morbidity and/or adult disease. Semin Reprod Med. 2008;26(5):436–445 [DOI] [PubMed] [Google Scholar]
- 14. Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128(3):301–311 [DOI] [PubMed] [Google Scholar]
- 15. Rinaudo PF, Giritharan G, Talbi S, Dobson AT, Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril. 2006;86(suppl 4): 1265: 1252–1265, e1–e36 [DOI] [PubMed] [Google Scholar]
- 16. Fernandez-Gonzalez R, de Dios Hourcade J, López-Vidriero I, et al. Analysis of gene transcription alterations at the blastocyst stage related to the long-term consequences of in vitro culture in mice. Reproduction. 2009;137(2):271–283 [DOI] [PubMed] [Google Scholar]
- 17. Giritharan G, Talbi S, Donjacour A, et al. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction. 2007;134(1):63–72 [DOI] [PubMed] [Google Scholar]
- 18. Delle Piane L, Lin W, Liu X, et al. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod. 2010;25(8):2039–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adjaye J, Huntriss J, Herwig R, et al. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23(10):1514–1525 [DOI] [PubMed] [Google Scholar]
- 20. Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci U S A. 1975;72(12):5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imbeaud S, Graudens E, Boulanger V, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33(6):e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansis C, Grifo JA, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol Hum Reprod. 2000;6(11):999–1004 [DOI] [PubMed] [Google Scholar]
- 23. Barker CS, Griffin C, Dolganov GM, et al. Increased DNA microarray hybridization specificity using sscDNA targets. BMC Genomics. 2005;6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giritharan G, Li MW, De Sebastiano F, et al. Effect of ICSI on gene expression and development of mouse preimplantation embryos. Hum Reprod. (2010);25(12):3012–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rossant J, Chazaud C, Yamanaka Y. Lineage allocation and asymmetries in the early mouse embryo. Philos Trans R Soc Lond. 2003;358(1436):1341–1348; discussion 1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nichols J, Davidson D, Taga T, et al. Complementary tissue-specific expression of LIF and LIF-receptor mRNAs in early mouse embryogenesis. Mech Dev 1996;57(2):123–131 [DOI] [PubMed] [Google Scholar]
- 27. Home P, Ray S, Dutta D, et al. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284(42):28729–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts RM, Ezashi T, Das P. Trophoblast gene expression: transcription factors in the specification of early trophoblast. Reprod Biol Endocrinol. 2004;2:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Constancia M, Hemberger M, Hughes J, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417(6892):945–948 [DOI] [PubMed] [Google Scholar]
- 30. Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62(6):1526–1535 [DOI] [PubMed] [Google Scholar]
- 31. Sutcliffe AG, Peters CJ, Bowdin S, et al. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod. 2006;21(4):1009–1011 [DOI] [PubMed] [Google Scholar]
- 32. Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda). 2005;20:218–224 [DOI] [PubMed] [Google Scholar]
- 33. Peters JL, Dufner-Beattie J, Xu W, et al. Targeting of the mouse Slc39a2 (Zip2) gene reveals highly cell-specific patterns of expression, and unique functions in zinc, iron, and calcium homeostasis. Genesis. 2007;45(6):339–352 [DOI] [PubMed] [Google Scholar]
- 34. Russ AP, Wattler S, Colledge WH, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404(6773):95–99 [DOI] [PubMed] [Google Scholar]
- 35. Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7(3):185–199 [DOI] [PubMed] [Google Scholar]
- 36. Boyle K, Robb L. The role of SOCS3 in modulating leukaemia inhibitory factor signalling during murine placental development. J Reprod Immunol. 2008;77(1):1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pant D, Keefer CL. Expression of pluripotency-related genes during bovine inner cell mass explant culture. Cloning Stem Cells. 2009;11(3):355–365 [DOI] [PubMed] [Google Scholar]
- 38. Xu RH, Chen X, Li DS, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20(12):1261–1264 [DOI] [PubMed] [Google Scholar]
- 39. Pampfer S. Apoptosis in rodent peri-implantation embryos: differential susceptibility of inner cell mass and trophectoderm cell lineages—a review. Placenta. 2000;21(suppl A):S3–S10 [DOI] [PubMed] [Google Scholar]
- 40. Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2(7):538–548 [DOI] [PubMed] [Google Scholar]
- 41. Eaves IA, Wicker LS, Ghandour G, et al. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res. 2002;12(2):232–243 [PubMed] [Google Scholar]
- 42. Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5(7):585–587 [DOI] [PubMed] [Google Scholar]