Abstract
The proton pores of F1Fo-ATP synthases consist of a ring of c-subunits, which rotates driven by downhill proton diffusion. An essential carboxylate side chain in the c-subunit provides a proton-binding site. In all the structures of c-rings reported to date, these sites are in a closed, ion-locked state. Structures are presented of the c10 ring from Saccharomyces cerevisiae determined at pH 8.3, 6.1 and 5.5, at resolutions of 2.0 Å, 2.5 Å and 2.0 Å, respectively. The overall structure of this mitochondrial c-ring is similar to known homologues, except that the essential carboxylate, Glu59, adopts an open extended conformation. Molecular dynamics simulations reveal that opening of the essential carboxylate is a consequence of the amphiphilic nature of the crystallization buffer. We propose that this new structure represents the functionally open form of the c-subunit, which facilitates proton loading and release.
Keywords: Mitochondrial bioenergetics, membrane protein complex, proton transport, membrane protein, proton pore, subunit 9, subunit c
Mitochondrial F1Fo-ATP synthases are molecular motors that utilize the proton gradient across the inner mitochondrial membrane to phosphorylate ADP, releasing ATP into the matrix. The enzyme is composed of two tightly coupled protein subcomplexes: the water-soluble F1 contains the active site, and consists of subunits α3β3γδε1; the membrane-bound Fo subcomplex, which is minimally composed of subunits a, b, and a ring-like assembly of 8 or 10 c-subunits, functions as a proton pore2–4. The mechanism of this proton pore resembles that of a turbine, with the c-subunit ring spinning within the membrane, driven by proton movement down an electrochemical gradient. The c-ring is also in contact with the central stalk of F14–6, which consists of subunits γδε. Rotation of the stalk within the core of F1 effects sequential conformational changes within the three active sites, which ultimately catalyze ATP synthesis and release7,8.
High-resolution structures of either a complete mitochondrial ATP synthase or an isolated mitochondrial c-ring have not yet been determined. However, high-resolution crystal structures of F1 have been obtained, from yeast and bovine heart1,4–6,9–12, as well as of the peripheral stalk (subunits b, d and OSCP)9. Various structures have also been reported of subcomplexes of the ATP synthase, namely F1c10 from S. cerevisiae and F1c8 from bovine heart4–6. In addition, several medium to high-resolution structures have been solved of isolated c-rings from other species, both from H+ and Na+-driven ATP synthases (and a related V-type ATPase)13–20.
Current models of the proton-conducting mechanism21–23 postulate that a strictly conserved Glu or Asp in the c-subunit (Glu59 in S. cerevisiae) plays an essential role, acting as the proton acceptor and donor at the interface of the c-ring with subunit-a. Crystal structures of crings clearly demonstrate that this side chain provides an ion-binding site (for H+ or Na+) at each of the subunit-c:c interfaces. However, in all published structures, these sites are observed in the closed, ion-locked conformation, which optimizes the coordination of the ion, presumably countering the energy cost of its dehydration14. Indeed, molecular dynamics simulations of these c-rings indicate that this closed state is the most favored energetically when the c-subunit binding sites face the hydrophobic core of the lipid bilayer, as the c-ring rotates in the membrane22.
Nevertheless, simulations also indicate that local hydration of the binding sites, which is believed to occur at the a:c interface24,25, would promote a transition to an open conformation, in which protonation and deprotonation would be facilitated22. This hypothetical open conformation has not yet been experimentally observed in an intact c-ring, but it was thought to resemble that observed in crystal structures of two bacterial rings after covalent modification of the essential carboxyl side chain with dicyclohexylcarbodiimide (DCCD)19,22. However, these structures may reflect an artifact caused by DCCD, which is much too large to fit within the closed binding site. Thus, it remains uncertain whether the c-subunit undergoes a transition from a closed to open state in the proton translocation pathway, and if so, whether this exchange entails local or global structural changes26,27. Further, the hypothesis that a hydrated microenvironment is sufficient to promote opening and reversible protonation of the binding site22 has not been experimentally verified. The high resolution structures of the intact mitochondrial c10 ring presented here, at pH 5.5, 6.1, and 8.3, as well as with bound DCCD at pH 5.5, reveal theproton-binding sites in the open state and at different pH values, and provides answers to these mechanistic questions.
Results
Crystallization of S. cerevisiae mitochondrial ATP synthase c10 ring
The conditions used to crystallize the yeast mitochondrial ATP synthase c-ring differed radically from those used previously. The primary difference is that the c-ring was crystallized in a buffer containing 2-methyl-2,4-pentandiol (MPD) and propylene glycol (PG) (MPD:PG, 34%:4% (v/v) initially, 67%:8% (v/v) in the reservoir), rather in a buffer containing detergent with polyethylene glycol as the primary precipitant (Methods). Crystals were obtained within a week, and subsequently grew for three or more weeks, reaching up to 200 μm in the largest dimension. Native data sets were obtained from single crystals that diffracted to a resolution of 2.0 Å, 2.5 Å and 2.0 Å, at pH 8.3, pH 6.1 and pH 5.5, respectively (Table 1). The crystallographic space group of the crystals was P4222, with 40 c-subunits in the unit cell and 5 subunits per asymmetric unit. The 5 subunits in the asymmetric unit form half of a complete c-ring. The crystal lattice resembles Type I packing of integral membrane proteins with stacked head-to-head 2D layers28. The solvent content, 47%, while normal for crystals formed from a water-soluble protein, is rather low as compared to crystals formed from integral membrane proteins. This tight packing may be responsible for the high-resolution diffraction qualities of these crystals. The structure was solved by molecular replacement using half a c-ring from the low-resolution structure of yeast F1c10 (PDB entry 2XOK)4. The resulting electron density map was clearly traceable and yielded an unambiguous model of the 5 c-subunits, each containing 74–75 amino acids, without any outliers in the Ramachandran plot. The refinement did not use non-crystallographic constraints or restraints and thus each of the 5 c-subunit structures is unique, although they are nearly identical, as evidenced by an average pair wise RMSD of just 0.18 Å. The average B-factors throughout structure were remarkably low for a membrane protein. For example, for the structure solved from crystals at pH, 5.5, the average B-factor was 23.2 Å2, including 85 assigned waters. In general, the magnitude of the B-factors for the backbone atoms showed a gradient distribution, with the atoms that would lie on the mitochondrial matrix space having the lowest B-factor and the atoms closer to what would be the intermembrane space, having the highest B-factor (Supplement Fig. 1). There are residual densities within the central opening of the ring, which might be due to lipids or detergent molecules though the shape of the density was not distinct and could not be modeled. The absence of lipid or detergent densities at such high resolution underscores the drastic effect of the MPD:propylene glycol solution, which apparently replaces lipid and detergent molecules from the hydrophobic surface of the c-ring.
Table 1.
Data collection and refinement statistics.
| pH 8.3 | pH 6.1 | pH 5.5 | pH 5.5 + DCCD | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P4222 | P4222 | P4222 | P4222 |
| Cell dimensions | ||||
| a, b, c (Å) | 54.2, 54.2, 246.0 | 54.1, 54,1, 244.8 | 54.2, 54,2, 245.4 | 54.6, 54.6, 244.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0 90.0 |
| Resolution (Å) | 50-2.0 (2.07-2.0)* | 50-2.5 (2.59-2.5) | 50-2.0 (2.07-2.0) | 50-2.0 (2.07-2.0) |
| Rmerge | 0.08 (0.276) | 0.164 (0.687) | 0.08 (0.217) | 0.112 (0.634) |
| I/σI | 13.8 (3.2) | 4.6 (3.5) | 15.8 (3.3) | 12.6 (2.7) |
| Completeness (%) | 98.1(96.4) | 99.9 (100.0) | 95.0 (92.9) | 95.7 (91.3) |
| Redundancy | 8.6 (7.7) | 6.8 (6.9) | 4.3 (4.2) | 3.9 (2.8) |
| Matthews Coeff. (Å3/Da) | 2.32 | 2.3 | 2.31 | 2.28 |
| Solvent Content (%) | 46.9 | 46.6 | 46.8 | 46.0 |
| Refinement | ||||
| Resolution (Å) | 50-2.0 (2.05-2.0) | 50-2.5 (2.57-2.5) | 50-2.0 (2.06-2.0) | 50-2.0 (2.05-2.0) |
| No. reflections | 24,159 | 12,750 | 23,261 | 23,303 |
| Rwork/Rfree | 0.192/0.216 | 0.192/0.227 | 0.200/0.221 | 0.224/0.254 |
| No. atoms | ||||
| Protein | 2643 | 2655 | 2644 | 2624 |
| Water | 67 | 42 | 85 | 55 |
| B-factors | ||||
| Protein | 25.4 | 26.4 | 23.0 | 39.5 |
| Water | 38.1 | 31.0 | 33.9 | 43.9 |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.011 | 0.016 | 0.008 | 0.012 |
| Bond angles (°) | 1.042 | 1.454 | 1.035 | 1.213 |
Values in parentheses are for highest-resolution shell. The data was collected from one crystal at each pH/condition.
Architecture of the c10 ring
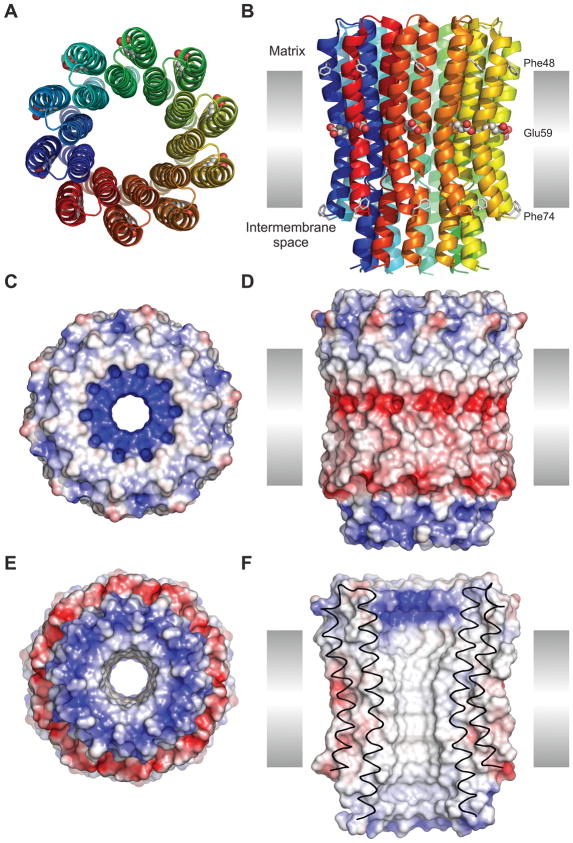
The overall structure of the c10 ring is shown in Fig. 1. Each subunit-c fold into a hairpin of two transmembrane-helices, connected by a short, partially structured loop (Supplement Fig. 2). The N-terminal helix (residues 3–39) is interrupted by a two-turn 310 helix, which includes an interhelical water molecule H-bonded with Thr16 (residues 15–18), while the C-terminal α-helix runs continuously across the membrane (residues 44–73). At the end of the N-terminal helix, Asn40 forms a sharp turn leading into another short 310 helix (residues 41–43) that precedes the C-terminal transmembrane span. Together with Arg39, Pro41, and Ser42, these form a highly conserved region, which apparently form an interaction motif with subunits γ, δ, and ε of the central stalk of the F1 complex4,6,29,30. Consequently, the N- and C-termini are located in the intermembrane mitochondrial space.
Figure 1. Structure of the c10 ring from yeast ATP synthase.
This figure is derived from the data obtained from the crystals grown at pH 8.3, but the results do not differ for the data sets from crystals at pH 5.5 or 6.1. A: Cartoon representation of the ring, viewed from the mitochondrial matrix, with each c-subunit in a different color. The conserved carboxyl side chain of Glu59 is shown with spheres. B: Same as A, viewed along the plane of the inner membrane (35 Å height). Phe48 and Phe74 (putative membrane limits) are represented with sticks. C: Electrostatic potential mapped on the surface of the c-ring, with positive areas in blue, negative in red, and neutral in white (color scale from −100 to 100 kBT/e). The ring is viewed from the matrix. D, E: Same as C, viewed along the plane of the membrane and from the inter-membrane space, respectively. F: Same as D, viewed in cross-section, illustrating the central hydrophobic opening of the ring.
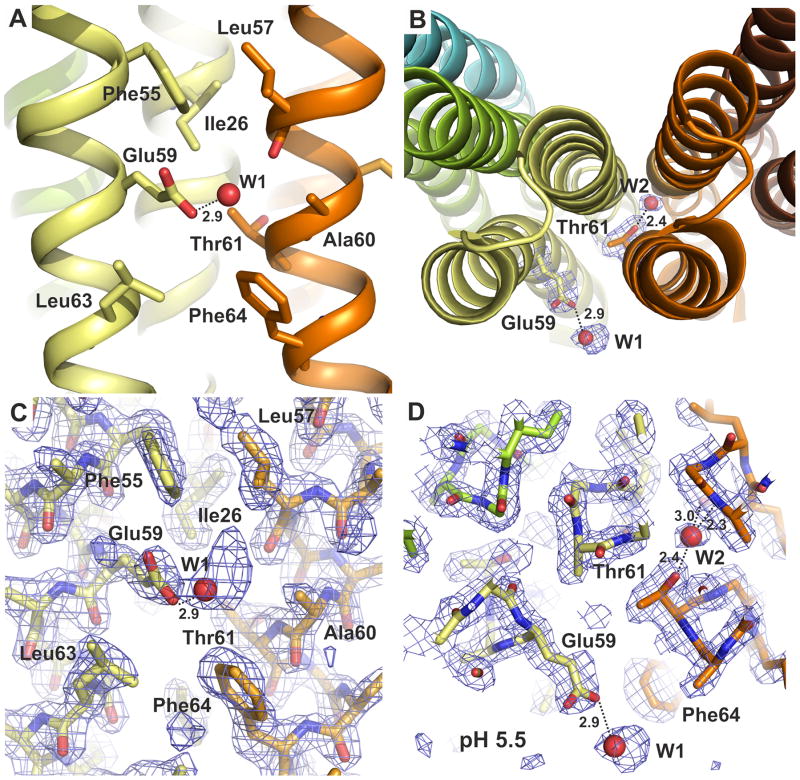
The assembled c-ring features the prototypical hourglass shape, with the N-terminal helices on the inside of the ring and the C-terminal helices facing the outside. Both the inner and outer rings of N-terminal and C-terminal helices are held together mostly by hydrophobic contacts, with few hydrogen-bonding interactions (Supplement Fig. 3). A second interhelical water molecule is observed, in this case associated with Thr61 and bridging adjacent helices (Fig. 2), much like the water molecule seen near Thr16 (Supplement Fig. 2). As in the bacterial c-rings of known structure, the inner and outer rings are staggered relative to each other.
Figure 2. Structure of the c10 ring proton-binding site.
A: Close-up of the proton-binding site, viewed along the plane of the membrane. Residues in the vicinity are indicated, including the key carboxyl side chain, Glu59, which projects away from the binding site, and appears to form a hydrogen-bond with a solvent molecule – probably a water or the hydroxyl group of an MPD molecule. B: The proton-binding site, viewed from the matrix side of the membrane. Note that additional water molecules bridge interactions between helices, e.g. near Thr61. C, D: 2Fo-Fc electron density maps in the region of the proton-binding sites, viewed as in A, B. The maps are contoured at 1.2σ.
Given that the width of the mitochondrial membrane is ~35 Å, the transmembrane span of the c-ring would extend from Phe48 to Phe74 in the outer helix and from Ile10 to Ile34 in the inner helix (Fig. 1). In this region, with the exception of Glu59, the surface of the c-ring is hydrophobic. On the matrix side of the protein surface, Arg39 creates a ring of strong positive electrostatic potential, which likely mediates the interaction of the c-ring with the central stalk of F1. On the opposite side facing the intermembrane space, the surface features two concentric rings of both positive and negative electrostatic potentials. The positive potential is due to the exposed backbone and side-chain amine dipoles of Gln2 and the N-formyl methionine at the N-terminus, while the negative region is primarily due to the charged C-terminus. As an aside, direct evidence for N-formyl methionine as the initiator amino acid in mitochondrial proteins has been rather limited. This high-resolution structure of the yeast c10 ring is the first crystallographic evidence demonstrating the formylation of the N-terminus of a mitochondrial protein31–36.
Conformation and protonation state of the binding site
A remarkable feature of the crystal structure is the conformation of Glu59, the key carboxylate in the outer helix of each c-subunit. In all published crystal structures of proton binding c-rings, this side chain is oriented towards the adjacent c-subunit, and forms a hydrogen-bond (as donor) with its backbone, either directly16 or mediated by a water molecule18,37. A similar conformation has been observed in Na+ binding crings14,15. By contrast, in this structure the carboxyl side chain of Glu59 projects away from the cring, and does not engage in any interactions with other residues within the c-ring (Fig. 2). Instead, electron density for a solvent molecule is observed within 2.9 Å of the carboxyl group, with which it seems to form a H-bond (Fig. 2C, D). This electron density is visible in all 5 c-subunits in the asymmetric unit, and was modeled as a water molecule, although there are other possibilities, such as a hydroxyl group of MPD; the electron density is particularly well defined in the structure grown at pH 6.1.
A water molecule has been observed to contribute to ion-coordination in the binding sites of other c-rings, in the closed state13,18,37. In the open conformation of the binding sites in the S. cerevisiae c10 ring, the corresponding region appears to be empty or any bound water is disordered. Nevertheless, it is possible that in the closed state a water molecule is part of the ion interaction network, as previously proposed6. A high-resolution structure of the closed conformation will be required to definitively resolve this question.
The pKa value of Glu59 in the S. cerevisiae c-ring has not been determined experimentally. However, indirect measurements have been made for the analogous side chain in bacterial c-subunits. For example, the pKa of Asp61 in the Escherichia coli c-subunit was determined by NMR to be 7.1 in a solvent composed of CHCl3:CH3OH:H2O (4:4:1), whose approximate dielectric constant is 2638. In comparison, the estimated dielectric constant of 67% MPD (v/v) in water is 43. Thus it is reasonable to estimate that the pKa value for Glu59 in the yeast c10 ring is similar, but not greater than 7.1, under the crystallization conditions used in this study. It is therefore worth noting that in these new structures, the proton binding sites are observed in the same open conformation at pH 5.5, 6.1 and 8.3, and therefore Glu59 is represented, to some degree, in both the ionized and protonated forms. In all cases, the crystal unit cells are nearly identical (Table 1), as are the structures of the ring, with pair wise RMSD of the Cα atoms of only 0.14 – 0.28 Å. This strongly indicates that the outward facing conformation of Glu59 is not a result of its deprotonation. Although it is clear that in the closed state the key carboxylate must be protonatated22 the open conformation revealed here appears to be compatible with either the protonated or deprotonated forms of Glu59.
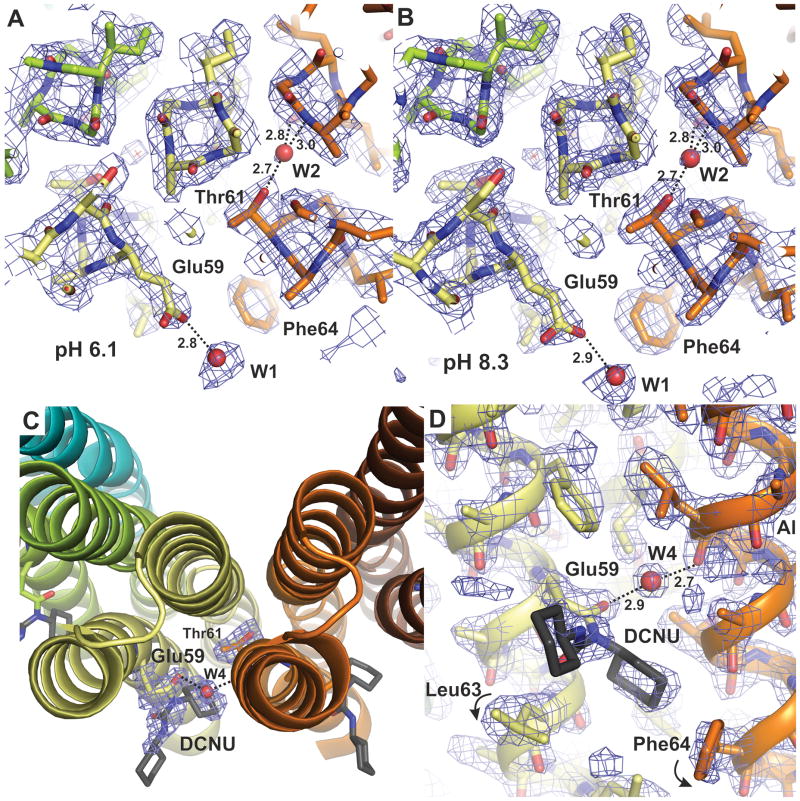
To confirm that Glu59 can retain the bound H+ in the open conformation, crystals grown initially at pH 6.1 were incubated with dicyclohexylcarbodiimide (DCCD, 50 mM), a chemical that modifies only protonated carboxyl groups39,40, at pH 5.5 (i.e. the same conditions as the pH 5.5 structure). The presence of DCCD modification was determined by x-ray crystallography at a resolution of 2.0 Å. Indeed, the measurements reveal electron density corresponding to a DCCD bound to Glu59 (Fig. 3), forming dicyclohexyl-N-acylurea (DCNU)41. Importantly, the data shows no indication of DCCD modification of the terminal carboxylate or that of Asp45, which, as the structure reveals, forms a salt-bridge with Lys44 and is therefore in the deprotonated form. The electron density of the first DCNU cyclohexyl group (atoms C2-C7) is particularly apparent in all c-subunits, while the second cyclohexyl group is less well defined. The first cyclohexyl group fits into a hydrophobic pocket that was previously occupied by Phe64 and has hydrophobic interactions with Ala60, Leu63 and Phe64 (Fig. 3). The displacement induced by DCCD on the side chains of Leu63 and Phe64, both of which adopt alternate rotamers, provides further evidence of the occupancy of DCCD at Glu59 (Fig. 3). Although there is some residual density that might be ascribed to unmodified Glu59 and the original rotamers of Leu63 and Phe64, the electron density strongly indicates that DCCD occupancy at position 59 is almost complete. In summary, the reactivity of Glu59 with DCCD demonstrates that the binding sites in the c10 ring can adopt an open conformation while in the protonated state.
Figure 3. Protonation state of the binding site.
A, B: Atomic structure of the proton binding sites in the c10 ring at pH 6.1 and 8.3, respectively, viewed as in Fig. 2D. Note the structures are essentially identical to each other, and to that at pH 5.5 (Fig. 2D). C, D: Structure of the binding site after modification with DCCD at pH 5.5, viewed either from the matrix side or along the membrane plane, respectively. The product of the reaction (DCNU) is shown in dark grey. The only significant difference in the structure of the c-ring after DCCD modification is the rotation of Leu63 and Phe64 (c.f. Fig. 2C). All 2Fo-Fc electron density maps are displayed at 1.2σ.
Environment controls binding-site conformation
As mentioned, the atomic structure of the mitochondrial c10 ring reported here differs from those previously determined from other ATP synthases, in the conformation of the proton-binding site. The cause for this difference is not immediately apparent. Those c-rings differ from this one in the c-subunit stoichiometry (13–15, versus 10) and primary sequence, but their structures were also resolved using different crystallization conditions.
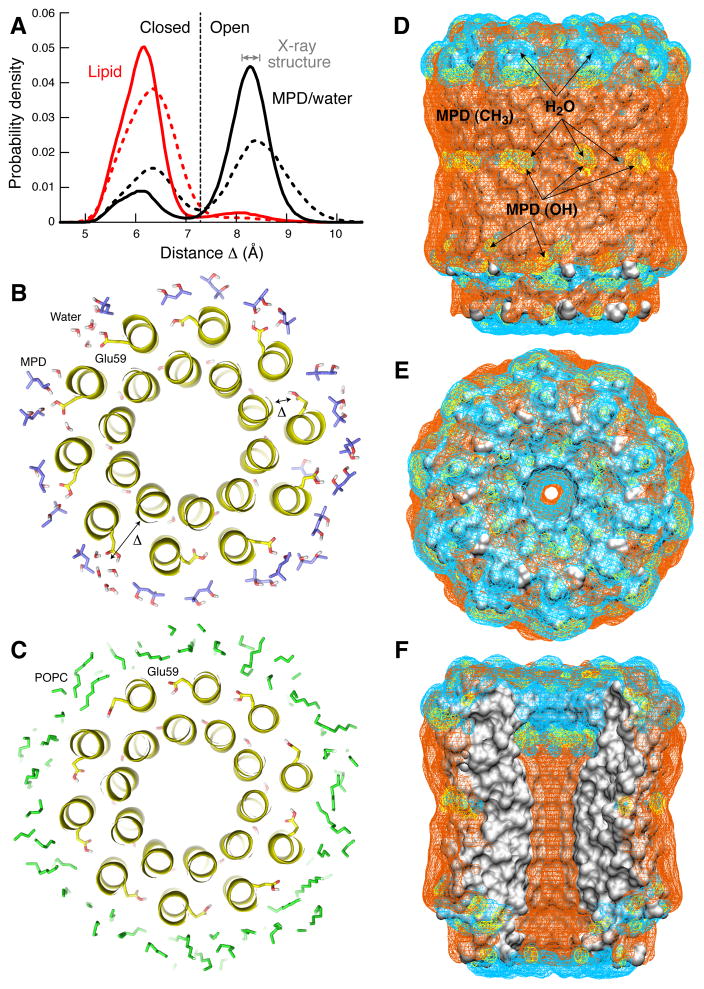
To examine this question in detail, we carried out a series of molecular dynamics simulations of the c10 structure in a model lipid membrane as well as in a solution that mimics the crystallographic buffer used here (see Methods). The results, summarized in Fig. 4, show that the MPD buffer stabilizes the open state, regardless of the conformation of the binding site at the start of the simulation (Fig. 4A). In this outward orientation, Glu59 is able to establish multiple H-bonds with MPD and water molecules (Fig. 4B). By contrast, the environment of the lipid membrane induces the (protonated) binding sites to adopt a closed conformation, analogous to that observed in crystal structures and comparable simulations of bacterial c-rings. That is, Glu59 retracts into the binding site and engages the adjacent c-subunit, through direct or water-mediated interactions (Fig. 4C).
Figure 4. Simulations of the c10 ring in the lipid membrane and in the crystallization buffer.
A: Probability of the open or closed conformation of the binding site, depending on whether the environment of the c-ring is the MPD:water solvent in the crystal (black) or a POPC phospholipid bilayer (red). Open and closed states are distinguished by the variable Δ, defined as the distance between the Cα atom of Ala22 and the center-of-mass of the COOH group of Glu59. Solid lines refer to simulations (100ns) in which the protein backbone is weakly constrained to the structure observed in the crystal, while side-chains are free to move. Dashed lines correspond to subsequent simulations (60ns) with no structural constraints. In the MPD:water solution, open and closed states interconvert, but the open conformation is the most likely. In the lipid membrane, the closed state is by far the most probable. B: Representative simulation snapshot of the c-ring in the MPD:water:salt solvent, viewed in cross-section; seven out of ten binding sites are open. C: Simulation snapshot of the c-ring in the lipid membrane; nine out of ten binding sites are closed. D-F: Interactions of the c10 ring structure with the crystallization buffer. All panels show 3D probability maps (mesh) for either water molecules (cyan); MPD molecules H-bonded to the protein surface via one or two of the MPD hydroxyl groups (yellow); and MPD molecules engaged in van der Waals interactions with the protein, via one or more of the MPD methyl groups (orange). The ring is viewed (D) along the plane of the membrane, (E) from the matrix side; and (F) in cross-section, illustrating the central hydrophobic opening. The MPD:water mixture provides stabilizing interactions for both polar and apolar regions on the protein surface. Note the binding sites interact either with water or with MPD hydroxyl groups.
In sum, these simulations demonstrate that the conformation of the c-subunit binding sites in crystal structures is dependent on their specific microenvironment, which probably holds true also in the context of the Fo complex. As shown in Fig. 4D–F, in the crystal of the yeast mitochondrial c10, MPD is predicted to coat the hydrophobic surface of the c-ring, both on the outer face of the ring and the inner pore, establishing van der Waals interactions with protein side chains mediated by its three methyl groups. However, MPD can also engage polar side chains in the protein, and in particular Glu59, with its hydroxyl groups. The polar surfaces on the yeast cring are also stabilized by water, which readily permeate the MPD layer. This is in contrast with, e.g. the crystal structure of the c15 ring from the cyanobacterium S. platensis, which at 1.8 Å resolution22 has a strictly hydrophobic environment in the transmembrane domain including in the vicinity of its proton binding sites. The organic solvent used here also protects the transmembrane hydrophobic surface of the protein, but its water content and the ability of MPD to form H-bonding interactions enabled us to observe a conformational state of the c-subunit.
Discussion
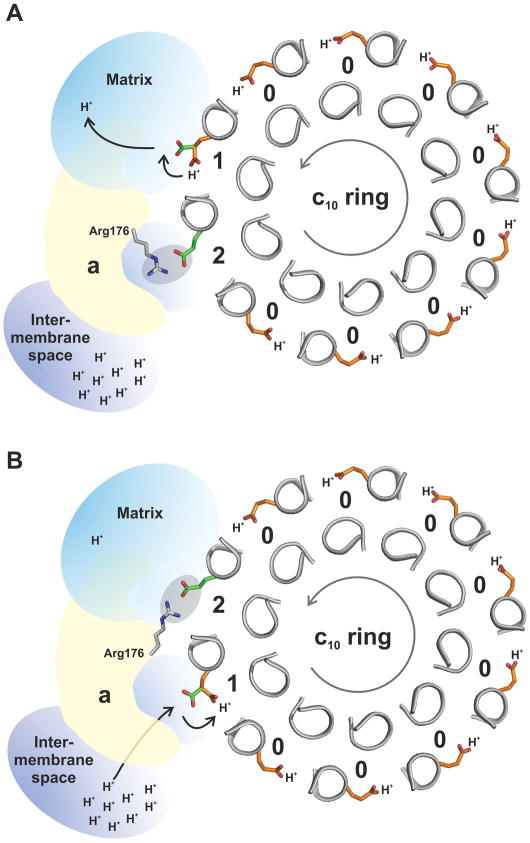
Although the rotary mechanism of the ATP synthase membrane motor is not understood in detail, it is widely accepted that during this mechanism the proton-binding sites in the c-ring become exposed to two distinct environments, to which they react very differently (Fig. 5). At the interface with subunit-a, a protonated binding site must be able to release its H+ towards one side of the membrane (e.g. the mitochondrial matrix), and subsequently protonate with a H+ from the other side (e.g. the intermembrane space). By contrast, the binding site of a c-subunit facing the lipid bilayer must preserve the protonated state until it eventually reaches the a-subunit interface. Provided that the sites of deprotonation and protonation at the subunit-a:c interface are spatially offset, and that the pathways for entry and exit of protons are disconnected, these basic features will result in a mechanism whereby stochastic rotations of the c-ring become directionally biased by the transmembrane proton gradient21,23.
Figure 5. Hypothetical rotary mechanism of proton translocation.
The stator subunit-a lies adjacent to the rotor c10 ring, and provides two half channels for protons – one leading to the matrix and the other to the intermembrane space. Subunit-a also interacts with the c-ring via a conserved arginine in its fourth transmembrane helix, Arg176. When facing the membrane, the c-subunit binding sites are in the closed state and Glu59 is constitutively protonated (color orange) (State 0). The direction of rotation during ATP synthesis is counter-clockwise, viewed from the matrix (grey arrow.). A: Upon entering the subunit-a interface, a site can adopt an open conformation, and Glu59 (color green) can release a proton into the channel leading to the matrix (State 1). Glu59 in the adjacent c-subunit (in the counter-clockwise direction) is initially paired with Arg176 (State 2), but becomes free when Arg176 switches to the trailing, now deprotonated Glu59. B: The release of this interaction enables the unengaged Glu59 site to load another proton from the intermembrane space, and ultimately to re-enter the membrane.
Previous crystal structures of isolated bacterial c-rings show the ion-binding sites in the closed, ion-locked state14–16,18. Because the local environment of the c-ring in these crystals is a hydrophobic detergent layer22, it is generally accepted that this is the conformation adopted by the c-subunit when facing the lipid bilayer. This notion is consistent with molecular dynamics simulations of c-rings in model phospholipid membranes37,42. In the structure of the mitochondrial c10 ring reported here, by contrast, the proton-binding site is seen in a open conformation, in which the key carboxylate side chain of Glu59 projects outwards from the site. Molecular dynamics simulations of this structure indicate that this alternate conformation is not a result of differences in the c-subunit primary sequence or the ring stoichiometry or its architecture. Instead, the simulations show that it is the hydrophilic environment around the proton-binding sites in the c10 crystal that induces their opening. In a hydrophobic environment, such as a lipid membrane or a detergent buffer, the prediction is that the c-subunit binding sites would adopt the closed, proton-locked conformation as previously observed. In regard to the current crystal structure, we conclude that the c-ring faces an environment that is also mechanistically relevant. In fact, water-accessibility studies of the E. coli Fo complex strongly indicate that c-subunits facing subunit-a encounter a certain amount of hydration24,25. Consistent with previous simulation studies of a bacterial homologue22, the experimental structure of the yeast mitochondrial c10 ring confirms that a hydrated, polar environment is sufficient to promote the opening of the c-subunit binding sites.
Based on NMR data from isolated c-subunits from E. coli, solubilized in a medium with a dielectric constant comparable that of our crystallization buffer, one can estimate the pKa of Glu59 to be around 7. Thus, the similarity in the structures obtained at pH 5.5, 6.1, and 8.3 leads us to conclude that the open-conformation observed here is compatible with both the protonated and deprotonated states of Glu59. The additional structure of the yeast c-ring at pH 5.5 in which Glu59 is unequivocally covalently labeled with DCCD demonstrates that this open conformation can retain a bound proton. By analogy, the hydrated environment of the subunit-a:c interface enables not only the opening of the proton-binding site, but also the exchange between protonated and deprotonated forms of the key carboxylate – which is thermodynamically unfeasible in the context of the lipid membrane22.
In sum, our interpretation of the structures of the c10 ring presented here is that they capture the c-subunit proton-binding sites in a conformational state that is characteristic of the interface with subunit-a. In its extended orientation, Glu59 would be able to bind and release H+ from either of the half water-channels that are thought to exist within the Fo subcomplex (Fig. 5). Also, in this conformation, Glu59 from an adjacent c-subunit would be able to transiently engage with the guanidinium group of the conserved arginine in subunit-a (Arg176 in S. cerevisiae excluding the 10 amino acid presequence35). Arg176 is hypothesized to play a crucial mechanistic role by coordinating deprotonation and protonation events21,43,44 and by preventing uncoupled proton leakage45.
Evidently, these events must take place in a concerted fashion, allowing downhill H+ transport to be coupled to the directional rotation of the c-ring, and thus ATP synthesis. Unfortunately, the lack of precise structural information on subunit-a precludes a detailed understanding of structural basis for these events. Nevertheless, the current crystal structures of the c10 ring, taken together with those previously determined (Supplement Fig. 4), seem to rule out large-scale conformational changes in the c-ring. In particular, it seems unlikely that access pathways for either H+ or Na+ are formed within the c-ring itself, or that the transmembrane helices rotate individually within the membrane. Despite the markedly different environments in the existing c-ring crystals, the variability among c-subunit structures seems to be mostly restricted to the ion-binding site.
In conclusion, we propose that the structure presented here represents the open conformation of the c-subunit, formed when facing subunit-a. It is this open conformation in which Glu59 is both protonated and deprotonated. When deprotonated, Glu59 engages Arg176 in subunit-a and only after reprotonation, and the subsequent rotation of the c-subunit away from subunit-a and into the lipid bilayer, does the ion-binding site revert to the closed conformation.
Methods
Purification and crystallization
Yeast F1Fo ATP synthase with an ε-GFP fusion (Green Fluorescent Protein) was purified as described29 in buffer containing DDM (0.05%). The ATP synthase from the Superose 6 column (4 ml, 8–12 mg protein) was concentrated to a volume, V1, (about 100 μl) using a Vivaspin 100K MWCO centrifugal concentrator (Sartorius Stedim, Aubagne, France) and diluted to 600 μl with a Buffer A solution (50 mM sodium bicine, pH 8.35, 0.3 M NaCl, 0.1 M sodium malonate, pH 7.0, 5 mM p-aminobenzoic acid, 2 mM MgCl2, 0.1mM thymol) containing DMPC:DHPC (3:1 molar ratio) bicelles46 with the percent, P1, (w/v) bicelle in the solution adjusted to a ratio that is equal to 0.1%/mg total protein. The solution was incubated on ice for 60 min. and then x mg of Biobeads (Biorad Co., Hercules, CA) (where × = V1(μl) *0.5 mg/μl) was added and the mixture rocked at 4°C for 60 min. The solution was removed from the cold, rocked at room temperature for 15 min., and the precipitated suspension was carefully separated from the Biobeads. The suspension was centrifuged at room temperature for 10 min. at 21,000 × g, which produced a translucent pellet with a green color from GFP. The pellet was dissolved at room temperature in a minimal amount (typically 100 μl) of Buffer A solution containing DMPC:CHAPSO (2:1 molar ratio) bicelle with 3% (w/w) 1,1′,2,2′-tetramyristoyl cardiolipin at a concentration defined by P1 from above. The protein concentration was adjusted to 6 mg/ml using Buffer A solution.
For crystallization at pH 6.1, Buffer A solution contained 50 mM sodium MES, 100 mM sodium malonate, pH 6.1 in place of the bicine and malonate used at pH 8.3. The crystals obtained at pH 5.5 were crystallized at pH 6.1 followed by overnight incubation of buffer containing 50 mM MES, pH 5.5 (see below).
The c-ring crystallized using the ATP synthase preparation at 6.0 mg/ml, which is an effective c-ring concentration of 0.8 mg/ml. An equal amount of reservoir solution containing 60–68% MPD, 6–8% propylene glycol, 2mM MgCl2 was added to the protein solution (typically 1 μl/1 μl) and the drop was incubated in a sitting drop format at 21°C. The crystals were visible within 1–2 weeks, fully formed in 1 month and stable for up to 6 months.
For crystals at pH 5.5, crystals obtained at pH 6.1 were incubated overnight at 21°C in a buffer containing 50 mM MES, pH 5.5, 68% MPD, 8% propylene glycol, 0.4 M NaCl, and 2 mM MgCl2. We were able to reduce the MPD concentration to 28% by stepwise reduction in the MPD concentration without ill effect on the diffraction quality. The structure determined from crystals soaked in 28% MPD did not differ significantly from the structures presented here for pH 8.3, 6.1, or 5.5 (data not shown). For modification with DCCD, the conditions were identical to those described for pH 5.5 except that the solution contained 50 mM DCCD.
The crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at Advanced Photon Source, GM/CA CAT, 23-ID or SER CAT, 22-ID, at 100K using a wavelength of 1.033Å or 1.000Å. The data were processed using HKL200047 and CCP4 program suite48. All models were validated using PROCHECK49. As determined with PROCHECK 98.3%–97.7% of all phi and psi geometries were in the most favored category, while the remaining were in the additional acceptable category, with no outliers of the Ramachandran statistics.
Crystallographic Data Processing
For the structure at pH 8.3, the initial phases were solved by molecular replacement using MOLREP50 with the search model consisting of chains K, L, M, N, O from the structure 2XOK4. The model was rebuilt using ArpWarp51 and further refined with REFMAC52, while using COOT53 for making minor corrections in the model, adding water molecules, and displaying electron density maps. In the final REFMAC cycles, we applied the TLS (Translation, Libration, Screw) procedure. The refinement did not use any non-crystallographic symmetry constraints or restraints. Several residues, including Ser15, Asn35 and Ser38, were refined with alternate side chain conformations as justified by the electron density maps.
For the structures at pH 6.1 and 5.5, residues 59–61 were omitted and the models were refined by simulated annealing in CNS54 to reduce possible model bias. Residues 59–61 were added using COOT and the same REFMAC refinement procedures used for the structure at pH 8.3 were applied. The TLS refinement had no effect on R factors and electron density maps for the structure at pH 6.1 at 2.5Å resolution.
The DCCD-modified carboxylate of Glu59, at pH 5.5, was apparent in all chains in the first electron density map after the rigid body refinement. Residue “GLH” from REFMAC library, which represents Glu59 modified with DCCD, was modeled with COOT in all five subunits. The same REFMAC refinement procedures used for the other structures were applied.
Molecular dynamics simulations
Molecular dynamics simulations of the complete c10 ring were carried out either with the protein structure embedded in a model POPC lipid membrane, or in a solvent that mimics the crystallization buffer. In the membrane system, the proton-binding sites in the c-ring were initially set in the open conformation observed in the X-ray structure. In the MPD buffer system, independent simulations were carried out with the binding sites set either in the open state or in the closed conformation observed in the membrane simulations. After the equilibration phase, the results of the two simulation series of the protein-MPD buffer system are statistically undistinguishable. For the membrane system, the phospholipid bilayer was prepared as previously described55. In the buffer system, MPD molecules were initially spaced uniformly in three dimensions, with waters and ions distributed randomly. These two environments were optimized through a series of simulations (approx. 100 ns in total) in which the c-ring was constrained to the X-ray structure, and gradually released. After this equilibration phase, a simulation of 160 ns was carried out for each system. During the first 100 ns, the protein backbone was still weakly constrained to the X-ray conformation; in the subsequent 60 ns, the structure was entirely free (both fractions are analyzed in Fig. 4). The simulation systems (along with further details) are included in Supplement Fig. 5.
All figures were produced using PYMOL56 (http://www.pymol.org/). Protein concentrations were determined as described using bovine serum albumin as the standard57. The electrostatic potential in Fig. 1 was computed with APBS58, using the PARSE force field59 and a homogenous dielectric constant of 1. The C-terminal carboxyl and the side chains of Asp, Glu, Lys and Arg and were set as the ionized form while the formylated N-terminus was set as neutral.
Supplementary Material
Acknowledgments
The work was supported by a grant from NIH R01GM66223 to DMM. TM and JFG are funded in part by the DFG Cluster of Excellence ‘Macromolecular Complexes’ (EXC115). Diffraction data were collected at GM/CA Collaborative Access Team (GM/CA-CAT) 23-ID and Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamlines at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Dept. of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number W-31-109-Eng-38. Computational resources were provided in part by the Jülich Supercomputing Center. We thank the reviewer for recognizing that residues 15-18 form a 310 helix.
Abbreviations and symbols
- CHAPSO
3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate)
- DCCD
N,N′-dicyclohexylcarbodiimide
- DCNU
dicyclohexyl-N-acylurea
- DDM
n-dodecyl-β-D-maltopyranoside
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- GFP
Green Fluorescent Protein
- MPD
2-methyl-2,4-pentandiol
- MES
2-(N-morpholino)ethanesulfonic acid
- OSCP
oligomycin sensitive-conferring protein
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
Footnotes
Accession Codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, under accession numbers 3U2F, 3U32, 3U2Y, 3UD0.
Competing financial interests
The authors declare no competing financial interests.
Author contributions:
J.S. collected and analyzed diffraction data and collaborated in writing the manuscript.
V.P. and D.O. designed and performed experiments and aided in collecting diffraction data.
J.D.F.-G. designed research, analyzed data, and collaborated in writing the manuscript.
A.K. performed research and analyzed data.
T.M. analyzed data and collaborated in writing the manuscript.
D.M.M. designed experiments, analyzed data, aided in collecting diffraction data, and collaborated in writing the manuscript.
References
- 1.Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE. The F1F0-ATPase complex from bovine heart mitochondria: The molar ratio of the subunits in the stalk region linking the F1 and F0 domains. Biochemistry. 1996;35:12640–12646. doi: 10.1021/bi960969t. [DOI] [PubMed] [Google Scholar]
- 3.Collinson IR, et al. Fo membrane domain of ATP synthase from bovine heart mitochondria: purification, subunit composition, and reconstitution with F1-ATPase. Biochemistry. 1994;33:7971–7978. doi: 10.1021/bi00191a026. [DOI] [PubMed] [Google Scholar]
- 4.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 5.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci U S A. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dautant A, Velours J, Giraud MF. Crystal structure of the Mg.ADP-inhibited state of the yeast F1c10-ATP synthase. J Biol Chem. 2010;285:29502–29510. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayalar C, Rosing J, Boyer PD. An alternating site sequence for oxidative phosphorylation suggested by measurement of substrate binding patterns and exchange reaction inhibitions. J Biol Chem. 1977;252:2486–2491. [PubMed] [Google Scholar]
- 8.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 9.Dickson VK, Silvester JA, Fearnley IM, Leslie AG, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Kabaleeswaran V, et al. Asymmetric structure of the yeast F1 ATPase in the absence of bound nucleotides. J Biol Chem. 2009;284:10546–10551. doi: 10.1074/jbc.M900544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees DM, Leslie AG, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci U S A. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meier T, et al. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J Mol Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 14.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 15.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 16.Pogoryelov D, Yildiz Ö, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 17.Vollmar M, Schlieper D, Winn M, Buchner C, Groth G. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J Biol Chem. 2009;284:18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. A new type of proton coordination in an F1Fo-ATP synthase rotor ring. PLoS Biol. 2010;8:e1000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizutani K, et al. Structure of the rotor ring modified with N,N′-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase. Proc Natl Acad Sci U S A. 2011;108:13474–13479. doi: 10.1073/pnas.1103287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krah A, Pogoryelov D, Meier T, Faraldo-Gómez JD. On the structure of the proton-binding site in the Fo rotor of chloroplast ATP synthases. J Mol Biol. 2010;395:20–27. doi: 10.1016/j.jmb.2009.10.059. [DOI] [PubMed] [Google Scholar]
- 21.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 22.Pogoryelov D, et al. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat Chem Biol. 2010;6:891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- 23.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 24.Steed PR, Fillingame RH. Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J Biol Chem. 2009;284:23243–23250. doi: 10.1074/jbc.M109.002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steed PR, Fillingame RH. Subunit a facilitates aqueous access to a membrane-embedded region of subunit c in Escherichia coli F1F0 ATP synthase. J Biol Chem. 2008;283:12365–12372. doi: 10.1074/jbc.M800901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- 27.Vincent OD, Schwem BE, Steed PR, Jiang W, Fillingame RH. Fluidity of structure and swiveling of helices in the subunit c ring of Escherichia coli ATP synthase as revealed by cysteine-cysteine cross-linking. J Biol Chem. 2007;282:33788–33794. doi: 10.1074/jbc.M706904200. [DOI] [PubMed] [Google Scholar]
- 28.Michel H. Crystallization of Membrane Proteins. CRC Press; 1990. [DOI] [PubMed] [Google Scholar]
- 29.Pagadala V, Vistain L, Symersky J, Mueller DM. Characterization of the mitochondrial ATP synthase from yeast Saccharomyces cerevisae. J Bioenerg Biomembr. 2011;43:333–347. doi: 10.1007/s10863-011-9364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraud MF, et al. Rotor architecture in the yeast and bovine F1-c-ring complexes of F-ATP synthase. J Struct Biol. 2012;177:490–497. doi: 10.1016/j.jsb.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Epler JL, Shugart LR, Barnett WE. N-formylmethionyl transfer ribonucleic acid in mitochondria from Neurospora. Biochemistry. 1970;9:3575–3579. doi: 10.1021/bi00820a011. [DOI] [PubMed] [Google Scholar]
- 32.Feldman F, Mahler HR. Mitochondrial biogenesis. Retention of terminal formylmethionine in membrane proteins and regulation of their synthesis. J Biol Chem. 1974;249:3702–3709. [PubMed] [Google Scholar]
- 33.Galper JB, Darnell JE. The presence of N-formyl-methionyl-tRNA in HeLa cell mitochondria. Biochem Biophys Res Commun. 1969;34:205–214. doi: 10.1016/0006-291x(69)90633-0. [DOI] [PubMed] [Google Scholar]
- 34.Halbreich A, Rabinowitz M. Isolation of Saccharomyces cerevisiae mitochondrial formyltetrahydrofolic acid:methionyl-tRNA transformylase and the hybridization of mitochondrial fMet-tRNA with mitochondrial DNA. Proc Natl Acad Sci U S A. 1971;68:294–298. doi: 10.1073/pnas.68.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michon T, Galante M, Velours J. NH2-terminal sequence of the isolated yeast ATP synthase subunit 6 reveals post-translational cleavage. Eur J Biochem. 1988;172:621–625. doi: 10.1111/j.1432-1033.1988.tb13934.x. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi N, et al. Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J Biol Chem. 1998;273:15085–15090. doi: 10.1074/jbc.273.24.15085. [DOI] [PubMed] [Google Scholar]
- 37.Leone V, Krah A, Faraldo-Gómez JD. On the question of hydronium binding to ATP-synthase membrane rotors. Biophys J. 2010;99:L53–L55. doi: 10.1016/j.bpj.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assadi-Porter FM, Fillingame RH. Proton-translocating carboxyl of subunit c of F1Fo H+-ATP synthase: The unique environment suggested by the pKa determined by 1H NMR. Biochemistry. 1995;34:16186–16193. doi: 10.1021/bi00049a034. [DOI] [PubMed] [Google Scholar]
- 39.Williams A, Ibrahim IT. Carbodiimide chemistry: recent advances. Chem Rev. 1981;81:589–636. [Google Scholar]
- 40.von Ballmoos C, Dimroth P. Two distinct proton binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]
- 41.Azzi A, Casey RP, Nalecz MJ. The effect of N,N′-dicyclohexylcarbodiimide on enzymes of bioenergetic relevance. Biochim Biophys Acta. 1984;768:209–226. doi: 10.1016/0304-4173(84)90017-x. [DOI] [PubMed] [Google Scholar]
- 42.Krah A, et al. Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase Fo rotors. Biochim Biophys Acta. 2010;1797:763–772. doi: 10.1016/j.bbabio.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Lightowlers RN, Howitt SM, Hatch L, Gibson F, Cox GB. The proton pore in the Escherichia coli F0F1-ATPase: a requirement for arginine at position 210 of the a-subunit. Biochim Biophys Acta. 1987;894:399–406. doi: 10.1016/0005-2728(87)90118-6. [DOI] [PubMed] [Google Scholar]
- 44.Cain BD, Simoni RD. Proton translocation by the F1F0ATPase of Escherichia coli. Mutagenic analysis of the a subunit. J Biol Chem. 1989;264:3292–300. [PubMed] [Google Scholar]
- 45.Mitome N, et al. Essential arginine residue of the Fo-a subunit in FoF1-ATP synthase has a role to prevent the proton shortcut without c-ring rotation in the Fo proton channel. Biochem J. 2010;430:171–177. doi: 10.1042/BJ20100621. [DOI] [PubMed] [Google Scholar]
- 46.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, MacArthur MW, Moss DS, Thorton JM. PROCHECK: a program software suite for macromolecular structure determination. J Applied Crystallogr. 1993;26:283–291. [Google Scholar]
- 50.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 51.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54 (Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gómez JD. GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J Chem Theor Comp. 2011;7:1167–1176. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLano WL. The PyMOL Molecular Graphics System. Delano Scientific; San Carlos: 2002. [Google Scholar]
- 57.Stoscheck CM. Quantitation of proteins. Methods in Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 58.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.