Abstract
The nutrient/target-of-rapamycin (TOR) pathway has emerged as a key regulator of tissue and organismal growth in metazoans. The signalling components of the nutrient/TOR pathway are well defined; however, the downstream effectors are less understood. Here, we show that the control of RNA polymerase (Pol) III-dependent transcription is an essential target of TOR in Drosophila. We find that TOR activity controls Pol III in growing larvae via inhibition of the repressor Maf1 and, in part, via the transcription factor Drosophila Myc (dMyc). Moreover, we show that loss of the Pol III factor, Brf, leads to reduced tissue and organismal growth and prevents TOR-induced cellular growth. TOR activity in the larval fat body, a tissue equivalent to vertebrate fat or liver, couples nutrition to insulin release from the brain. Accordingly, we find that fat-specific loss of Brf phenocopies nutrient limitation and TOR inhibition, leading to decreased systemic insulin signalling and reduced organismal growth. Thus, stimulation of Pol III is a key downstream effector of TOR in the control of cellular and systemic growth.
Keywords: Drosophila , growth, insulin, RNA polymerase III, TOR
Introduction
An important question in developmental biology concerns the mechanisms that control growth and final size in multicellular animals. Studies in different model organisms have identified many conserved cell–cell secreted factors and signalling pathways that control growth. One key regulator that has emerged from this work is the serine/threonine kinase, target-of-rapamycin (TOR; for reviews, see De Virgilio and Loewith, 2006; Wullschleger et al, 2006 and Foster and Fingar, 2010).
From yeast to mammals, TOR activity is cell-autonomously stimulated by an array of extracellular cues such as amino acids, glucose and oxygen to control growth and proliferation (Arsham and Neufeld, 2006; Dann and Thomas, 2006; Wullschleger et al, 2006; Hietakangas and Cohen, 2009; Wang and Proud, 2009; Foster and Fingar, 2010). In addition, in metazoans TOR can be activated by an endocrine insulin/insulin-like growth factor (IGF) signalling pathway (Oldham and Hafen, 2003; Grewal, 2008; Teleman, 2009). Insulins and insulin-like peptides bind to receptors on the surface of target cells. Ligand-receptor binding then triggers a conserved intracellular signalling cascade involving phosphoinositol 3-kinase (PI3K) and Akt, ultimately leading to increased TOR activity (Bhaskar and Hay, 2007; Efeyan and Sabatini, 2009). While these cell–cell and intracellular signalling inputs to TOR are well defined, the key downstream outputs by which TOR mediates its effects on metabolism and growth in vivo are less clear.
Considerable attention has focussed on the role of cellular protein synthesis as a regulator of cell growth. Extensive studies in mammalian cell culture have identified several mechanisms by which TOR can control mRNA translation (for reviews, see Proud, 2007; Ma and Blenis, 2009 and Sonenberg and Hinnebusch, 2009). For example, TOR can phosphorylate and inhibit the translational repressor eukaryotic initiation factor 4E-binding protein (4E-BP) leading to stimulation of protein synthesis (Thomas, 2002; Jastrzebski et al, 2007; Ma and Blenis, 2009). This translational mechanism is widely proposed as a key growth-regulatory target of TOR signalling (Dowling et al, 2010). These effects may not, however, account fully for the in vivo growth functions of TOR. For example, in Drosophila, TOR null mutants are lethal with severe growth defects (Oldham et al, 2000; Zhang et al, 2000) and overactivation of TOR signalling can promote considerable overgrowth; null mutants for 4E-BP, on the other hand, are viable with no effects on growth during development (Miron et al, 2001; Teleman et al, 2005). The regulation of ribosome synthesis is another TOR function important for protein synthesis and growth. Studies in yeast and mammalian cell culture have identified several mechanisms by which TOR can control the expression of ribosomal RNA (rRNA) and ribosome biogenesis genes (Mayer and Grummt, 2006). Moreover, recent work in Drosophila has emphasized the in vivo regulation of ribosome synthesis by TOR. For example, in larvae the insulin/TOR pathway controls the expression of ribosome synthesis genes via the transcription factors FOXO and Myc (Teleman et al, 2008; Li et al, 2010). In addition, the RNA polymerase I factor, TIF-IA, is required for rRNA synthesis and larval growth and is a downstream target of insulin/TOR signalling (Grewal et al, 2007).
In this paper, we explore the regulation of RNA polymerase (Pol) III-dependent transcription as a growth-regulatory output of insulin/TOR signalling in Drosophila. Pol III is responsible for the synthesis of small non-coding RNAs that are essential for mRNA translation (e.g., 5S rRNA and transfer RNAs—tRNAs). Thus, control of Pol III may therefore represent another mechanism by which TOR alters protein synthesis to regulate growth. Studies on TOR signalling and Pol III have been exclusively limited to work in yeast and mammalian cell culture studies. For example, the multisubunit transcription factor TFIIIB is essential for Pol III transcription initiation, and nuclear extracts from either nutrient-deprived or TOR-inhibited yeast show reduced TFIIIB activity in vitro (Dieci et al, 1995; Sethy et al, 1995; Clarke et al, 1996; Zaragoza et al, 1998). Furthermore, in cultured mammalian cells the Brf (TFIIIB-related factor) subunit of TFIIIB is regulated downstream of several growth-regulatory signalling pathways including the TOR cascade (Goodfellow and White, 2007; Woiwode et al, 2008). These effects on TFIIIB/Pol III-dependent transcription in yeast and mammalian cells may reflect the ability of TOR to phosphorylate and inhibit the Pol III repressor Maf1, thus promoting transcription (Upadhya et al, 2002; Lee et al, 2009; Wei et al, 2009; Kantidakis et al, 2010; Michels et al, 2010; Shor et al, 2010). Mammalian Brf activity can also be stimulated by direct interaction with oncogenes such as c-Myc (White, 2005). While these in vitro studies have provided important molecular details about the regulation of Pol III in vitro, they do not address questions about metabolism, growth and size control in a developing multicellular animal: How does regulation of Pol III influence cell and tissue growth? Is Pol III required for the in vivo functions of TOR? If so, what are the regulatory mechanisms involved?
Our approach has been to use Drosophila as a model system to examine the contribution of Pol III-dependent transcription to the control of cell and tissue growth in vivo. During Drosophila larval development, the period of the life cycle characterized by an immense increase in growth, the major function of TOR signalling is to couple dietary nutrition to cell and tissue growth (Britton et al, 2002). TOR activity is required to cell-autonomously control growth in all larval tissues. In addition, stimulation of TOR in specific tissues can also play a non-autonomous role in systemic growth. For example, in well-fed larvae, amino-acid import into fat cells activates TOR leading to relay of a signal to the brain to promote the release of several Drosophila insulin-like peptides (dILPs) from discrete neurosecretory cells (Ikeya et al, 2002; Geminard et al, 2009). These dILPs then circulate through the larval haemolymph and activate the insulin-signalling pathway to stimulate cell growth in all larval tissues. We show here that Brf is an essential effector of TOR in the control of both cell-autonomous and non-autonomous effects on growth and body size in Drosophila. Moreover, we present evidence for a prominent role for dMaf1, but only a limited role for Drosophila Myc (dMyc), in the control of Pol III by nutrient-TOR signalling in developing animals.
Results
Brf is required for both cellular and organismal growth in Drosophila larvae
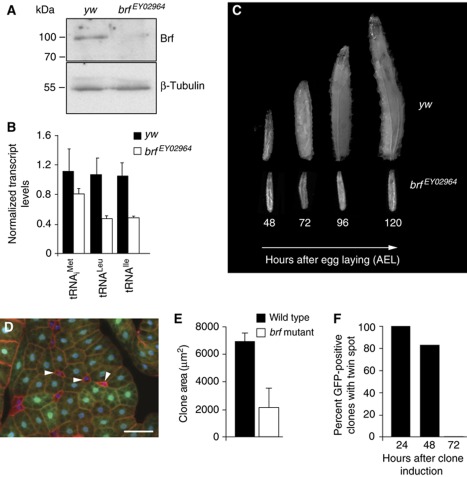
Brf, a conserved component of the TFIIIB complex, is limiting for Pol III-dependent transcription in yeast and mammals (Geiduschek and Kassavetis, 2001; Marshall et al, 2008). We therefore investigated if Brf is involved in controlling Pol III-dependent transcription and growth in Drosophila larvae. For these experiments, we analysed two publicly available lines (Bloomington Stock Center) carrying P-element insertions in the brf locus (brfEY02964 and brfc07161). Homozygous brfEY02964 flies were lethal and this lethality could be rescued by ubiquitous GAL4-dependent expression of a UAS-brf transgene. Homozygous brfEY02964 larvae also had reduced levels of both Brf protein (Figure 1A) and Pol III-dependent transcripts (Figure 1B) compared with control, wild-type larvae at the same developmental stage. Furthermore, levels of 7SL RNA were lower in brf mutants compared with controls; however, we did not detect any changes in the levels of 5S rRNA or the Pol I-dependent transcript, pre-rRNA (Supplementary Figure S1). Phenotypically, brfEY02964 larvae progressed through embryogenesis but arrested as second instar larvae, surviving for several days (Figure 1C). A similar phenotype was seen in flies transheterozygous for brfEY02964 and a deficiency that uncovers the brf locus (Df(3R)BSC565), suggesting that brfEY02964 is either a null or strong hypomorphic loss-of-function allele of brf. We therefore used this line as a brf mutant. Flies that were homozygous for the second P-element line, brfc07161, also exhibited lethality, but this could not be rescued by ubiquitous GAL4-dependent expression of a UAS-brf transgene. Hence, this P-element line must also be mutated in another essential gene, and so we did not study it any further. The growth inhibitory effects seen in homozygous brf mutant larvae could be phenocopied by expression of a UAS-brf RNAi construct using the ubiquitous daughterless (da)-GAL4 driver (Supplementary Figure S2). Reducing Brf protein levels in this manner also decreased rates of Pol III-dependent transcription (Supplementary Figure S2A and B) and reduced larval growth rates (Supplementary Figure S2C). Expression of brf RNAi in either the salivary gland (patched (ptc); Supplementary Figure S2D and E) or eye imaginal discs (eyeless (ey); data not shown) also led to a reduction in tissue growth. Importantly, the growth inhibitory effects of the brf RNAi transgene were reversed by overexpression of UAS-brf, indicating that the RNAi-mediated effects were specifically due to Brf knockdown (Supplementary Figure S2F and G). In contrast to the effects of Brf inhibition, we found that overexpression of Brf alone was not sufficient to stimulate Pol III activity or affect organismal growth (data not shown). Thus, Brf, probably through its role in driving Pol III-dependent transcription, is essential for both tissue and organismal growth in Drosophila larvae.
Figure 1.
Loss of Brf function leads to severe growth defects in Drosophila larvae. (A) Brf protein levels were reduced in brf mutant (brfEY02964) larvae compared with controls (yw) 48 h after egg laying (AEL), as determined by immunoblot. (B) Levels of Pol III-dependent transcripts were significantly decreased in brf mutant larvae compared with control larvae 48 h AEL, as measured by qRT–PCR (P<0.05, Student's t-test). Error bars indicate s.e.m. (C) brf mutant larvae are growth arrested. Images of control and brf mutant larvae throughout larval development (48–120 h AEL) are shown. (D) brf clones (non-GFP, arrowheads) were induced by flp/FRT-mediated recombination during embryogenesis and visualized in the fat body 120 h AEL. Blue, DAPI staining; red, actin; green, GFP. Scale bar, 100 μm. (E) brf mutant clones in wing discs were induced 72 h AEL and clone areas measured 120 h AEL (n=100 twin spots). (F) brf mutant clones were induced and scored in wing imaginal discs at the times indicated post clone induction. The viability of mutant clones was assessed by counting the percentage of wild-type clones that were still paired with a brf mutant twin spot. Genotypes used in (D–F): hsflp122;+; FRT82B, brfEY02964/FRT82B, ubi-GFP.
We examined whether these growth defects observed following the loss of Brf function were due to inhibition of cellular growth. Most of the mass increase in developing larvae occurs in endoreplicating cells that make up the bulk of larval organs, such as the Drosophila fat body. Using mosaic analysis, we found that brf mutant cells (Figure 1D, GFP negative) in the larval fat body showed a marked decrease in size compared with surrounding heterozygote and wild-type cells (Figure 1D, GFP positive). We also created mosaic brf clones in the mitotically dividing cells of the larval wing imaginal disc. At 48 h following the clone induction, brf clones showed a growth defect and were approximately half the size of sister wild-type twin spots (Figure 1E). We also used flow cytometry to measure the size of dissociated wing disc cells at this time point, and found that brf cells showed little change in size compared with wild-type and brf heterozygous cells (data not shown). Together, these data suggest that loss of Brf leads to a coordinated decrease in both cellular growth (mass increase) and cell division in the wing disc, resulting in fewer, slower dividing cells that maintain a normal size. We also found that the viability of the brf mutant clones decreased with time following the clone induction. Approximately 80% of wild-type clones still had sister brf mutant twin-spot clone at 48 h after clone induction; however, at 72 h post induction none of the wild-type clones we examined still had a brf mutant sister clone (Figure 1F). This result suggests that slower growing and dividing brf mutant cells are outcompeted and eliminated by their faster growing neighbours. Consistent with this interpretation, we could rescue the viability of brf mutant cells by genetically reducing the growth rate of surrounding cells by making them heterozygous for a dominant Minute (M) allele of ribosomal protein S3. In this case, we could recover brf mutant clones at 72 h after clone induction, a time point at which all clones are normally eliminated (Supplementary Figure S3A and B). These rescued brf clones still showed a strong growth defect compared with wild-type cells. Moreover, we found that overexpression of the baculovirus anti-apoptotic protein p35 in the brf mutant wing disc cells rescued clone viability at 72 h post clone induction. These rescued brf mutant clones were, however, still smaller than wild-type clones (Supplementary Figure S3C–E; clones of each genotype are GFP positive). Together, these data show that Brf function is required for Pol III-dependent transcription and cell and organismal growth during Drosophila development.
Brf activity in fat cells is required to maintain systemic insulin signalling and organismal growth
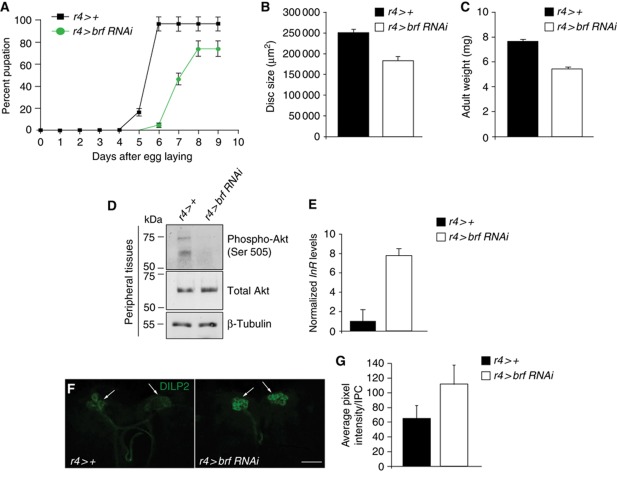
The brf mutant phenotype is reminiscent of the phenotype of larvae raised in nutritionally poor conditions. In Drosophila larvae, the function of nutrient sensing is performed by the fat body, an insect organ that performs both endocrine and nutrient storage functions similarly to the vertebrate liver and adipose tissue. In nutrient-rich conditions, the fat body signals to the brain to promote release of several dILPs, thereby promoting systemic insulin signalling and growth. We therefore explored whether changes in Pol III activity play a role in the Drosophila fat body in controlling organismal growth in a cell non-autonomous manner. To address this, we expressed a brf RNAi transgene specifically in the Drosophila fat body and examined the effects on the growth and development of the larvae. We found that silencing brf in the fat body (r4>brf RNAi) reduced larval growth rates and delayed pupation, with ∼15% of larvae failing to pupate and remaining as third instar larvae (Figure 2A). At wandering third instar stage, r4>brf RNAi larvae had significantly smaller wing imaginal discs than control larvae (Figure 2B). Subsequently, we found that r4>brf RNAi adults were smaller than controls and weighed less (Figure 2C). Similar but stronger effects were seen using another fat body driver (cg-GAL4); cg>brf RNAi larvae were significantly smaller than controls and failed to progress into the pupal stage (Supplementary Figure S4A).
Figure 2.
Fat body-specific reduction in Brf activity has cell non-autonomous effects on organismal growth and development. (A) Fat body-specific reduction in Brf levels (r4>brf RNAi, green trace) delayed pupariation when compared with controls (r4>+, black trace). The data are represented as a percentage of larvae that develop to pupal stage. Error bars indicate s.e.m. (B) Fat body loss of Brf function results in smaller wing imaginal discs, compared with controls (n>20 per genotype). Disc size was quantified in wandering third instar larvae using the histogram tool (Adobe Photoshop). Error bars indicate standard error. (C) Reduced Brf in the fat body reduced adult weight compared with control adults. Error bars indicate standard error. (D–G) Silencing of brf specifically in the Drosophila fat body decreased peripheral insulin signalling. (D) brf silencing in the fat body abolished Akt phosphorylation at serine 505 in peripheral tissues while total Akt protein levels remained constant. Levels of β-tubulin were measured to ensure equal loading. (E) Decreasing Brf levels specifically in the fat body using the r4-GAL4 driver significantly increased dInR mRNA levels in peripheral tissues of these animals when compared with controls (P<0.05, Student's t-test; n=32 per genotype). All qRT–PCR error bars represent s.e.m. (F) There is an accumulation of dILP2 protein in the IPCs of r4>brf RNAi larval brains compared with controls (r4>+). (G) Quantified pixel intensities of DILP2 staining in IPC clusters in the larval brain (r4>+, n=12 and r4>brf RNAi, n=21; error bars represent standard deviation; P=000174). For (D–G), larvae were analysed at 96 h AEL.
We explored whether these organismal growth effects caused by inhibiting Brf in the fat body were a consequence of reduced systemic insulin signalling. To do so, we first examined phospho-Akt levels in the peripheral tissues of r4>brf RNAi animals by immunoblotting. Akt is a key downstream effector of the insulin pathway, and Akt activity can be measured by assaying for phosphorylation of a carboxy terminus serine residue at position 505. We found that phosphorylation of Akt at serine 505 was reduced in r4>brf RNAi larvae peripheral tissues (larval carcasses devoid of fat body), even though total Akt was still present at levels comparable to control animals (Figure 2D). Similarly, brf mutant larvae also had lower levels of Akt phosphorylation at serine 505 compared with age-matched wild-type whole larvae (Supplementary Figure S4B). To further confirm that the inhibition of larval growth caused by fat body silencing of Brf was due to reduced systemic insulin signalling, we measured the levels of dInR mRNA. Transcription of this gene is negatively regulated by the insulin/PI3K pathway through the activation of FOXO (Puig and Tjian, 2005), and hence levels of dInR mRNA act as an additional readout of insulin signalling (Delanoue et al, 2010). When we used r4-gal4 to drive brf RNAi in the fat body, we found an increase in dInR mRNA levels in peripheral tissues, consistent with a suppression of peripheral insulin signalling (Figure 2E). We saw a similar increase in dInR mRNA in both peripheral tissues from cg>brf RNAi (Supplementary Figure S4C), and also in brf mutant animals when compared with control animals (Supplementary Figure S4D). Finally, we examined whether these changes in systemic insulin signalling following the knockdown of Brf might be explained by either reduced expression or release of brain dILPs. Previous reports have shown that mRNA levels of dilp5, but not dilp2, are suppressed upon amino-acid starvation (Geminard et al, 2009). We saw no change in dILP mRNA expression levels in r4>brf RNAi larvae (Supplementary Figure S4E). In contrast, we saw reduced expression levels of the mRNAs encoding dilp2 and dilp5 in cg>brf RNAi larvae (Supplementary Figure S4F). We also found that dilp5 mRNA levels were reduced in brf mutants (Supplementary Figure S4G). Amino-acid deprivation also leads to reduced release of dILPs from the brain and hence dILP proteins are retained in the neurosecretory insulin producing cells (IPCs) of starved animals (Geminard et al, 2009). Using immunostaining, we also found that dILP2 protein was retained in the IPCs of brains from r4>brf RNAi larvae compared with controls (Figure 2F and G). Taken together, these data suggest that Brf function and hence Pol III-dependent transcription is required in the fat body to maintain normal systemic insulin signalling and growth.
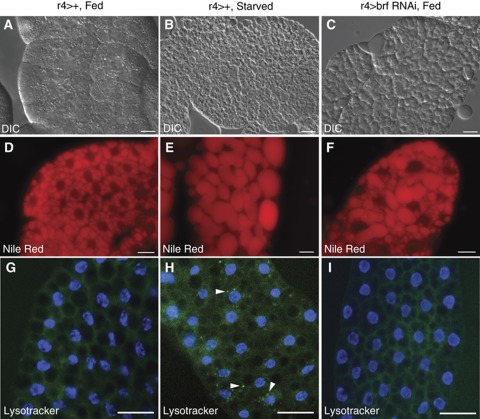
Given the organismal effects we observed following brf knockdown in fat cells, we examined whether Brf might be required for nutrient-dependent effects on fat body metabolism. To do so, we compared the fat bodies of starved larvae with those from larvae in which Brf had been specifically silenced in the fat body by expression of brf RNAi using the fat body driver r4-GAL4. Nutrient-deprivation/TOR inhibition induces marked changes in lipid metabolism (Colombani et al, 2003), which can be observed as an increase in lipid droplet size. Using both Differential Interference Contrast (DIC) microscopy and Nile Red staining, we observed an increased lipid droplet size in r4>brf RNAi larvae compared with control animals (Figure 3A, D versus C, F). These effects were similar to changes in lipid droplets in fat bodies dissected from either amino acid-deprived (Figure 3B and E) or tor mutant larvae (Zhang et al, 2000). Similar effects were seen when we expressed the UAS-brf RNAi transgene with another fat body driver, cg-GAL4 (Supplementary Figure S5A and B). Starvation for amino acids also stimulates a rapid induction of autophagy, a response that is required for organismal survival. We found that fat bodies from 4 h starved larvae showed a marked increase in autophagasomes by using lysotracker staining (Figure 3H). In contrast, we found that r4>brf RNAi fat bodies showed no induction of autophagy (Figure 3I), similarly to fat bodies dissected from fed larvae (Figure 3G). These results suggest that Brf and Pol III-dependent transcription in the Drosophila fat body are required for some but not all of the metabolic effects of nutrient availability.
Figure 3.
The fat body-specific loss of Brf function phenocopies some aspects of the starvation response. Fat bodies were dissected from 72 h larvae and stained with Nile Red to visualize lipid droplets or lysotracker green to vizualize autophagosomes. (A–C) DIC and (D–F) Nile Red images of fat bodies isolated from control (r4>+) fed (A, D) and control 24 h starved larvae (B, E). (C, F) Fed larvae with fat body-specific reduction in Brf levels (r4>brf RNAi) are shown. (G–I) Lysotracker green images of fat bodies isolated from control fed (G) and r4>brf RNAi (I) larvae show no lysotracker staining. Starved control larvae exhibit a punctuate staining pattern (H, arrowheads) caused by the formation of autophagosomes. Images were all taken at the same exposure. Scale bars, 100 μm.
Pol III transcription is stimulated by the TOR pathway
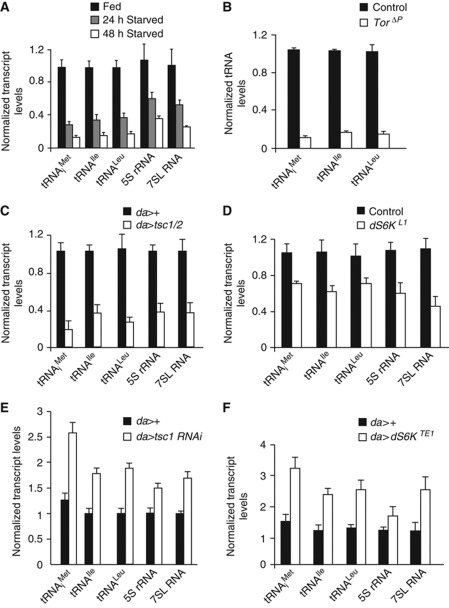
The cell and organismal changes in metabolism, physiology and growth that we described for loss of Brf function are similar to those seen following the inhibition of TOR signalling. We therefore explored whether TOR regulates Pol III-dependent transcription in Drosophila, and whether this regulation is required for the in vivo functions of TOR signalling. To address this, we first measured levels of Pol III-dependent transcripts, by qRT–PCR, in Drosophila larvae following the modulation of the TOR pathway. Starvation for dietary protein leads to inhibition of TOR activity in larvae (Oldham et al, 2000; Zhang et al, 2000). We found that larvae starved in 20% sucrose/PBS had reduced levels of several Pol III-dependent transcripts such as the tRNAs, 5S rRNA and 7SL RNA (Figure 4A). To further investigate the involvement of the TOR pathway in Pol III regulation in vivo, we performed gene expression analyses in larvae in which we genetically manipulated TOR signalling. We first found that tor null mutants had significantly reduced levels of Pol III-dependent transcripts compared with age-matched control larvae, (Figure 4B). We also found that cultured Drosophila S2 cells treated with the TOR-specific inhibitor, rapamycin, also had reduced levels of Pol III-dependent products (Supplementary Figure S6). Similarly, we found that overexpression of negative regulators of the TOR pathway, TSC1/2 using the UAS-GAL4 system, resulted in a substantial reduction in Pol III-dependent transcript levels compared with control larvae (Figure 4C). Finally, we found levels of Pol III-dependent transcripts were reduced in homozygous mutants for S6 kinase, a key TOR effector (Figure 4D). We next asked if overactivation of the TOR pathway can increase Pol III-dependent transcription. To address this, we first ubiquitously expressed a tsc1 RNAi transgene using the da-GAL4 driver and found that these larvae had significantly increased levels of each of the Pol III-dependent transcripts measured (Figure 4E). We then examined larvae expressing a constitutively active form of the downstream TOR effector S6K, and found that levels of Pol III-dependent transcripts were significantly elevated in these larvae compared with controls (Figure 4F). Taken together, these data demonstrate that the TOR pathway is necessary and sufficient to stimulate Pol III-dependent transcription in developing larvae, in part through activation of S6 kinase.
Figure 4.
TOR signalling regulates Pol III-dependent transcription in Drosophila larvae. (A) Pol III-dependent transcripts were significantly decreased in wild-type (yw) larvae starved for dietary protein for 24 or 48 h compared with wild-type fed larvae. (B) tRNA levels were significantly decreased in torΔP homozygous mutant larvae when compared with control (yw) larvae 48 h AEL (P<0.05, Student's t-test). (C) Pol III-dependent transcript levels were significantly decreased in larvae ubiquitously overexpressing tsc1/2 (da>tsc1/2) compared with controls (da>+) larvae 48 h AEL (P<0.05, Student's t-test). (D) Levels of Pol III-dependent transcripts were significantly reduced in S6K homozygous (dS6KL1) mutant larvae when compared with control (yw) larvae 48 h AEL (P<0.05, Student's t-test). (E) Levels of Pol III-dependent transcripts were elevated in whole larvae following the ubiquitous expression of a tsc1 RNAi transgene by da-GAL4 (da>tsc1 RNAi) compared with controls (da>+, P<0.05, Student's t-test) at 72 h AEL. (F) Ubiquitous expression of a constitutively active form of S6K (da>S6KTE1) significantly increased Pol III-dependent transcript levels in whole larvae compared with control (da>+) larvae 72 h AEL (P<0.05, Student's t-test). Each experiment was independently performed three times with n=32 per genotype. For each qRT–PCR error bars indicate s.e.m.
Brf is required for TOR-induced cell growth
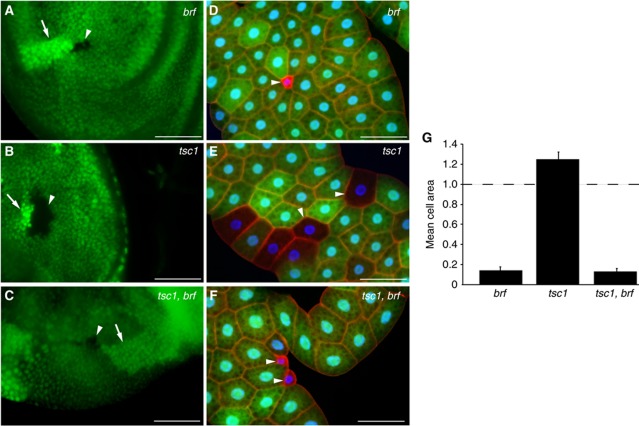
Given that Pol III-dependent transcription is regulated by the insulin/TOR pathway in vivo in Drosophila larvae, we next wanted to examine whether Brf functions downstream of TOR in the control of growth. TSC1 and TSC2 function together as negative regulators of TOR signalling (Ito and Rubin, 1999; Gao and Pan, 2001; Potter et al, 2001; Tapon et al, 2001). As a consequence, loss of TSC1 or TSC2 function leads to a TOR-dependent increase in cellular growth in larval tissues. We examined whether the overgrowth induced by the loss of TSC1 function was dependent on Brf. We used flp/FRT-mediated recombination to generate mutant clones in the wing imaginal discs (Figure 5A–C). As described above, brf mutant clones were growth defective and as a consequence were smaller than their wild-type twin spots (Figure 5A). In contrast, tsc1 mutant clones were significantly larger than their wild-type twin spot, consistent with the growth promoting effects of increased TOR signalling (Figure 5B). However, we found that brf, tsc1 double mutant clones were similar in size to the brf mutant clones (Figure 5C). The small clone size of either brf mutants or brf, tsc1 double mutants could not be rescued by the expression of P35. Therefore, the small clone sizes were not merely a direct consequence of apoptosis (Supplementary Figure S7A–F). We performed similar mosaic clonal experiments in the larval fat body (Figure 5D–G). As in the wing discs, we found that brf mutant cells were smaller than controls (Figure 5D) and that brf was epistatic to tsc1 (Figure 5F). Thus, tsc1 cells exhibited a growth increase compared with surrounding cells (Figure 5E) while brf, tsc1 double mutant cells phenocopied brf cells, and were severely growth impaired (Figure 5F and G). These data suggest that Brf is required for TOR-induced growth in both endoreplicating and mitotically dividing and cells of the larvae.
Figure 5.
Brf is required for TOR-induced cell growth in both mitotically dividing and endoreplicating tissue in Drosophila larvae. (A–F) brf, tsc1 or tsc1,brf double mutant clones were induced in both wing discs (A–C) and fat body (D–G). Mutant clones, arrowheads; wild-type sister clones, arrows. Blue, DAPI staining; red, actin; green, GFP. (G) The areas of both mutant and wild-type cells in the fat body were measured and presented here as mean cell area compared with control. Genotypes: (A, D) hsflp122; +; FRT82B, brfEY02964/FRT82B, ubi-GFP; (B, E) hsflp122; +; FRT82B, tsc1Q87X/FRT82B, ubi-GFP; (C, F) hsflp122; +; FRT82B, brfEY02964, tsc1Q87X/FRT82B, ubi-GFP. (A–C) Scale bar, 50 μm. (D–F) Scale bar, 100 μm.
Drosophila Maf1 is the predominant regulatory link between nutrition/TOR and Pol III activity
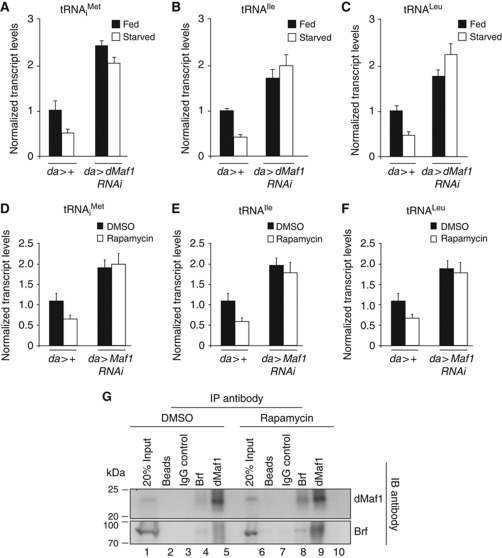
We next wanted to address the mechanism by which TOR regulates Pol III-dependent transcription. We found that inactivation of TOR, by starving larvae for dietary protein, did not affect Brf protein levels (Supplementary Figure S8). One candidate we considered to be involved in controlling Pol III-dependent transcription downstream of TOR activity was the transcriptional repressor Drosophila Maf1 (dMaf1, CG40196). In both yeast and mammalian cells, Maf1 represses Pol III activity and this repression, in turn, is reversed by nutrient/TOR signalling. We therefore examined if this mode of controlling Pol III was conserved in Drosophila. In feeding larvae, when insulin/TOR signalling is high, we found that ubiquitous expression of a dMaf1 RNAi transgene using the da-GAL4 driver led to elevated levels of tRNAs compared with control larvae consistent with a role for dMaf1 as a repressor (Figure 6A–C). Similar effects on Pol III-dependent transcription were seen using another dMaf1 RNAi transgene that targets an overlapping but smaller region of dMaf1 (Supplementary Figure S9A–C). Furthermore, the elevated tRNA levels seen in da>Maf1 RNAi larvae could be restored to wild-type levels by the expression of a UAS-dMaf1 transgene (Supplementary Figure S9D and E), allowing us to conclude that these dMaf-RNAi effects on Pol III-dependent transcription were specific to loss of dMaf1. We found that inhibition of dMaf1 had no effect on transcript levels of components of the Pol III machinery, such as Brf, Trf1 or RPIII128 (Supplementary Figure S9F). We also found that inhibition of dMaf1 had no effect on levels of both pre-rRNA and Ribosomal protein 49 mRNA. Thus, in contrast to a previous report on human Maf1 (Johnson et al, 2007), we find that Drosophila Maf1 has no effect on either Pol I- or Pol II-dependent transcription, and is probably a specific regulator of Pol III. As described above, starvation for dietary amino acids leads to reduced insulin/TOR signalling and consequently tRNA synthesis was suppressed (Figure 6A–C, compare fed versus starved in da>+ animals). In starved da>dMaf1 RNAi larvae, however, we found that tRNA levels remained elevated (Figure 6A–C). Similar effects were seen when we used rapamycin feeding, instead of starvation, as a more specific way to inhibit TOR signalling (Figure 6D–F). Finally, we also found that activation of TOR signalling (by expression of a tsc1 RNAi transgene) only modestly augmented the effects of dMaf1 RNAi on tRNA levels (Supplementary Figure S9F). Together, these data argue that, in Drosophila, nutrient/TOR signalling stimulates Pol III and tRNA synthesis via inhibition of dMaf1. Maf1 is thought to function as a repressor by interacting with Brf1 and/or Pol III, sequestering these factors away from tRNA gene promoters. Using Drosophila cultured S2 cells, we found under normal conditions, when TOR activity was high, dMaf1 and Brf showed a weak association as measured by co-immunoprecipitation (Figure 6G). This interaction was, however, enhanced following the inhibition of TOR by rapamycin treatment (Figure 6G). Therefore, one mechanism by which dMaf1 may repress Pol III-dependent transcription under low TOR activity is to bind and sequester Brf away from Pol III promoters. Together, these data suggest that during larval development nutrient-dependent insulin/TOR signalling stimulates Pol III-dependent transcription through the inhibition of dMaf1.
Figure 6.
Drosophila Maf1 is the regulatory link between TOR and Pol III activity. (A–F) qRT–PCR analyses of RNA extracted from whole larvae following the ubiquitous expression of a dMaf1 RNAi transgene (da>Maf1 RNAi) compared with control (da>+) larvae. TOR activity was modulated either by starving the larvae of dietary protein (A–C) or by feeding larvae rapamycin (D–F). (A–F) tRNA levels were significantly elevated following loss of dMaf1 (da>Maf1 RNAi) compared with controls (da>+) when TOR activity was high under normal fed or DMSO-treated conditions (P<0.05, Student's t-test). tRNA levels remain elevated in da>Maf1 RNAi animals even under starved or rapamycin-treated conditions when control tRNA levels are normally reduced (P<0.05, Student's t-test). For qRT–PCR error bars indicate s.e.m. (G) Immunoprecipitation of Brf and dMaf1 from Drosophila cultured S2 cell extracts revealed an enhanced association between Brf and dMaf1 following rapamycin treatment (compare lane 4 with 9 and lane 5 with 10).
dMyc activates Pol III transcription in vivo by two distinct mechanisms, but is not the major mediator of nutrition/TOR signalling
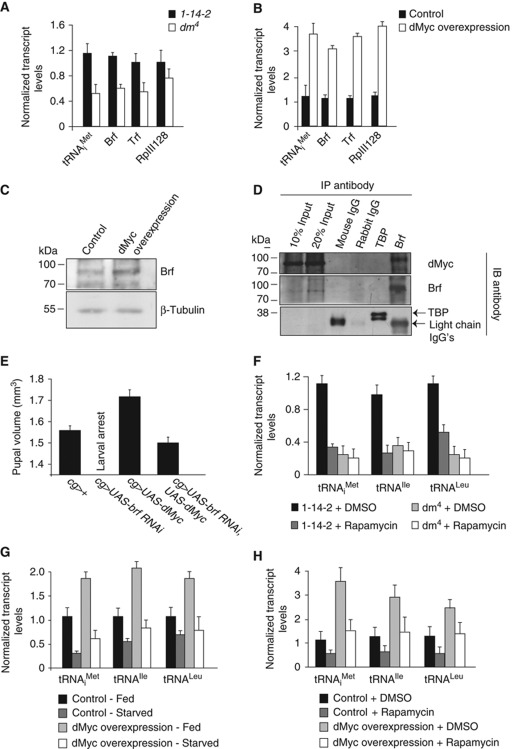
In both mammalian and Drosophila cultured cells, Myc can interact with Brf and stimulate Pol III-dependent transcription (Gomez-Roman et al, 2003; Steiger et al, 2008). We therefore explored the involvement of dMyc in regulating Pol III-dependent transcription in vivo. First, we examined dMyc null mutants and found that both tRNA levels and mRNA levels of components of the Pol III machinery—Brf, Tata Binding Protein (TBP)-related factor (Trf) and RNA polymerase III subunit 128 (RpIII128)—were lower than in control larvae (Figure 7A). Conversely, we found that when we overexpressed a UAS-dMyc transgene using the flp-out technique we observed significantly higher levels of tRNA and Brf, Trf and RpIII128 mRNAs (Figure 7B). We also preformed immunoblot analysis of larval extracts and found an increase in Brf protein levels following dMyc overexpression (Figure 7C). Finally, we performed a co-immunoprecipitation experiment in larval extracts, and using antisera directed against Brf, we identified an association between dMyc and Brf (Figure 7D). In contrast, mouse or rabbit immunoglobulins or antisera against TBP, which does not participate in initiating Pol III-dependent transcription in Drosophila (Takada et al, 2000), did not pull down dMyc (Figure 7D). Together, these data suggest that during Drosophila development, dMyc can promote Pol III-dependent transcription by at least two mechanisms: by controlling the levels of components of the Pol III apparatus and by directly associating with Brf. We explored whether these effects on Pol III were required for dMyc-induced growth. Previous studies have shown that, like TOR activity, overexpression of dMyc in the larval fat body can promote systemic growth and hence increase body size (Delanoue et al, 2010). We found that the increased pupal volume seen following the fat body expression of dMyc (cg>dMyc) was suppressed by co-expression of brf RNAi (Figure 7E). Given that dMyc levels are also controlled by nutrient/TOR signalling (Teleman et al, 2008), dMyc may also be a possible candidate for the regulatory link between TOR and Brf. We found that the reduced tRNA levels seen in dMyc null mutants were not decreased further upon rapamycin treatment, suggesting that both TOR and dMyc may function in a linear pathway to control Pol III-dependent transcription (Figure 7F). To further investigate this, we asked whether maintaining high dMyc activity in Drosophila larvae could bypass the starvation induced decrease in tRNA synthesis. Our approach was to drive a UAS-dMyc transgene expression using the flp-out system and measure tRNA levels in both fed and starved larvae. As expected, we found that starvation led to a decrease in tRNA levels in control larvae (Figure 7G). Overexpression of dMyc stimulated tRNA synthesis in fed larvae, but only produced a modest increase in tRNA levels in starved animals (Figure 7G). This was not due to the fact that dMyc was less active in starved animals, since PPAN, a dMyc target gene, was as strongly induced in starved animals as in fed animals (Supplementary Figure S10A). Similar results on both tRNA and PPAN mRNA levels were obtained when we specifically inhibited TOR by rapamycin treating larvae overexpressing dMyc (Figure 7H; Supplementary Figure S10B). These findings with dMyc contrast with our findings with dMaf1 suppression, which was sufficient to completely bypass either the starvation- or rapamycin-induced inhibition of tRNA synthesis (see Figure 6). Together, our data suggest a model for Pol III regulation in Drosophila that is outlined in Figure 8.
Figure 7.
dMyc activates Pol III-dependent transcription in vivo by two distinct mechanisms. (A) tRNAiMet, Brf and Trf mRNA levels were significantly decreased in dMyc homozygous mutants (dm4) compared with controls (1-14-2, P<0.05; Student's t-test) 48 h AEL. (B) tRNA synthesis and TFIIIB mRNA levels were significantly elevated in larvae following dMyc overexpression using the flp-out technique (hsflp/+;+;UAS-dMyc/act>CD2>Gal4) compared with controls (hsflp/+;+;act>CD2>Gal4/+, P<0.05; Student's t-test) at 120 h AEL following a 24-h induction in dMyc gene expression. (C) Immunoblotting of whole larval protein extracts with antibodies specific to Brf and β-tubulin revealed an increase in Brf protein levels following dMyc overexpression (using the flp-out technique) compared with controls at 120 h AEL following a 24-h induction in dMyc gene expression. (D) Co-immunoprecipitation studies revealed a specific association between Brf and dMyc proteins in wild-type (yw) Drosophila larval extracts 96 h AEL. (E) Fat body expression of dMyc (cg>UAS-dMyc) increases pupal volume compared with controls (cg>+). This effect is abolished when Brf levels are decreased in the larval fat body (cg>UAS-brf RNAi, UAS-dMyc) and pupae are similar in size to controls. cg>brf RNAi larvae fail to progress to the pupal stage. (F) Rapamycin treatment of dMyc mutants failed to decrease Pol III-dependent transcription further at 48 h AEL in whole larvae. (G, H) Overexpression of dMyc (using the flp-out technique) failed to reverse the decrease in tRNA synthesis in starved animals (G) and rapamycin-treated animals (H), as was measured by qRT–PCR of RNA extracted from whole larvae 72 h AEL. For all qRT–PCR experiments error bars indicate s.e.m.
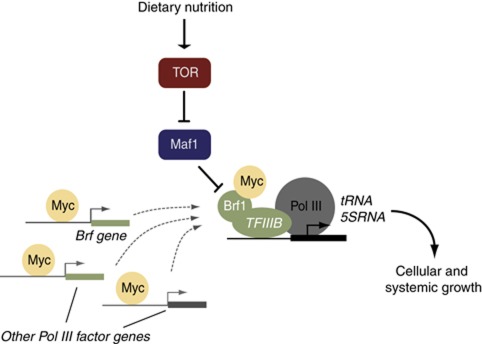
Figure 8.
A model for nutrient/TOR regulation of Pol III in Drosophila. Our data suggest the predominant mechanism by which nutrition/TOR controls Pol III is via Maf1 repression, since Maf1 inhibition completely reverses the decrease in tRNA synthesis caused by TOR inhibition. Myc is sufficient and necessary for Pol III transcription, through controlling levels of Pol III factors (such as Brf) and through interaction with Brf. TOR can control Myc protein levels (Parisi et al, 2011; Teleman et al, 2008—dashed arrow in model figure). But these effects probably do not play a major role in how TOR activates Pol III since our data show that—unlike Maf1 inhibition—maintaining Myc levels and activity cannot reverse the decrease in tRNA synthesis caused by TOR inhibition.
Discussion
The TOR kinase is one of the best-established growth regulators (Wullschleger et al, 2006). In virtually all animals, TOR activity can be stimulated by extracellular cues such as growth factors, nutrients and oxygen (Wang and Proud, 2009) to control cell, tissue and organismal growth.
Despite the knowledge of the signalling inputs to TOR, we know little about the mechanisms that allow TOR to modulate cell metabolism and drive growth. Most studies on metabolic functions modulated by TOR have been confined to yeast and mammalian cell culture. These studies have been important in defining roles for TOR in protein synthesis, nutrient uptake and metabolism and autophagy (De Virgilio and Loewith, 2006; Wullschleger et al, 2006). But they leave open the question of what mechanisms operate in vivo to control tissue and organ growth during animal development. Genetic studies in Drosophila have been pivotal in this regard (Grewal, 2008; Hietakangas and Cohen, 2009; Teleman, 2009). Here, we show that the ability of the TOR pathway to control transcription through Pol III governs cell, tissue and ultimately organismal growth in Drosophila. Given that Pol III drives transcription of several non-coding RNAs required for mRNA translation, we suggest that the stimulation of Pol III by TOR enhances the protein synthetic capacity of cells. We previously showed that Drosophila TOR also controls synthesis of rRNA synthesis, via the RNA polymerase I factor, TIF-IA (Grewal et al, 2007). Moreover, recent studies in Drosophila larvae demonstrated that the insulin/TOR pathway regulates the expression of ribosome biogenesis genes via the transcription factors FOXO and Myc (Teleman et al, 2008; Li et al, 2010). Thus, in Drosophila, tissue and organismal growth relies on the ability of TOR to regulate all three nuclear RNA polymerases to ultimately promote protein synthesis. Given that regulation of all three polymerases is a conserved function for TOR, we suggest that these mechanisms may also underlie tissue and organ growth in mammalian development.
The Pol III transcription factor Brf is an essential component of the TFIIIB complex responsible for recruiting Pol III to gene promoters (Geiduschek and Kassavetis, 2001). Our work indicates that Brf activity is required for Drosophila development. Patterning and cell fate specification appear normal in brf embryos. However, once these mutants hatch as larvae they fail to grow. Our data suggest that this growth arrest phenotype reflects a role for Brf activity downstream of TOR. We found that Brf is cell-autonomously required for growth in both endoreplicating cells, which make up the bulk of larval mass, and the mitotically dividing cells of the imaginal discs. In particular, we find that brf mutant wing disc cell clones are outcompeted by wild-type neighbours. This cell competition phenotype is seen in mutants for other genes required for protein synthesis, such as the ribosomal proteins and Myc (Johnston, 2009). An important finding was that the overgrowth caused by loss of TSC1 (and hence increased TOR activity) was blocked in brf mutant cells. In mammalian cells, Brf activity is induced by cues that promote cell growth (e.g., during hypertrophic growth of cardiac cells) whereas cell differentiation leads to inhibition of Brf (Goodfellow and White, 2007; Athineos et al, 2010). In fact, overexpression of Brf alone can promote proliferation and transformation in immortalized fibroblasts (Marshall et al, 2008), while loss of Brf inhibits these processes (Johnson et al, 2007; Marshall et al, 2008). Mutations in tumour suppressors such as TSC are common in cancer and lead to elevated TOR activity and promotion of tumour growth. Based on our data, we suggest that Brf is required in vivo for both normal tissue growth and TOR-induced tumour growth.
Our data indicate the predominant mechanism by which nutrition/TOR controls Pol III is via Maf1 repression, since Maf1 inhibition completely reverses the decrease in tRNA synthesis caused by reducing TOR activity. These findings extend those observed in both yeast and mammalian cell culture, and suggest an important role for dMaf1 in vivo in developing tissues. The exact mechanism by which Maf1 functions is not clear, but it may involve inhibition of Brf and Pol III recruitment to genes, possibly by direct binding or association with Brf/Pol III (Desai et al, 2005; Vannini et al, 2010). Indeed, we could see an enhanced association between dMaf1 and Brf1 upon TOR inhibition. We also explored the role of dMyc as a potential link between nutrient-TOR signalling and Pol III. We found that dMyc was both necessary and sufficient for the control of Pol III activity during development. As previously reported in both mammalian and Drosophila culture, we were able to identify an interaction between dMyc and Brf (Gomez-Roman et al, 2003; Steiger et al, 2008). In addition, we identified a role for dMyc in controlling the levels of components of the Pol III machinery, including both Trf and Brf which form part of the TFIIIB complex. Thus, dMyc likely has both direct and indirect effects on Pol III activity in Drosophila. These effects are necessary for both dMyc-induced cell growth (Steiger et al, 2008) and, as we show here, for the non-autonomous increases in body size caused by dMyc in fat cells. Previous studies have shown that, in Drosophila, TOR controls Myc protein levels (Teleman et al, 2008; Parisi et al, 2011). But these effects on Myc probably do not play major role in how TOR activates Pol III since our data show that, unlike inhibition of Maf1, maintaining Myc levels and activity cannot reverse the decrease in tRNA synthesis caused by TOR inhibition. Moreover, if Myc protein levels were limiting for TOR-dependent control of Pol III, then we would not expect that knockdown of Maf1 could completely reverse the effects of rapamycin/starvation. Given that Maf1 inhibition did not influence levels of Pol III factors, pre-rRNA or RP gene mRNA—transcripts that are upregulated by dMyc—it is unlikely that Maf1 influences Myc function. We did find that rapamycin feeding could not exacerbate the reduction of tRNA levels seen in dMyc null mutants. This result in principle may suggest that TOR signalling does not exert any dMyc-independent effects on Pol III function. But, we suggest this finding probably occurs because in the absence of Myc, Pol III activity may be approaching basal levels and cannot be significantly decreased much further. Taken together, although our data may not completely rule out some contribution of Myc to TOR-dependent control of Pol III, they do indicate that it is not the major contributor.
It is clear that both TOR and Myc are essential regulators of Pol III. But, it is likely that while TOR can control Myc levels, both TOR and Myc can also function in parallel and independently of each other. Teleman et al (2008) previously showed that overactivation of TOR signalling could not promote growth when Myc was inhibited, but at the same time Myc overexpression could not promote growth when TOR was inhibited. These findings and our data suggest that TOR and Myc cannot necessarily be placed in a simple, linear pathway. Recent studies in Drosophila have emphasized how other conserved growth-regulatory pathways, particularly those that control growth of the imaginal tissues (such as Wingless, EGF/Ras, the Hippo-Yorkie pathway and Bantam RNAi) function via control of dMyc (Johnston et al, 1999; Prober and Edgar, 2002; Herranz et al, 2011; Neto-Silva et al, 2011; Ziosi et al, 2011). Thus, dMyc may play a role in coupling these pathways to the control of Pol III activity to stimulate cell growth and proliferation.
It is interesting to speculate as to which Pol III targets are important for growth control. Pol III regulates the expression of several short non-coding RNAs, such as the tRNAs, 5S rRNA and 7SL RNA. Regulation of 5S rRNA production by Brf could influence ribosome synthesis and hence growth. However, we found that loss of Brf did not inhibit Pol I activity or alter levels of rRNA, suggesting that Brf probably does not directly influence ribosome numbers. One attractive possibility is that levels of the tRNAs may be limiting for translation and growth. In support of this notion, a recent paper showed that overexpression of Brf increased tRNA levels and promoted proliferation and transformation of cultured mammalian fibroblasts (Marshall et al, 2008). These effects of Brf were phenocopied by just increasing levels of tRNAiMet, and were associated with augmented mRNA translation and increased protein levels of growth promoters such as c-Myc and cyclin D1. We did not see a consistent increase in tRNAs when we simply overexpressed Brf in larvae, perhaps because levels of other components of the TFIIIB complex are limiting in flies. Nevertheless, by controlling Brf activity and tRNA synthesis, TOR could promote translation of growth regulators and drive larval growth. In fact, a recent paper indicated that TOR signalling in Drosophila regulates dMyc protein levels, but not dMyc mRNA levels, consistent with a possible role for translational control (Teleman et al, 2008).
One interesting result of our work was the identification of a non-cell autonomous role for Brf in organismal growth. Specifically, we found that Brf activity in the fat cells of Drosophila larvae could influence larval growth and final size. Elegant work by Leopold and colleagues has outlined a role for TOR in the fat body as a relay to control peripheral insulin signalling. In feeding larvae, amino-acid input into fat cells activates TOR, leading to transmission of a secreted signal from fat to brain to increase dILP expression and release from brain IPCs (Colombani et al, 2003; Geminard et al, 2009). Our data suggest that stimulation of Pol III activity may be an important downstream effector of this adipose function of TOR. Thus, adipose-specific silencing of Brf led to reduced peripheral insulin signalling, slower larval growth rate and reduced final body size. We found that, as in starved larvae, loss of brf led to reduced expression of dilp mRNA (seen in both brf mutants and cg>brf RNAi larvae) and reduced dILP release from the brain. Moreover, given that levels of phospho-Akt are lower, and levels of dInR (a FOXO target) are higher in tissues from both brf mutant and r4>brf RNAi larvae it is clear that systemic insulin signalling is reduced when Brf is inhibited in the fat body. We also found that another fat phenotype associated with starvation and loss of TOR, accumulation of lipid droplets, was phenocopied by loss of Brf. However, the autophagy phenotype of starved larval fat bodies was not phenocopied by loss of Brf. Therefore, Brf and Pol III function in the Drosophila fat body may mediate some, but not all of TOR's effects on growth and metabolism. The exact nature of the fat-to-brain secreted factor that controls insulin release in flies is not yet known, but perhaps translation of this signal, if it is a peptide or secreted protein, is influenced by changes in tRNA synthesis and translation rates. Indeed, Leopold et al showed that dMyc activity in the fat body was also important for controlling systemic insulin signalling, growth and body size (Delanoue et al, 2010). This effect of dMyc correlated with elevated expression of ribosome biogenesis genes and increased nucleolar size, an index of ribosome synthesis. We find that dMyc overexpression can also stimulate Pol III and tRNA levels, and that the increase in body size caused by fat body overexpression of dMyc is reversed by knockdown of Brf. These data suggest that regulation of mRNA translational capacity is a key step downstream of TOR and dMyc in fat cells to control signalling to IPCs.
Together, these data suggest that mRNA translational control may underlie a role for the fat body as an endocrine organ. A similar theme is emerging in mouse models. Mammalian adipose tissue is known to secrete adipokines and leptin to influence organismal metabolism and growth (Waki and Tontonoz, 2007). The secretion of many of these factors is influenced by diet, suggesting a regulatory role for TOR signalling. Genetic inhibition of either TOR and S6K in mice leads to alterations in metabolic activity in adipose tissue (Um et al, 2004, 2006; Polak et al, 2008; Cybulski et al, 2009). Moreover, loss of the translational repressors, 4E-BP1 and 4E-BP2, both of which are downstream TOR effectors, alters lipid and glucose metabolism in mice (Le Bacquer et al, 2007). To date, there are no mouse models of Pol III. However, it is interesting to speculate that changes in Pol III and tRNA synthesis are involved in mediating effects of TOR in adipose tissue in mice. Regulation of Pol III by TOR may also be important in the metabolic control of other processes. For example, TOR is a conserved regulator of organismal stress responses and lifespan (Kapahi et al, 2010). These stress responses rely on TOR's ability to control translation. We suggest that regulation of Pol III and tRNA synthesis may also be a mode of control. Further organismal studies, using genetic modulation of Pol III function, should provide additional insights into these points.
Materials and methods
Fly stocks
UAS-brf RNAi (NIG, Japan), dtorΔP (Zhang et al, 2000); dS6KL1 (Teleman et al, 2005); UAS-dS6KTE1, UAS-dMaf1 RNAi, UAS-tsc1 RNAi/Tm6B, UAS-tsc1/2, UAS-dMyc, 1-14-2 (Pierce et al, 2004); dm4 (Pierce et al, 2004); brfEY02964; FRT82B, brfEY02964; FRT82B, tsc1Q87X (Tapon et al, 2001); Df(3R)BSC565, da-GAL4, ptc-GAL4, ey-GAL4, r4-GAL4 (Lee and Park, 2004) and cg-GAL4 were used (see FlyBase for further information: http://flybase.org).
All flies were reared and maintained at 25°C on standard Drosophila media (150 g agar, 1500 g cornmeal, 315 g yeast, 675 g sucrose, 1875 g D-glucose, 240 ml propionic acid per 34.5 l water).
Egg collection
Adult flies were allowed to lay eggs on grape juice agar plates supplemented with yeast paste for 4 h at 25°C. Twenty-four hours after egg laying (AEL), the plates were precleared of larvae and then larvae that hatched within the next 4 h were placed in food vials in groups of 50 and allowed to develop.
Starvation
Larvae were collected for starvation 72 h AEL and starved in sterile 20% sucrose in PBS, for 24 h unless stated otherwise in the figure legends. Following starvation, whole larvae were collected.
Rapamycin treatment of Drosophila S2 cells
Drosophila S2 cells (a kind gift from Edan Foley) were cultured at 25°C in Schneider's medium (Gibco; 11720-034) supplemented with 10% fetal bovine serum (Gibco; 10082-139), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco; 15140). At 90% confluency, cells were treated with either 20 nM rapamycin (Calbiochem; 80054-246) or DMSO (Sigma; D2650) for 1 h, following which cells were washed twice with ice-cold PBS. Cells were then scraped into either TriZOL or protein lysis buffer (both procedures are detailed below) to prepare RNA and protein extracts, respectively.
Collection of material for RNA and protein extractions
Whole larvae or peripheral tissues were collected at the time points AEL as indicated in the figure legends. In the case of r4>brf RNAi experiments, peripheral tissues were prepared by stripping whole larvae of fat body. dMyc overexpression was performed using the heat-shock flp-out method (Elliott and Brand, 2008). For dMyc starvation and rapamycin treatment experiments, transgene expression was induced by incubating larvae at 37°C for 1, 48 h AEL. Controls lacking the UAS transgene were similarly heat shocked and treated as below. At 72 h AEL, fed control larvae were harvested while starved larvae were placed in 20% sucrose/PBS for a further 24 h after which they were also taken for RNA extractions. Rapamycin treatment took place in 35 mm petri dishes and involved placing larvae on a mixture of 3 g of prepared instant Drosophila media formula 4–24 (California Biologic Supply Company), 1 g of liquid inactivated yeast food and either 0.1% DMSO (Sigma; D2650) or 20 μM rapamycin (Calbiochem; 80054-246). Larvae were transferred to this food at 72 h AEL for a period of 24 h before being taken for RNA extractions. This rapamycin treatment method was used for the dMyc mutant and dMaf1 RNAi experiments at the times indicated above however in the case of the dMyc mutant experiments larvae were transferred to the DMSO/rapamycin containing food at 24 h AEL for a period of 24 h prior to RNA extractions. In each qRT–PCR experiment, a minimum of four groups of 8–10 larvae were collected. Each collection was independently performed a minimum of three times.
Quantitative RT–PCR
Total RNA was extracted using TRIzol according to manufacturer's instructions (Invitrogen; 15596-018). RNA samples (1 μg per reaction) were DNase treated according to manufacturer's instructions (Ambion; 2238G) and reverse transcribed using Superscript II (Invitrogen; 100004925). The generated cDNA was used as a template to perform qRT–PCRs (BioRad Laboratories; MyIQ PCR machine using SyBr Green PCR mix) using specific primer pairs (sequences available upon request). PCR data were normalized to the average fold change of either β-tubulin1 or tak1 mRNA levels, both of which were unchanged in response to a variety of environmental and genetic manipulations (Li et al, 2010). Each experiment was independently repeated a minimum of three times. All data were analysed by Student's t-tests.
Preparation of protein extracts, immunoblotting and antibodies
Whole larval protein extracts were prepared by washing material twice in ice-cold PBS before being homogenized in the appropriate volume of lysis buffer (20 mM Hepes (pH 7.8), 450 mM NaCl, 25% glycerol, 50 mM NaF, 0.2 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 1 mM DTT, 1 × Protease Inhibitor Cocktail (Roche, 04693124001)) using a motorized pestle. Following incubation on ice for 10 min, the lysates were cleared by centrifugation for 10 min at 10 000 r.p.m. at 4°C. Drosophila S2 cell extracts cells were prepared as previously described (Goodfellow and White, 2007). Protein (15 μg) was resolved by SDS–PAGE and immunoblotting performed as previously described (Marshall et al, 2008). Antibodies used were against a C-terminal fragment of Drosophila Brf (Takada et al, 2000), TBP (Santa Cruz Biotechnology Inc; 58C9), β-tubulin (E7, Drosophila Studies Hybridoma Bank), dMyc (Prober and Edgar, 2002) phospho-Drosophila Akt Ser505 (Cell Signaling Technology; 4054) and Akt (Cell Signaling Technology; 9272). Peptide antiserum against dMaf1 was raised by immunizing rabbits with synthetic peptide NNSQSGDEGITLC, corresponding to residues 74–87.
Immunoprecipitation
Drosophila S2 whole cell or larval extract (500 μg) was incubated at 4°C for 3 h on a rotating wheel with 25 μl protein A-sepharose beads (Sigma; P9424) that had been preincubated with antiserum against Brf, dMaf1, or a rabbit IgG control (Santa Cruz Biotechnology; sc-2027). Bound material was resolved by SDS–PAGE and specific proteins detected by immunoblotting as previously described (Marshall et al, 2008). Antibodies used for immunoprecipitation and immunoblot are described above.
Mitotic recombination, clone and cell size analysis
Mitotic recombination was performed using the flp/FRT method. For the fat body cell analysis, we performed a 6-h egg collection followed by a 1-h heat shock at 37°C. Larvae were transferred to food 24 h after heat shock. DAPI and phalloidin staining of fat bodies was performed on inverted and 4% paraformaldehyde fixed 120 h AEL larvae, following which fat bodies were dissected and mounted in Vectashield (Vector Laboratories Inc; H-1000). For analysis of twin-spot clones in the wing imaginal discs in Figure 3A–C, larvae were heat shocked for 20 min at 37°C, 60 h AEL. Wing discs were dissected at 120 h AEL and mounted in Vectashield for visualization. For Figure 1F clones were induced and wing discs were analysed at 24, 48 and 72 h after clone induction for counting. Viability of mutant clones in the wing imaginal discs was assessed by counting the percentage of wild-type clones that were still paired with a brf twin spot. Clone and cell sizes were calculated using Adobe Photoshop using the histogram tool. To induce wing disc clones for fluorescence-activated cell sorting (FACS) analysis, larvae were heat shocked at 37°C for 1, 72 h AEL and discs dissected and trypsinized 120 h AEL.
Flow cytometry
FACS analysis was performed on dissociated wing imaginal discs as previously described (Johnston et al, 1999).
Microscopy
All images were obtained on a Zeiss Observer Z1 microscope using Axiovision software. Microscopy and image capture were performed at room temperature and captured images were processed using Photoshop 7.0 (Adobe).
Nile red staining
Nile red staining of lipid droplets was performed as described previously (Grönke et al, 2005).
Lysotracker staining
Lysotracker staining was performed on dissected fat bodies from 72 h AEL larvae. Larval fat bodies were dissected in PBS then incubated in 1 μM Hoescht (Invitrogen; H3570), 100 μM lysotracker green (Molecular Probes; L7526) in 80% glycerol, on a coverslip for 15 min prior to image capture.
Pupation rates
Larvae were collected 24 h AEL and placed in food vials in groups of 50 per vial. The number of pupae formed on the side of the food vial was counted every 24 h and presented as a percentage of the total of number of pupae formed for each genotype.
Adult weight measurement
Following eclosion, adult flies were transferred to food vials in groups of 50 and aged for 3 days The average weight of adult males was calculated by weighing flies in groups of 10 with a precision balance (Sartorius). Data are presented as average weight calculated from at least five independent groups.
dILP2 immunostaining
dILP2 antibody staining of larval brains at 96 h AEL was as previously described (Geminard et al, 2009).
Pupal volume
Pupal volume was calculated as previously described (Delanoue et al, 2010).
Statistics
For all experiments, error bars represent s.e.m., and P-values are the results of a Student's t-test provided by Microsoft Excel.
Supplementary Material
Acknowledgments
We thank Tom Neufeld, Eugenia Piddini, Michael Pankratz, Jae Park, Iswar Hariharan, Robert Eisenman and the NIG (Japan), VDRC (Vienna) and Bloomington Stock Centres for providing flies; Edan Foley for kindly supplying the Drosophila S2 cells, Shinako Takada for providing the Brf antibody, Robert Eisenman for kindly providing the dMyc antibody and Ernst Hafen for kindly providing the dILP2 antibody. The β-tubulin (E7) antibody, developed by Michael Klymkowsky, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. We thank William Brook, Jeb Gaudet and Jim McGhee for comments on the manuscript and S Salgia for technical assistance. This work was supported by a Canadian Institutes of Health Research (CIHR) grant (MOP-86622) and an Alberta Cancer Foundation New Investigator grant to SS Grewal. L Marshall was supported by postdoctoral fellowships from Alberta Innovates Health Solutions and Alberta Cancer Foundation. E Rideout was supported by postdoctoral fellowships provided by the Natural Sciences and Engineering Research Council of Canada, Alberta Innovates Health Solutions and the CIHR Training Program in Genetics, Child Development and Health (Alberta Children's Hospital Research Institute for Child and Maternal Health).
Author contributions: LM drafted and SG revised the manuscript. LM, ER and SG conceived and designed the experiments. LM, ER and SG performed the experiments, analysed the data and prepared the figures. All authors read and approved the final manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arsham AM, Neufeld TP (2006) Thinking globally and acting locally with TOR. Curr Opin Cell Biol 18: 589–597 [DOI] [PubMed] [Google Scholar]
- Athineos D, Marshall L, White RJ (2010) Regulation of TFIIIB during F9 cell differentiation. BMC Mol Biol 11: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N (2007) The two TORCs and Akt. Dev Cell 12: 487–502 [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA (2002) Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell 2: 239–249 [DOI] [PubMed] [Google Scholar]
- Clarke EM, Peterson CL, Brainard AV, Riggs DL (1996) Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J Biol Chem 271: 22189–22195 [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P (2003) A nutrient sensor mechanism controls Drosophila growth. Cell 114: 739–749 [DOI] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN (2009) mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci USA 106: 9902–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann SG, Thomas G (2006) The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett 580: 2821–2829 [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R (2006) The TOR signalling network from yeast to man. Int J Biochem Cell Biol 38: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Delanoue R, Slaidina M, Leopold P (2010) The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev Cell 18: 1012–1021 [DOI] [PubMed] [Google Scholar]
- Desai N, Lee J, Upadhya R, Chu Y, Moir RD, Willis IM (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem 280: 6455–6462 [DOI] [PubMed] [Google Scholar]
- Dieci G, Duimio L, Peracchia G, Ottonello S (1995) Selective inactivation of two components of the multiprotein transcription factor TFIIIB in cycloheximide growth-arrested yeast cells. J Biol Chem 270: 13476–13482 [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM (2009) mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Brand AH (2008) The GAL4 system: a versatile system for the expression of genes. Methods Mol Biol 420: 79–95 [DOI] [PubMed] [Google Scholar]
- Foster KG, Fingar DC (2010) Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 285: 14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Pan D (2001) TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 15: 1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA (2001) The RNA polymerase III transcription apparatus. J Mol Biol 310: 1–26 [DOI] [PubMed] [Google Scholar]
- Geminard C, Rulifson EJ, Leopold P (2009) Remote control of insulin secretion by fat cells in Drosophila. Cell Metab 10: 199–207 [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ (2003) Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, White RJ (2007) Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 6: 2323–2326 [DOI] [PubMed] [Google Scholar]
- Grewal SS, Evans JR, Edgar BA (2007) Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J Cell Biol 179: 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS (2008) Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int J Biochem Cell Biol 41: 1006–1010 [DOI] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP (2005) Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 1: 323–330 [DOI] [PubMed] [Google Scholar]
- Herranz H, Hong X, Perez L, Ferreira A, Olivieri D, Cohen SM, Milan M (2011) The miRNA machinery targets Mei-P26 and regulates Myc protein levels in the Drosophila wing. EMBO J 29: 1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM (2009) Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43: 389–410 [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E (2002) Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol 12: 1293–1300 [DOI] [PubMed] [Google Scholar]
- Ito N, Rubin GM (1999) gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell 96: 529–539 [DOI] [PubMed] [Google Scholar]
- Jastrzebski K, Hannan KM, Tchoubrieva EB, Hannan RD, Pearson RB (2007) Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25: 209–226 [DOI] [PubMed] [Google Scholar]
- Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL (2007) Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell 26: 367–379 [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P (1999) Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA (2009) Competitive interactions between cells: death, growth, and geography. Science 324: 1679–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ (2010) mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA 107: 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L (2010) With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab 11: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N (2007) Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 117: 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Park JH (2004) Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM (2009) Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem 284: 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Edgar BA, Grewal SS (2010) Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cis-regulatory element. BMC Cell Biol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ (2008) Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133: 78–89 [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I (2006) Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391 [DOI] [PubMed] [Google Scholar]
- Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N (2010) mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 30: 3749–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N (2001) The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol 3: 596–601 [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA (2011) Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E (2003) Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol 13: 79–85 [DOI] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E (2000) Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev 14: 2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F, Riccardo S, Daniel M, Saqcena M, Kundu N, Pession A, Grifoni D, Stocker H, Tabak E, Bellosta P (2011) Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Britton JS, Loo LW, Flynn EM, Edgar BA, Eisenman RN (2004) dMyc is required for larval growth and endoreplication in Drosophila. Development 131: 2317–2327 [DOI] [PubMed]
- Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN (2008) Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8: 399–410 [DOI] [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T (2001) Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357–368 [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA (2002) Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev 16: 2286–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG (2007) Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J 403: 217–234 [DOI] [PubMed] [Google Scholar]
- Puig O, Tjian R (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy I, Moir RD, Librizzi M, Willis IM (1995) In vitro evidence for growth regulation of tRNA gene transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J Biol Chem 270: 28463–28470 [DOI] [PubMed] [Google Scholar]
- Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K (2010) Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem 285: 15380–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger D, Furrer M, Schwinkendorf D, Gallant P (2008) Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Takada S, Lis JT, Zhou S, Tjian R (2000) A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101: 459–469 [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK (2001) The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM (2005) 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev 19: 1844–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Teleman AA (2009) Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem J 425: 13–26 [DOI] [PubMed] [Google Scholar]
- Thomas G (2002) The S6 kinase signaling pathway in the control of development and growth. Biol Res 35: 305–313 [DOI] [PubMed] [Google Scholar]
- Um SH, D’Alessio D, Thomas G (2006) Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab 3: 393–402 [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205 [DOI] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143: 59–70 [DOI] [PubMed] [Google Scholar]
- Waki H, Tontonoz P (2007) Endocrine functions of adipose tissue. Annu Rev Pathol 2: 31–56 [DOI] [PubMed] [Google Scholar]
- Wang X, Proud CG (2009) Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol 19: 260–267 [DOI] [PubMed] [Google Scholar]
- Wei Y, Tsang CK, Zheng XF (2009) Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J 28: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ (2005) RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL (2008) PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol 28: 4204–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Zaragoza D, Ghavidel A, Heitman J, Schultz MC (1998) Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol 18: 4463–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP (2000) Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14: 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S, Pession A (2011) dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet 6 (9): pii: e1001140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.