Abstract
Autophagy is a catabolic process by which cytoplasmic components are sequestered and transported by autophagosomes to lysosomes for degradation, enabling recycling of these components and providing cells with amino acids during starvation. It is a highly regulated process and its deregulation contributes to multiple diseases. Despite its importance in cell homeostasis, autophagy is not fully understood. To find new proteins that modulate starvation-induced autophagy, we performed a genome-wide siRNA screen in a stable human cell line expressing GFP–LC3, the marker-protein for autophagosomes. Using stringent validation criteria, our screen identified nine novel autophagy regulators. Among the hits required for autophagosome formation are SCOC (short coiled-coil protein), a Golgi protein, which interacts with fasciculation and elongation protein zeta 1 (FEZ1), an ULK1-binding protein. SCOC forms a starvation-sensitive trimeric complex with UVRAG (UV radiation resistance associated gene) and FEZ1 and may regulate ULK1 and Beclin 1 complex activities. A second candidate WAC is required for starvation-induced autophagy but also acts as a potential negative regulator of the ubiquitin-proteasome system. The identification of these novel regulatory proteins with diverse functions in autophagy contributes towards a fuller understanding of autophagosome formation.
Keywords: FEZ1, SCOC, siGenome screen, UVRAG, WAC
Introduction
Macroautophagy (here autophagy) is an essential, conserved degradative pathway that has a role in cell homeostasis in normal conditions, eliminating damaged organelles or misfolded proteins. Autophagy-deficient mice die immediately after birth (Kuma et al, 2004), and defects in autophagy have been linked to multiple diseases including neurodegenerative disorders such as Huntington's disease, cancer and immune diseases (Mizushima et al, 2008). Autophagy is also induced in response to amino-acid deprivation or external stress, recycling macromolecules and restoring metabolic functions (Kuma and Mizushima, 2010). Decreased amino acids in the cytosol causes inactivation of mTOR (mammalian target of rapamycin), a major regulator of cell growth, and induces autophagy (Jung et al, 2010).
Many essential mammalian autophagy (Atg) proteins, most originally discovered in yeast, have been identified, leading to our current basic understanding of the process (Mizushima et al, 2011). A double bi-layered membrane, the isolation membrane or phagophore, encloses cytoplasmic macromolecules and organelles, forming an autophagosome. Autophagosome content degradation occurs upon fusion with the lysosome, releasing amino acids and macromolecules. The molecular basis for the initiation of autophagy is the activation of ULK1 (UNC-51-like) kinase after its dissociation from mTOR complex 1 (mTORC1) (Chan and Tooze, 2009), and recruitment of the Beclin 1 phosphatidylinositol 3-kinase complex (Vps34, p150, Beclin 1 and Atg14) to the nascent phagophore (Simonsen and Tooze, 2009). Further expansion of the phagophore requires two Ubiquitin-like (UBL) conjugation reactions; one produces an Atg12-5-16 conjugate complex through the activity of the E1- and E2-like enzymes Atg7 and Atg10. The other produces MAP1LC3B (here referred to as LC3) covalently attached to phosphatidylethanolamine, mediated by the E1- and E2-like enzymes Atg7 and Atg3 after C-terminal processing by Atg4. Another key player in autophagy is mammalian Atg9 (mAtg9), which is proposed to shuttle membrane to the autophagosome (Webber and Tooze, 2010b) and is regulated by the p38α MAPK signalling pathway (Webber and Tooze, 2010a).
Many aspects of initiation, formation and expansion of phagophores and autophagosomes, as well as the regulation of the autophagic process in particular in response to amino-acid starvation remain poorly understood (Kuma and Mizushima, 2010; Yang and Klionsky, 2010). Large-scale siRNA screens represent one approach to identify proteins whose functions may provide additional insight. ULK1 was identified in an siRNA screen using a kinome library (Chan et al, 2007). Recently, modulators of basal autophagy in a neuroblastoma cell line were uncovered in an siRNA genome screen (Lipinski et al, 2010). We developed our previous screen strategy using amino-acid starvation-dependent formation of GFP–LC3-positive autophagosomes (Chan et al, 2007) to a genome-wide screen and we discovered nine novel proteins that regulate starvation-induced autophagy. Of these, we validated and characterised two positive regulators of autophagy, SCOC (short coiled-coil protein, also called SCOCO) and WAC (WW domain containing adaptor with coiled-coil). In depth analysis of SCOC, a Golgi protein, reveals it is required for formation of an ULK1 complex containing FEZ1 (fasciculation and elongation protein zeta 1). Furthermore, SCOC forms a complex with UVRAG (UV radiation resistance associated gene), which is associated with the Beclin 1 complex, and FEZ1; SCOC shows a starvation-sensitive interaction with UVRAG that is stabilised by overexpression of FEZ1. In fact, further analysis reveals that FEZ1 and SCOC form a trimeric complex with UVRAG. Analysis of the function of WAC supports the notion that while required for autophagy, WAC may also be regulating proteasomal degradation. Our genome-wide screen has revealed key roles for two novel proteins in starvation-induced autophagy.
Results
Genome-wide siRNA screen identified 51 genes modulating starvation-induced autophagy
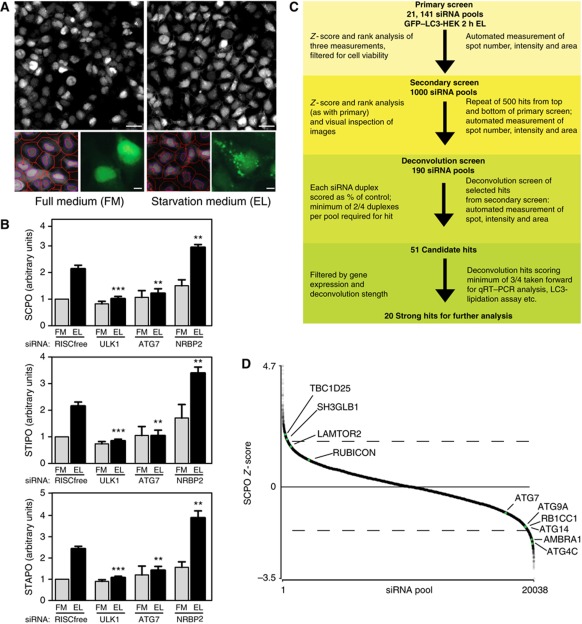
We previously used siRNA knockdown in a human HEK293 cell line stably expressing EGFP-tagged rat LC3B (hereafter GFP–LC3-HEK cells) to identify ULK1 as a key regulator of autophagy (Chan et al, 2007). We developed this platform further by quantitative analysis of the GFP–LC3 spots formed when autophagy is induced by culturing the cells in medium devoid of amino acids and including the lysosomal protease inhibitor leupeptin (Figure 1A). The addition of leupeptin allows us to distinguish the effects on autophagy initiation versus autophagosomal maturation (Rubinsztein et al, 2009); we chose to use it rather than for example bafilomycin A1, which inhibits the vacuolar-type H+-ATPase found in endosomes and lysosomes, as leupeptin is an effective inhibitor of lysosomal protease activity and autophagosome degradation (Chan et al, 2007). Knockdown of known autophagy genes such as ULK1 and ATG7 decreased the three measured parameters SCPO (spot count per object), STIPO (spot total intensity per object) and spot total area per object (STAPO) whereas NRBP2, formerly LOC340371 (Chan et al, 2007) increased the three parameters (Figure 1B). To identify novel modulators of starvation-induced autophagy, we adopted a stringent screening procedure with multiple rounds of screening and validation steps to produce a small list of very high-confidence hits (Figure 1C). We first screened for the effect of 21 141 siRNA pools of the human genome in triplicate on GFP–LC3 spots after autophagy was induced by incubation for 2 h in starvation medium containing leupeptin.
Figure 1.
Genome-wide screen for starvation-induced autophagy. (A) Cellomics images (top) of GFP–LC3-HEK cells in full medium (FM) or after 2 h in starvation medium (ES) with leupeptin (EL). Scale bars=40 μm. Optimised automated analysis (bottom left) programme detecting hoechst-labelled nuclei (purple line), cell outline (red line) and identifies GFP–LC3 spots (red spots) in FM and EL. Enlarged GFP–LC3 images in FM and EL (bottom right). Scale bars=10 μm. (B) SCPO, STIPO and STAPO after indicated siRNA treatment of GFP–LC3-HEK cells incubated as in (A). Error bars represent s.e.m. Significance was determined using a two-tailed paired t-test compared with RISCfree (RF) in EL: SCPO siULK1, ***P=0.0002; siATG7, **P=0.0078; siNRBP2, **P=0.0088. STIPO siULK1, ***P<0.0001; siATG7, **P=0.0060; siNRBP2, **P=0.0041. STAPO siULK1, ***P<0.0001; siATG7, **P=0.0037; siNRBP2, **P=0.0028. The experiments were performed: RISCfree (n=5), siULK1 (n=5), siATG7 (n=3), siNRBP2 (n=3). (C) Overview of screening strategy. (D) Scatter plot of median SCPO Z-scores from siRNA pools in the primary screen that met the minimum object count (OC) cutoff. Dashed lines represent approximate cutoff for 500 best increasers and decreasers. RP of known autophagy genes (ATG4C, ATG14, RB1CC1, ATG9A, ATG7) are given in Supplementary Table S2; AMBRA1 (FLJ20294), TBC1D25 (OATL1) and SH3GLB1 (BIF-1) in Supplementary Table S1; LAMTOR2 has a RP=708.7 and RUBICON has a RP=1126.
After the cells were fixed, multiple images of GFP–LC3 spots and nuclei were collected from each well and analysed for the three parameters; the normalised scores for these parameters were then ranked and these three ranks combined to create a rank product (RP). The top 500 spot increasers and 500 spot decreasers were determined from this RP (Supplementary Table S1). A number of essential autophagy genes had a strong negative effect on the three spot parameters underscoring their critical nature (Supplementary Table S2). Knockdown of FIP200 (RB1CC1; a component of the ULK1 complex), ATG4C, ATG9A and ATG14 significantly reduced spot count (Figure 1D), as well as AMBRA1 (FLJ20294), which is required for starvation-induced autophagy (Fimia et al, 2007). Other genes that are known negative regulators of autophagy increased GFP-LC3 spots after knockdown include TBC1D25 (OATL1) (Itoh et al, 2011), SH3GLB1/Bif-1 (Takahashi et al, 2011), LAMTOR2 (p14) (Sancak et al, 2010) and RUBICON (KIAA00226) (Matsunaga et al, 2009; Zhong et al, 2009).
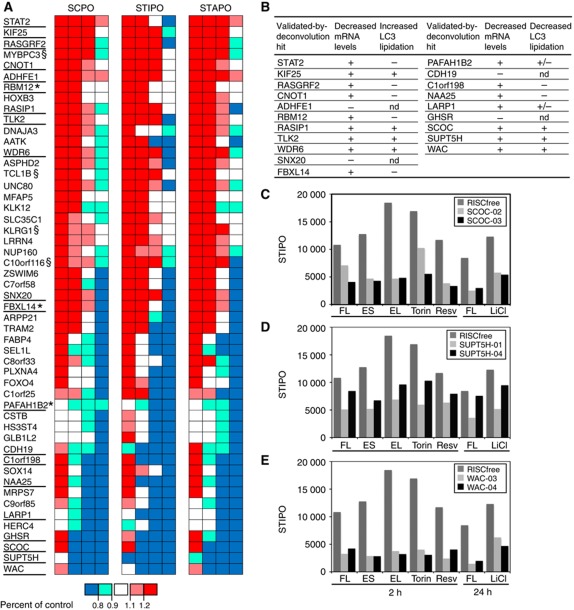
To increase confidence in the primary screen hits, we repeated the assay on the 1000 siRNA pools and used visual curation of the images to discard any obvious miscalling or other extreme cellular phenotypes. Using this method and excluding known autophagy genes, 190 siRNA pools were scored as hits (Supplementary Table S3) and taken forward for deconvolution analysis in which we tested the individual component oligonucleotide duplexes that make up the siRNA pools. We required that a minimum of two out of the four individual duplexes gave a significant effect on any of the spot parameters, and applied additional criteria to hit selection. For example, if two duplexes repeated the siRNA pool phenotype but two duplexes had the opposite effect for more than one spot parameter we did not pass this gene as a hit (see Supplementary Table S3). The 51 genes identified by this analysis (Figure 2A) map to diverse biological pathways and many of them are relatively uncharacterised (Supplementary Table S4).
Figure 2.
In all, 51 hits including SCOC and WAC are validated by deconvolution and additional assays. (A) SCPO, STIPO and STAPO for the 51 genes validated by two out of four individual siRNA duplexes reproducing the GFP–LC3 spot phenotype, above a cutoff of greater or less than 20% of control, and passing additional criteria (Supplementary Table 3). Red boxes indicate an increase >1.2; pink boxes an increase between 1.1 and 1.2; white boxes are between 0.9 and 1.1; light blue boxes a decrease between 0.9 and 0.8; dark blue boxes a decrease to <0.8. Hits underlined were taken forward as three out of four4 hits, (§) not chosen because of restricted tissue expression, (*) are three out of four when alternate method of normalisation is used. (B) Table summarising results from further validation of 20 genes. mRNA levels scored as (+) or (−) indicate efficient or poor knockdown, respectively (see Supplementary Figures S1 and S2). Increasers (left) or decreasers (right) assayed for LC3 lipidation were scored (+) for yes, (−) for no, (+/−) subtle change and (nd) not done (Supplementary Figure S3). (C–E) STIPO results after incubation under different autophagy-inducing conditions following (C) SCOC knockdown (D) SUPT5H knockdown and (E) WAC knockdown. siRNA-treated cells were incubated for the indicated time in FL: full medium plus leupeptin; ES; EL: ES plus leupeptin; Torin: 250 nM Torin1 in FL; Resv: 128 μM Resveratrol in FL; LiCl: 10 mM LiCl in FM.
Nine new genes regulate autophagy
We characterised further the genes that had three out of four validated siRNA duplexes in at least one of the three spot parameters recorded in the deconvolution screen; these 20 are underlined in Figure 2A. For these genes we tested the ability of the two best siRNA duplexes from the deconvolution screen to decrease mRNA levels of the target gene in GFP–LC3-HEK cells (Supplementary Figures S1 and S2) in parallel with GFP–LC3 spot counting from confocal images. We validated the effects of the siRNA duplexes on autophagy in a different cell line, HeLa cells, by assaying the extent of endogenous LC3 lipidation after incubation in full growth medium, starvation medium and starvation medium plus leupeptin (Supplementary Figure S3). These data are summarised in Figure 2B. Note for LC3-lipidation assays, we continued to use leupeptin instead of Bafilomycin A1 (BafA1) to recapitulate the screen conditions.
Loss of 11 of the 20 validated-by-deconvolution genes resulted in an increase in spots in GFP–LC3-HEK cells. This increase can result from a loss of a negative regulator, as a consequence of a stress response, or from a defect in maturation of autophagosomes. Of these increasers, seven did not pass the extended stringent validation because the siRNA did not decrease the target mRNA message levels (Supplementary Figure S1) or did not increase LC3 lipidation in HeLa cells (results are summarised in Figure 2B). Four genes showed an increase in GFP–LC3 spots, an increase of endogenous LC3 lipidation and a decrease in the target mRNA: Kinesin family member 25 (KIF25) (Supplementary Figure S3A), Ras-interacting protein 1 (RASIP1) (Supplementary Figure S3B), tousled-like kinase (TLK2) (Supplementary Figure S3C) and WD repeat domain 6 (WDR6) (Supplementary Figure S3D). siRNA duplexes against KIF25, RASIP1 and WDR6 also led to increased LC3 lipidation in full medium and thus are potential regulators of basal autophagy. We conclude that the four genes we validated are negative regulators but it remains to be determined how loss of these genes increases autophagy.
Loss of nine genes resulted in decreased spots in GFP–LC3-HEK cells. Four did not pass further validation because the mRNA message did not decrease (Supplementary Figure S2) or their knockdown did not inhibit LC3 lipidation (results are summarised in Figure 2B). Two genes showed a decrease in mRNA levels with individual duplexes, a corresponding decrease in GFP–LC3 spots and a subtle decrease in lipidation of LC3 in starvation conditions in HeLa cells: platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30 kDa) (PAFAH1B2) (Supplementary Figure S3E) and La ribonucleoprotein domain family, member 1 (LARP1) (Supplementary Figure S3F). Three gene knockdowns were effective and had a robust effect on LC3 lipidation: SCOC (Supplementary Figure S3G), suppressor of Ty5 homologue (Saccharomyces cerevisiae) (SUPT5H) (Supplementary Figure S3H) and WAC (Supplementary Figure S3I).
We concluded that these three proteins are required for starvation-induced autophagy and then tested if they were also required for GFP–LC3 spot formation stimulated by other known autophagy inducers. Indeed, GFP–LC3 STIPO was strongly inhibited after knockdown of SCOC (Figure 2C), SUPT5H (Figure 2D) and WAC (Figure 2E) after induction of autophagy with Torin1 (Thoreen et al, 2009), Resveratrol (Scarlatti et al, 2008; Trincheri et al, 2008) and LiCl (Sarkar et al, 2005). We chose to study SCOC and WAC since existing literature suggested they would provide new insight into the process of autophagy. Our choice of SCOC and WAC was reinforced by the subsequent detection of both proteins in the autophagy interaction network (AIN) (Behrends et al, 2010).
SCOC interacts with FEZ1 to regulate autophagy
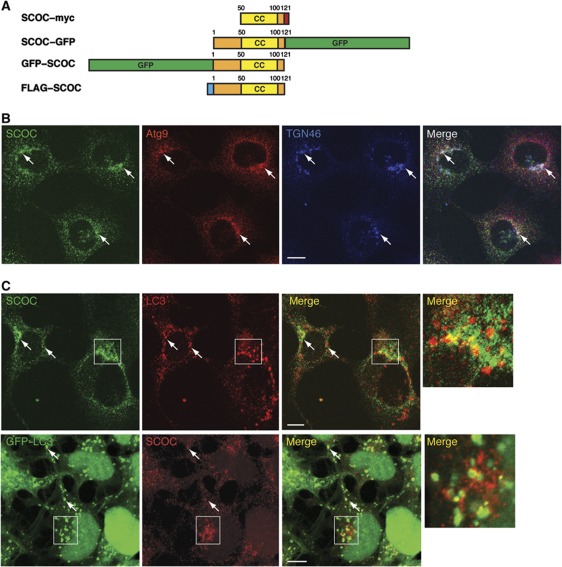
SCOC, a small protein with at least five isoforms, is widely expressed in human tissues and reported to localise to the Golgi (Van Valkenburgh et al, 2001). The five isoforms contain the conserved coil–coil domain but different N-terminal domains arising from alternative splicing. We obtained a truncated form of SCOC Q9UIL1-2 (isoform 3) tagged with myc (Van Valkenburgh et al, 2001) and subcloned SCOC isoform Q9UIL1-3 (isoform 5) with either FLAG or EGFP at the N- or C-terminal (Figure 3A). Human SCOC and its yeast homologue Slo1 have been shown to interact with ARL1 (Van Valkenburgh et al, 2001) and Arl3p (Panic et al, 2003), respectively, suggesting a function in Golgi transport. UNC-69, the orthologue of SCOC in Caenorhabditis elegans, controls axon extension through its interaction with UNC-76, a kinesin heavy chain adaptor and the orthologue of mammalian FEZ1 (Su et al, 2006) and in a yeast-two-hybrid screen FEZ1 was shown to bind SCOC (Assmann et al, 2006). In HEK293 cells, we could confirm that SCOC colocalises with TGN46, a trans-Golgi network protein (Ponnambalam et al, 1996), and mAtg9, a transmembrane autophagy protein found in the Golgi and endosomes (Young et al, 2006) (Figure 3B). In starved HEK293 cells, we observed a partial colocalisation of SCOC with endogenous LC3 and in GFP–LC3-HEK cells with GFP–LC3 (Figure 3C).
Figure 3.
Localisation of SCOC with the Golgi, Atg9 and LC3. (A) Schematic of SCOC constructs. Numbers represent amino acids; CC is coiled-coil domain. SCOC–myc was constructed with clone AF330205 (Van Valkenburgh et al, 2001). SCOC–GFP, GFP–SCOC and FLAG–SCOC were constructed with DNA corresponding to Uniprot isoform Q9UIL1-3, Entrez gene isoform 5. (B) Endogenous SCOC shows colocalisation with Atg9 (red) and TGN46 (blue). HEK293 cells were fixed and permeabilised in saponin. Colocalisation indicated with arrows. (C) Endogenous SCOC (green top, red bottom) partially colocalises with endogenous LC3 (red, top) and GFP–LC3 (green, bottom) in HEK293 and GFP–LC3-HEK cells, respectively, after 2 h amino-acid starvation. Boxed region is at higher magnification in merge panel on right. Scale bars=10 μm.
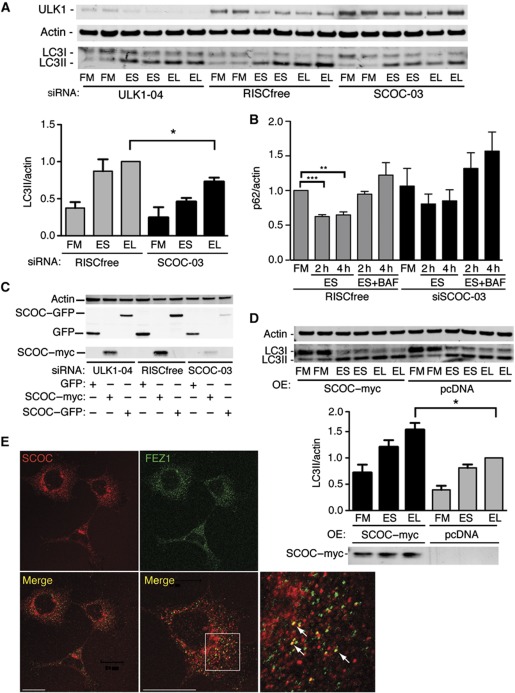
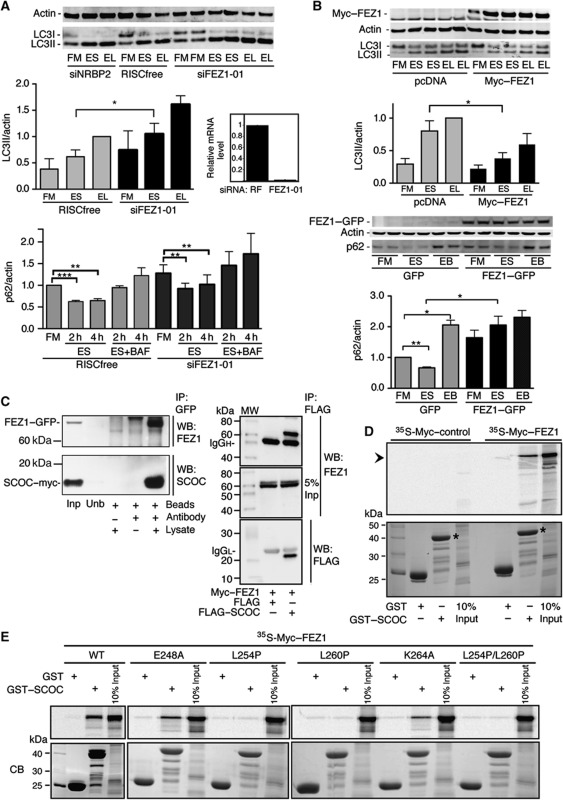
A single siRNA duplex targeting all isoforms was used to knockdown SCOC in HEK293 cells, resulting in reduced endogenous LC3 lipidation and flux using leupeptin (Figure 4A) and no significant degradation of p62 (Figure 4B; Supplementary Figure S5). p62/SQSTM1 is an LC3- and ubiquitin-binding protein that is selectively degraded by autophagy (Pankiv et al, 2007) and p62 levels are used as a read-out for autophagy. BafA1 was used for a more potent inhibition of lysosomal degradation. As the available SCOC antibody detects the endogenous protein by indirect immunofluorescence but not by western blotting (Van Valkenburgh et al, 2001), SCOC depletion was shown by detection of overexpressed GFP- and myc-tagged SCOC constructs whose levels were reduced after SCOC siRNA treatment (Figure 4C). In line with the requirement of SCOC for autophagy, overexpression of SCOC–myc in HEK293 cells increased LC3 lipidation under starvation conditions (Figure 4D).
Figure 4.
SCOC is required for autophagy. (A) SCOC is required for autophagy in HEK293 cells. Anti-ULK1, -Actin and -LC3 blot after siRNA treatment in HEK293 cells in FM, ES or EL. Representative blot; quantification of LC3II/actin of averaged duplicates; error bars represent s.e.m. (n=3). Significance was determined using a two-tailed paired t-test: RF EL versus siSCOC-03 EL, *P=0.0419. (B) SCOC is required for p62 degradation. p62 levels were determined by western blot (Supplementary Figure S5) after incubation in FM, ES for 2 or 4 h, or ES with BafilomycinA1 (ES+BAF) for 2 and 4 h. Quantification of averaged duplicates; error bars represent s.e.m. (n=3); RISCfree FM versus RISCfree ES 2 and 4 h, ***P>0.0001 and **0.0014, respectively. (C) SCOC–GFP and SCOC–myc levels are reduced by SCOC siRNA duplex-03. Anti-GFP, -Myc and -Actin blots of HEK293 cells after treatment with ULK1, RISCfree or SCOC siRNA and overexpression of GFP, SCOC–myc or SCOC–GFP. (D) Overexpression of SCOC–myc increases autophagy. Anti-Actin, -LC3 and -Myc blots of HEK293 cells after transfection of pcDNA or SCOC–myc. Representative blot; quantification of LC3II/actin of averaged duplicates; error bars represent s.e.m. (n=5). SCOC–myc EL versus pcDNA EL, *P=0.0215. (E) Endogenous SCOC colocalises with endogenous FEZ1 in HEK293 cells. Arrows show partial colocalisation. Boxed region is at higher magnification in merge panel on bottom right. Scale bars=20 μm.
To explore the function of SCOC, we investigated the role of the mammalian homologue of the reported SCOC-interactor FEZ1 in autophagy. Using indirect immunofluorescence, we saw colocalisation of a small percentage of endogenous SCOC and endogenous FEZ1 (Figure 4E). In our primary screen, knockdown of FEZ1 modestly increased the three parameters used to measure autophagosomes and we observed that knockdown of FEZ1 increased LC3 lipidation in HEK293 cells (Figure 5A; Supplementary Figure S6A). Conversely, overexpression of FEZ1 decreased LC3 lipidation (Figure 5B; Supplementary Figure S6B). The effect of loss of FEZ1 on p62 was more complex: the levels of p62 were slightly increased after knockdown of FEZ1 (Figure 5A; Supplementary Figure S5) but p62 degradation was similar, although not as efficient, as the RISCfree control. However, overexpression of FEZ1 caused an accumulation of p62 in FM as well as an inhibition of p62 degradation in starvation (Figure 5B) in agreement with the inhibition of autophagy using LC3-lipidation assays.
Figure 5.
SCOC interacts with FEZ1. (A) Knockdown of FEZ1 enhances LC3 lipidation but does not affect p62 degradation. Anti-Actin and -LC3 blot after siRNA treatment in HEK293 cells. Quantification of LC3II/actin of averaged duplicates; error bars represent s.e.m. (n=3). Significance was determined using a two-tailed paired t-test: RISCfree ES versus siFEZ1-01 ES, P=0.034. Quantification of p62/actin (Supplementary Figure S5) performed as in Figure 4B; error bars represent s.e.m. (n=3); RISCfree FM versus RISCfree 2 and 4 h P>0.0001 and P=0.0014, respectively. qRT–PCR showing FEZ1 mRNA levels after indicated knockdown in GFP–LC3-HEK cells (error bars represent s.d.). (B) Overexpression of FEZ1 inhibits LC3 lipidation and p62 degradation. Anti-Myc, -Actin and -LC3 blot after transfection with pcDNA or Myc–FEZ1 in HEK293 cells. Quantification of LC3II/actin; error bars represent s.e.m. (n=3); pcDNA ES versus Myc–FEZ1 ES, P=0.0318. Anti-GFP, -Actin and -p62 blot after transfection with GFP or FEZ1–GFP and treatment with ES or ES plus Bafilomycin (EB) for 4 h. Quantification of averaged duplicates; error bars represent s.e.m. (n=3); GFP FM versus GFP ES, P=0.0077; GFP FM versus GFP EB, P=0.0212; GFP ES versus FEZ1–GFP ES, P=0.048. (C) SCOC and FEZ1 interact. Left panel shows anti-SCOC and -FEZ1 blots after co-expression in HEK293 cells and co-immunoprecipitation of SCOC–myc and FEZ1–GFP with anti-GFP antibody. Input lysate (Inp), unbound supernatant (Unb). Right panel shows anti-FEZ1 and -FLAG blots after co-expression in HEK293 cells and co-immunoprecipitation of FLAG–SCOC and Myc–FEZ1 with anti-FLAG antibody. (D) FEZ1 interacts with SCOC in vitro. GST-pulldown assays using in vitro translated 35S-labelled Myc–FEZ1 (right) or 35S-labelled Myc-control (left) and GST or GST–SCOC (asterisk in bottom panel). Bound proteins were detected by autoradiography following SDS–PAGE (top), protein loading by coomassie blue (bottom). (E) The FEZ1–SCOC interaction requires the evolutionary conserved residues L254 and L260 in the coiled-coil domain of FEZ1 as demonstrated in GST-pulldown assays with in vitro translated 35S-labelled WT or mutant Myc-FEZ1 and GST or GST-SCOC. Autoradiographs of 10% input and bound proteins (top panels) and Coomassie blue-stained gels of bead-bound GST and GST-SCOC proteins used (bottom panels).
To test if SCOC and FEZ1 interact, we co-expressed SCOC–myc and FEZ1–GFP in HEK293 cells and as shown by immunoprecipitation of FEZ1–GFP were able to co-immunoprecipitate the two proteins (Figure 5C). FLAG–SCOC was similarly able to immunoprecipitate Myc–FEZ1 (Figure 5C). Note that the C-terminal coiled-coil domain present in the two SCOC isoforms (Figure 3A) is sufficient for FEZ1–GFP co-immunoprecipitation. SCOC and FEZ1 also interacted in a GST-pulldown assay using GST–SCOC (isoform 5) and in vitro translated Myc–FEZ1 (Figure 5D). We mapped the SCOC-interacting region of FEZ1 and found that binding requires the conserved residues L254 and L260 in the FEZ1 coiled-coil domain (Figure 5E) as reported for UNC-69 and UNC-76 (Su et al, 2006).
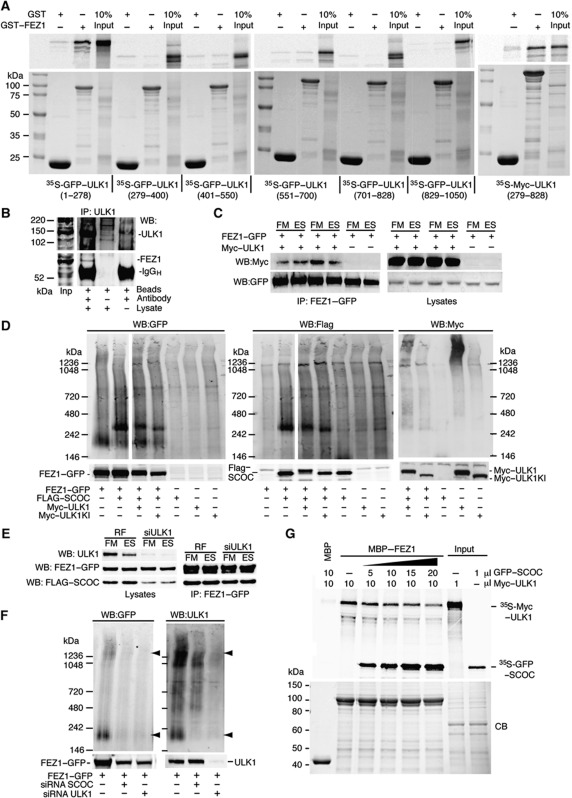
Interestingly, Drosophila melanogaster UNC-76 binds to and is regulated by UNC-51, the orthologue of mammalian ULK1 (Toda et al, 2008). We confirmed the interaction of the human proteins using GST–FEZ1 and either full length or pieces of GFP–ULK1. Unlike C. elegans in which the C-terminal domain of UNC-51 bound UNC-76, we found the N-terminal kinase domain and the middle spacer domain of ULK1 interact with FEZ1 (Figure 6A). Endogenous ULK1 could also immunoprecipitate endogenous FEZ1 (Figure 6B). We confirmed the interaction using co-immunoprecipitation and found that the interaction between FEZ1–GFP and Myc–ULK1 was not amino acid sensitive (Figure 6C). The mutations L254P/L260P in FEZ1 that abolish SCOC binding do not affect the interaction of FEZ1 with ULK1 (Supplementary Figure S7A). To investigate further the SCOC–FEZ1–ULK1 interactions, we used non-denaturing Blue-native–PAGE (BN–PAGE), which allows detection of protein complexes. FLAG-tagged SCOC was not detectable on western blots of lysates analysed by BN–PAGE when transfected alone although it was detected in SDS–PAGE denaturing gels (Figure 6D). Upon co-transfection of FEZ1–GFP with FLAG–SCOC, a complex was detected at a molecular weight of ∼300 kDa that contained both FEZ1–GFP and FLAG–SCOC, most likely associated in a 2:2 stoichiometric complex. FEZ1–GFP migrated at a molecular weight of about 200 kDa, which may correspond to a dimer (Assmann et al, 2006) and also as a higher molecular weight species. To test if the FEZ1–SCOC complex is regulated by ULK1, we co-expressed Myc–ULK1 wild type and kinase-inactive Myc–ULK1. While ULK1 wild type did not affect the complex, expression of kinase-inactive Myc–ULK1 increased its mobility (Figure 6D). As the migration of both proteins on denaturing gels appears to be influenced by the kinase-inactive ULK1 (Figure 6D, lower gel), we asked whether SCOC bound directly to ULK1. Recombinant GST–SCOC did not bind to ULK1 (Supplementary Figure S7B) although we cannot exclude low-affinity, transient interactions in vivo. The interaction between FLAG–SCOC and FEZ1–GFP was not sensitive to siRNA depletion of ULK1 (Figure 6E). However, the complexes formed by FEZ1–GFP and endogenous ULK1 that are detected at a molecular weight of ∼200 and 1200 kDa in BN–PAGE were sensitive to siRNA depletion of SCOC and as expected, depletion of ULK1 (Figure 6F). Further support for the regulation of the FEZ1–ULK1 complex by SCOC could be shown in competition assays with MBP–FEZ1 and Myc–ULK1 (Figure 6G): addition of increasing amounts of GFP–SCOC decreased the amount of 35S-labelled Myc–ULK1 bound to MBP–FEZ1.
Figure 6.
SCOC regulates FEZ1's interaction with the ULK1 complex. (A) FEZ1 interacts with both the kinase domain (1–278) and the proline–serine rich spacer of ULK1 (279–828). GFP- or Myc-tagged ULK1 fragments were in vitro translated with 35S-methionine and subjected to GST-pulldown assays using GST and GST–FEZ1 purified from E. coli (bottom). Bound proteins were detected by autoradiography following SDS–PAGE (top). (B) ULK1 interacts with FEZ1. ULK1 was immunoprecipitated from HEK293 lysates with anti-ULK1 and bound ULK1 and FEZ1 were detected with anti-ULK1 and anti-FEZ1 antibodies. Input lysate (Inp). (C) The FEZ1–GFP and Myc–ULK1 interaction is unaffected by amino-acid starvation. Anti-Myc and -GFP blots after HEK293 cells were co-transfected with FEZ1–GFP and Myc–ULK1, incubated in FM or ES for 2 h, harvested and the complex was immunoprecipitated using anti-GFP antibody. (D) BN–PAGE reveals SCOC and FEZ1 are in a complex. FEZ1–GFP, FLAG–SCOC and Myc–ULK1 wild type or kinase-inactive ULK1K46I (ULK1KI) were co-expressed in HEK293 cells. Anti-GFP, -FLAG and -Myc western blots are shown for the BN–PAGE gel (top) or the SDS–PAGE gel (bottom). In GFP and FLAG western blots, two different exposures of the same BN–PAGE blot are shown spliced together. (E) siRNA depletion of ULK1 does not affect the interaction of FEZ1 with SCOC. RF or ULK1 siRNA-treated cells were co-transfected with FEZ1–GFP and FLAG–SCOC, then incubated and immunoprecipitated as in (C). Western blot of lysates for ULK1, GFP and FLAG, and immunoprecipitates with GFP and FLAG antibodies. (F) siRNA depletion of SCOC reduces FEZ1–ULK1 complex. HEK293 cells treated with either SCOC or ULK1 siRNAs were transfected with FEZ1–GFP. FEZ1–GFP was detected with anti-GFP in both BN–PAGE and SDS–PAGE. Endogenous ULK1 was detected with anti-ULK1 antibody. Arrowheads indicate a FEZ1–GFP and endogenous ULK1 complex at 200 kDa and a larger MW complex >1236 kDa also detected in (D) in the triple transfection. The 200 KDa complex is sensitive to loss of SCOC and ULK1. (G) SCOC can reduce the interaction of ULK1 with MBP–FEZ1. Immobilised MBP–FEZ1 was incubated with in vitro 35S-methionine-labelled Myc–ULK1. Addition of increasing amounts of in vitro translated 35S-methionine SCOC reduced the amount of Myc–ULK1 bound. Top and bottom panels are as described in (A).
Amino-acid starvation sensitive interaction of SCOC and UVRAG is modulated by FEZ1
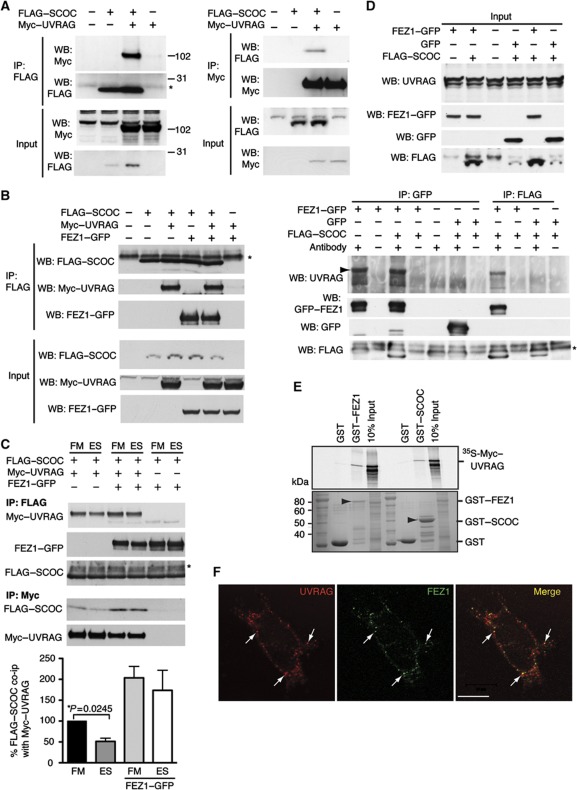
The interaction of SCOC with two proteins in the Beclin 1 AIN subnetwork, NRBF2 (nuclear receptor binding factor 2) and UVRAG (Behrends et al, 2010), suggested that SCOC may also function through the Beclin 1–autophagy complexes. To validate these interactions, we performed co-immunoprecipitation with FLAG–SCOC and Myc–NRBF2 or Myc–UVRAG. We efficiently co-precipitated Myc–NRBF2 with FLAG–SCOC but no FLAG–SCOC was co-immunoprecipitated with Myc–NRBF2 (Supplementary Figure S4B). We also validated the FLAG–SCOC and Myc–UVRAG interaction with reciprocal co-immunoprecipitations (Figure 7A). We next investigated what effect Myc–UVRAG had on the interaction of SCOC and FEZ1: FLAG–SCOC was able to interact with both Myc–UVRAG and FEZ1–GFP and all three proteins could be detected in a complex. The presence of FEZ1–GFP or Myc–UVRAG had no effect on either's ability to co-immunoprecipitate with FLAG–SCOC or FEZ1–GFP, respectively (Figure 7B). Given that UVRAG has been implicated in autophagy (Liang et al, 2006; Itakura et al, 2008), we asked if any of these interactions were affected by amino-acid starvation. Amino-acid depletion for 2 h significantly reduced the interaction between Myc–UVRAG and FLAG–SCOC (Figure 7C). While the interaction between FLAG–SCOC and FEZ1–GFP was not affected by starvation, the presence of FEZ1 inhibited the starvation-dependent dissociation and stabilised the complex. To validate the interaction of SCOC and UVRAG, we asked if we could detect endogenous UVRAG with FLAG–SCOC (Figure 7D). We were unable to detect UVRAG after immunoprecipitation of FLAG–SCOC; however, when FEZ1–GFP was present, we could detect a co-immunoprecipitation of FLAG–SCOC and UVRAG. In fact, GFP–FEZ1 was able to interact alone with endogenous UVRAG. In vitro pulldown experiments however demonstrated that both GST–SCOC and GST–FEZ1 were able to bind in vitro translated 35S-labelled Myc–UVRAG (Figure 7E). In addition, endogenous FEZ1 and endogenous UVRAG partially colocalise by indirect immunofluorescence (Figure 7F).
Figure 7.
SCOC interacts with UVRAG and the interaction is reduced in starvation. (A) SCOC interacts with UVRAG. HEK293 cells were transfected with FLAG–SCOC and Myc–UVRAG, lysed and incubated with anti-FLAG (left) or anti-Myc (right) antibodies. Co-immunoprecipitation of Myc–UVRAG (left) and FLAG–SCOC (right) was confirmed using anti-Myc and anti-FLAG antibodies for western blotting. Asterisk in (A–C) and (D) indicates non-specific bands. (B) Interaction of SCOC with UVRAG is unaffected by FEZ1–GFP. As in (A), co-immunoprecipitation was preformed in HEK293 cells transfected with FLAG–SCOC, Myc–UVRAG and FEZ1–GFP. Myc–UVRAG was detected as in (A), FEZ1–GFP was detected with anti-GFP antibody. (C) The SCOC and UVRAG interaction is reduced by starvation, and this reduction is inhibited by FEZ1–GFP. HEK293 cells were transfected as in (B) and 24 h later either treated with FM or ES for 2 h. FLAG–SCOC or Myc–UVRAG was immunoprecipitated with anti-FLAG or anti-Myc antibodies and the bound proteins were detected with anti-Myc, -GFP or -FLAG antibodies by western blotting. Quantification of the amount of FLAG–SCOC co-immunoprecipitated with Myc–UVRAG in FM or ES, without overexpressed FEZ1–GFP (left) or with overexpressed FEZ1–GFP (right). Significance was determined using a two-tailed paired t-test (n=3). (D) FEZ1 interacts directly with endogenous UVRAG and is required for SCOC interaction. HEK293 cells were transfected with FEZ1–GFP, GFP or FLAG–SCOC alone or in combination. Top panel input, bottom panel immunoprecipitation with anti-GFP or anti-FLAG. Western blot for endogenous UVRAG, GFP–FEZ1 GFP or FLAG–SCOC as indicated. Arrowhead: endogenous UVRAG. (E) UVRAG can interact with both FEZ1 and SCOC in vitro. GST, GST–FEZ1 or GST–SCOC were immobilised and incubated with 35S-labelled Myc–UVRAG as indicated. Arrowheads indicate GST–FEZ1 (left) and GST–SCOC (right). (F) Endogenous UVRAG partially colocalises with endogenous FEZ1 in HEK293 cells. Arrows show colocalisation. Scale bar=10 μm.
WAC is required for autophagy
Our second robust positive regulator of autophagy is WAC, which contains a conserved coiled-coil and WW domain and was first described to be a nuclear protein (Xu and Arnaout, 2002). It has also been shown by yeast-two-hybrid analysis to bind UBQLN4 (Lim et al, 2006), a UBQ–UBA-containing protein (Li et al, 2008). UBQLN4 is a member of the Ubiquilin family of which UBQLN1/Plic-1 is required for autophagy (N'Diaye et al, 2009) and its levels are regulated by chaperone-mediated autophagy (Rothenberg et al, 2010). In addition, a yeast-two-hybrid screen found an interaction between huntingtin (Htt) protein and WAC (Kaltenbach et al, 2007) and two microarray analyses of Huntington's disease patients showed WAC is upregulated in the symptomatic stages of the disease (Borovecki et al, 2005; Hodges et al, 2006). Recently, it has been described to partner with RNF20/40, which mediates histone H2B ubiquitination (Zhang and Yu, 2011). Furthermore, WAC associates with another protein VCIP135 to modulate its deubiquitinating activity (Totsukawa et al, 2011). These diverse roles of WAC have a common theme of regulating ubiquitin-dependent pathways.
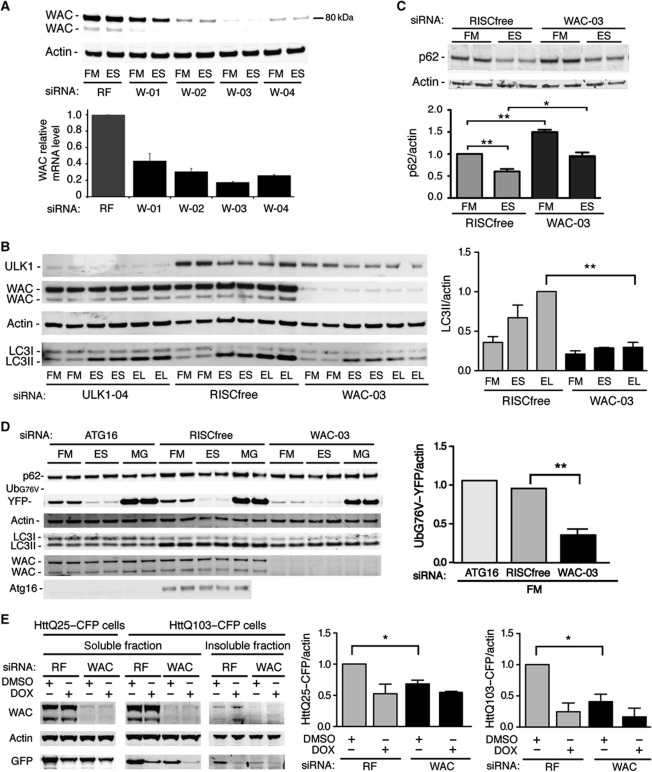
We raised an antibody against an N-terminal peptide of WAC that detects two endogenous isoforms of 81 and 72 kDa (Figure 8A). Western blotting of endogenous WAC in HEK293 cells revealed that all four siRNA duplexes reduce the protein and this reduction correlates with a reduction of WAC mRNA levels and GFP–LC3 spots in the deconvolution screen (Supplementary Table S3). WAC siRNA duplex-03 significantly reduces LC3 lipidation in HEK293 cells (Figure 8B). As expected after starvation of GFP–LC3-HEK cells, we see a marked reduction in p62 levels, which is inhibited by WAC siRNA. Basal levels of p62 are also higher after WAC knockdown (Figure 8C).
Figure 8.
WAC is required for autophagy and negatively regulates the UPS and degradation of mutant Htt. (A) Depletion of endogenous WAC. Anti-WAC and -Actin blot and WAC mRNA levels by qRT–PCR after treatment with RF or individual WAC siRNAs (W-01 to W-04) in HEK293 cells. qRT–PCR error bars represent standard deviation of triplicates (n=2). (B) WAC is required for autophagy. Anti-ULK1, -WAC, -Actin and -LC3 blots after siRNA treatment in HEK293 cells and incubation in FM, ES or EL. Representative blot; quantification of LC3II/actin of averaged duplicates; error bars represent s.e.m. (n=3). Significance was determined using a two-tailed paired t-test: RF EL versus siWAC-03 EL, **P=0.0079. (C) p62 degradation requires WAC. Anti-p62 and -Actin blot after siRNA treatment of GFP–LC3-HEK cells in FM or ES for 2 h in duplicate. Bars represent p62/actin levels normalised to RF FM; quantification of p62/actin; error bars represent s.e.m. (n=3): RF FM versus RF ES, **P=0.0057; RF FM versus siWAC-03 FM, **P=0.0025; RF ES versus siWAC-03 ES, *P=0.0386. (D) WAC depletion accelerates degradation of a UPS reporter. Anti-p62, -GFP, -Actin, -LC3, -WAC and -Atg16 blots after siRNA treatment in UbG76V–YFP MelJuSo cells and incubation conditions FM, ES or FM with 10 μM MG132 (MG) for 2 h. Quantification of normalised average GFP intensity in FM of RISCfree compared with WAC-03 siRNA treatment (n=3) and compared with ATG16 siRNA treatment (n=2). Error bars represent s.e.m.: RF FM versus siWAC-03 FM, **P=0.0082. (E) Soluble HttQ25–CFP degradation is increased after WAC depletion. Anti-WAC, -Actin and -GFP blots after indicated siRNA treatment (siWAC duplexes -02, -03, -04) of HttPolyQ-mutant HeLa cell lines and incubation conditions DMSO or DOX (Doxycyline) for 3 days. The insoluble fraction was obtained by pelleting the HttQ103 lysates (see Materials and methods). Quantification of normalised average HttQ25–CFP intensity in DMSO of RISCfree compared with WAC siRNA treatment; error bar represents s.e.m. (n=3): HttQ25–CFP/actin RF DMSO versus siWAC DMSO, *P=0.0350; soluble HttQ103–CFP/actin RF DMSO versus siWAC DMSO, *P=0.0380.
These results and our data in GFP–LC3-HEK cells (Figure 2E) and HeLa cells (Supplementary Figure S3I) demonstrated that WAC is required for GFP–LC3 spot formation, LC3 lipidation and p62 degradation during starvation-induced autophagy. However, based on reported interactions with UBQLN4 and Htt, we hypothesised that WAC may be also involved in autophagy of ubiquitinated proteins. Thus, we examined two substrates for the proteasome, p62 and UbG76V–GFP, of which p62 is also an autophagic cargo (Menéndez-Benito et al, 2005; Pankiv et al, 2007). Accumulation of p62 leads to inhibition of the ubiquitin-proteasome system (UPS) (Korolchuk et al, 2009), which can be measured by an accumulation of a proteasomal substrate UbG76V–GFP, and we asked if WAC depletion, which causes an increase in p62 in both FM and ES, altered turnover of UbG76V–YFP. To our surprise, WAC depletion resulted in a substantial decrease in the level of UbG76V–YFP in full medium, which was blocked by the proteasome inhibitor MG132; there was no significant change in ES (Figure 8D). Thus, WAC may negatively regulate the UPS. We next examined the clearance of mutant Htt proteins using inducible stable cell lines expressing non-aggregating HttQ25–CFP and aggregating HttQ103–CFP (Yamamoto et al, 2006). Addition of doxycycline to suppress expression leads to clearance of the soluble HttQ25 and the selective degradation of the aggregated insoluble HttQ103 in autolysosomes (Filimonenko et al, 2010). After WAC depletion, the levels of both HttQ25–CFP and the soluble fraction of HttQ103–CFP were decreased (Figure 8E) and this decrease was independent of doxycycline-mediated suppression of expression. While we have not investigated if WAC is required for autophagy of ubiquitinated proteins, our results suggest that WAC is required for autophagy, but also functions to negatively regulate the activity of the proteasome towards soluble substrates such as UbG76V–YFP and HttQ25–CFP.
Discussion
To identify novel autophagy regulators by a genome-wide siRNA screen, we relied on a cell model system that provides a robust read-out for autophagy: the quantifiable starvation-dependent formation of GFP–LC3-labelled autophagosomes. Our hit-selection strategy to identify both negative and positive regulators of starvation-induced autophagy was based on several rounds of screening (primary, secondary and deconvolution) using both automated counting and visual clarification and the use of multiple assays (GFP–LC3 spots, LC3 lipidation, qRT–PCR) in two cell lines, coupled with the elimination of candidates that did not satisfy strict criteria. In this way we have identified four proteins that are potential negative regulators and five positive regulators of starvation-induced autophagy, two of which we have investigated further. Three (SUPT5H, KIF25, WDR6) of the nine validated hits that remain to be characterised are classified as ‘druggable’ targets and thus potentially tractable therapeutic targets. In addition, relaxation of some of the thresholds in the deconvolution screen or further study of the hits with two out of four deconvoluted siRNA duplexes could well reveal more candidate genes in our data set.
Our screen results and candidate selection are supported by the identification of four (SCOC, WAC, SUPT5H and LARP1) (Supplementary Figure S4) out of the nine further validated hits independently in the AIN (Behrends et al, 2010). The AIN is based on a proteomic screen revealing 751 interactions between 409 candidate autophagy or autophagy-interacting proteins and identified 10 functional protein subnetworks involved in autophagy. Although not classified as high-confidence interacting candidates, SCOC was identified as interacting with UVRAG and NRBF2, members of the Beclin 1 subnetwork, while WAC was identified using Beclin 1 as bait (Behrends et al, 2010; Supplementary Table S3). We have validated the interaction of WAC with Beclin 1 by co-immunoprecipitation (Supplementary Figure S4), as well as the interaction of SCOC with UVRAG and NRBF2. An additional four of the 51 deconvoluted hits (DNAJA3, C8orf33, C1orf25, CSTB) (Supplementary Figure S4) were also found in the AIN although their confidence scores were significantly less than SCOC and WAC.
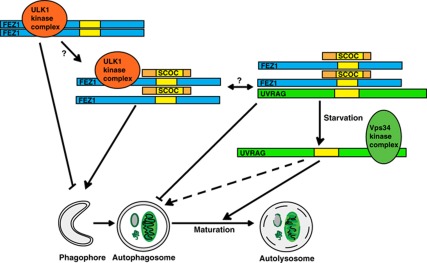
We propose SCOC functions to mediate crosstalk or regulate the interaction between the ULK1 and Beclin 1 complexes in the induction of autophagy (see Figure 9). This regulation depends on FEZ1, which binds directly to SCOC and ULK1. ULK1 expression appears to increase the size of the SCOC–FEZ1 complex on native gels and slightly decrease the mobility of both SCOC and FEZ1 on denaturing gels suggesting both may be targets of ULK1 or ULK1-regulated kinases (although SCOC does not directly interact with ULK1). Intriguingly, SCOC and FEZ1 both interact with UVRAG, a stable component of a Beclin 1 complex required for autophagy. Importantly, the interaction between SCOC and UVRAG is starvation sensitive and regulated by FEZ1. Our data suggest that during amino-acid starvation SCOC promotes autophagy. We speculate that this is through stabilisation of the ULK1–FEZ1–SCOC complex, concurrent with dissociation from UVRAG. FEZ1 appears to be a negative regulator of LC3 lipidation and when overexpressed, inhibits p62 degradation possibly through sequestration of ULK1. While the role of FEZ1 in the regulation of autophagy requires further investigation, we have provided evidence that the SCOC–FEZ1 complex is a likely target for ULK1 and that SCOC and FEZ1 associate with UVRAG to regulate autophagy.
Figure 9.
The role of SCOC and FEZ1 during amino-acid starvation-induced autophagy. We show that SCOC bound to FEZ1 is in a complex with ULK1 or UVRAG. Our results suggest that the inhibition of autophagy induction by FEZ1, possibly achieved through inhibition of ULK1 kinase activity or membrane targeting, is relieved by association of SCOC with the ULK1–FEZ1 complex. Also, amino-acid starvation causes the dissociation of UVRAG from the FEZ1–SCOC complex and this may allow UVRAG to associate with the Vps34 complex and promote autophagy, most likely at later stages compared with the Vps34 complex containing Atg14 (not shown here). We hypothesise that SCOC or the SCOC–FEZ1 complex may coordinate the regulation of both the ULK1 and the Vps34 kinase complexes.
The coiled-coil domain of SCOC is likely to be the functional region of the molecule, being responsible for both regulation of autophagy and binding to FEZ1. As SCOC was originally discovered to interact with ARL1 (Van Valkenburgh et al, 2001) and shown to be on Golgi membranes (where it colocalises with mAtg9), we speculate that SCOC may function as a membrane-proximal scaffold protein. The spatial regulation of the interactions between SCOC–FEZ1 and the ULK1 complex and the Beclin 1 complex remains to be explored; however, we hypothesise that SCOC functions at the site of autophagosome formation.
Furthermore, we identified the positive regulator WAC, a protein that may communicate between cargo, the proteasome and autophagosomes and potentially plays a role in Huntington's disease. Our screen assayed events downstream of amino-acid starvation but future studies on the role of WAC in autophagy induced by aggregated or misfolded proteins are clearly a priority. While our data show that WAC is a positive regulator of starvation-induced autophagy, it also suggests that WAC is a negative regulator of the UPS. Depletion of WAC accelerated clearance of a UPS reporter UbG76V–YFP and the soluble HttQ25–CFP protein. Importantly, WAC is reported to bind to UBQLN4, a protein that contains UBQ and UBA domains, and although this interaction remains to be verified it could serve as the mechanism to allow WAC to interact with UPS.
Our siGenome screen and the subsequent analysis of two validated candidates, SCOC and WAC, reveal their role in autophagy and suggest further additional connections between their function and autophagy with the Golgi complex, the cytoskeleton through FEZ1, and to Huntington's disease. Furthermore, our study provides a validated list of additional starvation-induced autophagy regulators and provides new avenues for further understanding of autophagy.
Materials and methods
siRNA screens
The siGENOME SMARTpool human siRNA genome library and deconvoluted siGENOME SMARTpools were from Dharmacon (Thermo Scientific). siRNA was used at a final concentration of 37.5 nM with 0.4 μl of Lipofectamine 2000 (Invitrogen) reagent and 4000 cells in a final volume of 100 μl/well in poly-D-lysine (Sigma)-coated black and clear bottom 96-well Optilux plates (Falcon). siRNA libraries were aliquoted on a Beckman FX liquid handling station and all other liquid additions performed with a Thermo/Matrix Wellmate. The plates were incubated at 37°C in 10% CO2 for 48 h, then fresh medium was added and incubation continued for 24 h (72 h total transfection). The plates were washed twice with PBS and incubated in amino acid-free ES (Earle's Buffered Saline Solution) supplemented with leupeptin (0.25 mg/ml; Roche) (EL) for 2 h at 37°C. Cells were fixed with 8% formaldehyde (Sigma) in PBS at 4°C (final concentration 4% formaldehyde) and stained with DAPI (1 μg/ml) for 15 min and stored at 4°C in PBS. Control siRNAs added to each plate included CONTROL RISCfree and non-targeting pool 1, ULK1, ATG13, ATG7 and LOC340371 all from Dharmacon.
Image capture and analysis
In the primary screen, a maximum of 10 images/well were captured in both DAPI and GFP channels on a Cellomics ArrayScan VTI (Thermo Scientific) using a × 20 0.45NA lens. Images were analysed simultaneously upon acquisition using the Cellomics SpotDetector V3 Bioapplication. The GFP channel (GFP–LC3 spots) was used to set the focus with fixed illumination time and spots were defined as having the following broad characteristics: SpotArea 1.75–18, SpotShapeBFR 0.33–1.4, SpotShapeBAR 1–3, SpotAvgInten 44–888, SpotTotalInten 250–13 000, in an area 22 microns surrounding and external to the nuclear region as defined by the DAPI stain (excluding any contribution from nuclear spots) thereby restricting the analysis to perinuclear and cytoplasmic GFP–LC3 spots. Several parameters were recorded including the number of nuclei (object count; OC) from the DAPI channel, the number of GFP–LC3 spots per cell (SCPO), the total intensity of GFP–LC3 spots per cell (STIPO) and the total area covered by GFP–LC3 spots per cell (STAPO). More detailed descriptions of the algorithm parameters are available on request. In the secondary screen and deconvolution screen, 14 and 16 fields were imaged per well, respectively. The induction screen (Figure 2C–E) was done in triplicate 96-well plates and ∼500 cells/well were analysed for GFP–LC3 spots.
Data analysis
We applied a filter to the raw data such that if <50 cells were counted per well, the spot data for that well did not contribute to the analysis. Raw data for the three individual parameters were normalised using the B-score method to correct for plate, row and column effects and then divided by the individual plate median absolute deviation, thus creating a Z-score (Malo et al, 2006). Replicates were summarised by taking the median Z-score for each siRNA pool and normalised scores for the three parameters were then combined into a ranked list using the RP between parameters. The top 500 spot increasers and top 500 spot decreasers were thereby determined. Deconvolution assay results are expressed as percent of control with respect to RISCfree control and designated a hit if they induced more than a 20% increase or decrease in spot parameters (SCPO, STIPO, STAPO).
Cell culture
HeLa, HEK293A and GFP–LC3-HEK cells and their culturing conditions have been described (Chan et al, 2009). UbG76V–YFP MelJuSo cells (Menéndez-Benito et al, 2005), HttQ25–CFP and HttQ103–CFP cells (Yamamoto et al, 2006) have been described. Lipofectamine2000 or Metafectene Pro (Biontex) were used for transfection experiments according to the manufacturers' protocols. To induce autophagy, cells were cultured in ES, EL or treated with 250 nM Torin1 (N Gray, MIT), 128 μM Resveratrol (Sigma) or 10 mM LiCl (Sigma). BafA1 (Calbiochem) was used at 100 nM.
siRNAs and knockdown determination for validation assays
Cells were knocked down as above in 96-, 12- or 24-well plates. siRNAs are siGenome Smartpools, individual siGenome duplexes or pools of three individual siGenome duplexes and were from Dharmacon. siATG16 is an ON-TARGET plus pool (L-021033-01; Dharmacon). siFEZ1 is duplex-01 and -04 (D-013010-01, D-013010-04; Dharmacon). RNA was purified, cDNA was made and qRT–PCR was performed (Chan et al, 2009). QuantiTect (Qiagen Biotech) primer sets were used for detection of all genes and the β-actin control.
Western blotting
Methods and reagents to detect the Myc-tag, FLAG-tag, Actin, LC3 and GFP by western blotting have been described (Chan et al, 2009). Actin, LC3 and p62 blots were developed using LICOR and quantified using Metamorph software or ImageJ. WAC rabbit polyclonal antibodies were raised and affinity purified using the peptide SKSHPSSGDHRHEKMRDAG(C). Atg16 rabbit polyclonal antibodies were raised and affinity purified using the peptide SSGLRAADFPRWKRHISEQ(C). Other antibodies included: ULK1 rabbit polyclonal (sc-33182; Santa Cruz), FEZ1 goat polyclonal (ab53562; Abcam), FEZ1 purified MaxPab (H00009638-B02P; Abnova), UVRAG (ab72073; Abcam), FLAG (M2) (200472-21; Stratagene) and p62 monoclonal (610832; BD Transduction).
BN–PAGE western blotting
Methods and reagents to perform BN–PAGE western blotting have been described (Chan et al, 2009). Anti-GFP and -FLAG were detected using LICOR; anti-Myc was detected with HRP-conjugated secondary antibodies and ECL.
Indirect immunofluorescence
HEK293 or GFP–LC3-HEK cells were fixed with 3% paraformaldehyde in PBS, permeabilised with 0.2% saponin, blocked and incubated in 10% goat serum and SCOC polyclonal antibody (Van Valkenburgh et al, 2001), mAtg9 monoclonal antibody (Webber and Tooze, 2010a), TGN46 (AHP500; AbD Serotec), LC3 (M152-3; MBL), FEZ1 or UVRAG, and Alexa secondary antibodies (Invitrogen) in 10% goat serum.
Immunoprecipitation
Co-immunoprecipitation was done as described (Chan et al, 2009) with the following exceptions: TNTE contained 1% Triton X-100, protein A-Sepharose beads were used with anti-GFP monoclonal 4E12 and anti-myc (ab9106; Abcam) antibodies, lysates were incubated for 2h or for Figure 5C incubated overnight, eluted with 2 × SDS sample buffer and heated to 95°C for 10min. Alternatively, RIPA buffer (50mM Tris–HCl, pH 7.5, 150mM NaCl, 1mM EDTA, 1% NP-40 (v/v), 0.25% Triton X-100 (v/v)) was used for lysis.
GST pulldown
GST and GST-tagged proteins were expressed in Escherichia coli, purified and immobilised on Glutathione-sepharose 4 FastFlow beads (Amersham). 35S-labelled proteins were made using the TNT T7 Quick Coupled Transcription/Translation system (Promega) in the presence of 35S-methionine (Amersham). The in vitro translated products were diluted 20 × in NET-N buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 (v/v)) containing Complete EDTA-free protease inhibitor cocktail (Roche) and incubated with GST or GST-fusion proteins. After removing unbound proteins by washing with NET-N buffer, bound proteins were eluted and separated by SDS–PAGE. The 35S-labelled proteins were detected using a Fujifilm BAS-5000 bio-imaging analyser.
Htt polyQ-mutant experiments
HttQ25–CFP and HttQ103–CFP cells were cultured as described (Filimonenko et al, 2010). Cells were transfected with RISCfree, Atg16 or WAC (Dharmacon duplex-02, -03, -04) siRNA pool, replated on day 3 and treated with DMSO or doxycycline (100 ng/ml). On day 6, cells were harvested in TNTE (Chan et al, 2009) plus protease inhibitors and spun at 13 000 r.p.m. for 10 min. Soluble supernatant was added to 5 × sample buffer and insoluble pellets were resuspended in 1 × sample buffer. Anti-GFP and -Actin blots were obtained using LICOR and quantified with Metamorph software.
Supplementary Material
Acknowledgments
We thank Richard Kahn (Emory University, USA) for SCOC affinity purified antibody and SCOC–myc, Caroline Whitehouse (King's College London, UK) for FEZ1–GFP, N Dantuma (Karolinska Institute, Sweden) for UbG76V–YFP MelJoSo cells, Ai Yamamoto (Columbia University, NY) for HttQ25–CFP and HttQ103–CFP cells, NS Gray (MIT, USA) for Torin1, Richard Mitter (BABS, LRI), Rachael Instrell and Ming Jiang (HTS, LRI) for assistance with the screens, Gry Evjen for technical assistance, and the Secretory Pathways Laboratory (LRI) for support. This work was supported by Cancer Research UK to SAT and by grants from the Norwegian Research Council and the Norwegian Cancer Society to TJ.
Author contributions: NCM developed the screen, performed the experiments, analysed the data and wrote the manuscript. HBJ performed the experiments and analysed the data. EAA performed the experiments. RES performed the statistical analysis of the screens. MH developed the screen, performed the experiments and analysed the data. TJ directed the research, performed the experiments and analysed the data. SAT directed the research, performed the experiments, analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Assmann EM, Alborghetti MR, Camargo ME, Kobarg J (2006) FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J Biol Chem 281: 9869–9881 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D (2005) Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA 102: 11023–11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol Cell Biol 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EYW, Kir S, Tooze SA (2007) siRNA screening of the kinome identifies ULK1 as a multi-domain modulator of autophagy. J Biol Chem 282: 25464–25474 [DOI] [PubMed] [Google Scholar]
- Chan EYW, Tooze SA (2009) Evolution and expansion of Atg1 function. Autophagy 5: 758–765 [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 38: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL et al. (2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet 15: 965–977 [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 192: 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Ro S-H, Cao J, Otto N, Kim D-H (2010) mTOR regulation of autophagy. FEBS Lett 584: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, Cha GH, Ukani L, Chepanoske CL, Zhen Y, Sahasrabudhe S, Olson J, Kurschner C, Ellerby LM, Peltier JM, Botas J et al. (2007) Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet 3: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC (2009) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N (2010) Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Sem Cell Dev Bio 21: 683–690 [DOI] [PubMed] [Google Scholar]
- Li X, Su V, Kurata WE, Jin C, Lau AF (2008) A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem 283: 5748–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh B-H, Jung JU (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8: 688–698 [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY (2006) A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125: 801–814 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, Xavier RJ, Yuan J (2010) A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Devl Cell 18: 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R (2006) Statistical practice in high-throughput screening data analysis. Nat Biotechnol 24: 167–175 [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11: 385–396 [DOI] [PubMed] [Google Scholar]
- Menéndez-Benito V, Verhoef LGGC, Masucci MG, Dantuma NP (2005) Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet 14: 2787–2799 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of atg proteins in autophagosome formation. Ann Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- N'Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ (2009) PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B, Whyte JR, Munro S (2003) The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol 13: 405–410 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Ponnambalam S, Girotti M, Yaspo ML, Owen CE, Perry AC, Suganuma T, Nilsson T, Fried M, Banting G, Warren G (1996) Primate homologues of rat TGN38: primary structure, expression and functional implications. J Cell Sci 109 (Part 3): 675–685 [DOI] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ (2010) Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet 19: 3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk VI, Kaushik S, Klionsky DJ (2009) In search of an ‘autophagometer’. Autophagy 5: 585–589 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC (2005) Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 170: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R (2008) Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ 15: 1318–1329 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA (2009) Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CW, Tharin S, Jin Y, Wightman B, Spector M, Meili D, Tsung N, Rhiner C, Bourikas D, Stoeckli E, Garriga G, Horvitz HR, Hengartner MO (2006) The short coiled-coil domain-containing protein UNC-69 cooperates with UNC-76 to regulate axonal outgrowth and normal presynaptic organization in Caenorhabditis elegans. J Biol 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang H-G (2011) Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy 7: 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda H, Mochizuki H, Flores R III, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T (2008) UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev 22: 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Kaneko Y, Uchiyama K, Toh H, Tamura K, Kondo H (2011) VCIP135 deubiquitinase and its binding protein, WAC, in p97ATPase-mediated membrane fusion. EMBO J 30: 3581–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincheri NF, Follo C, Nicotra G, Peracchio C, Castino R, Isidoro C (2008) Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis 29: 381–389 [DOI] [PubMed] [Google Scholar]
- Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA (2001) ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J Biol Chem 276: 22826–22837 [DOI] [PubMed] [Google Scholar]
- Webber JL, Tooze SA (2010a) Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J 29: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA (2010b) New insights into the function of Atg9. FEBS Lett 584: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Xu GM, Arnaout MA (2002) WAC, a novel WW domain-containing adapter with a coiled-coil region, is colocalized with splicing factor SC35. Genomics 79: 87–94 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE (2006) Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 172: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ (2010) Eaten alive: a history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Chan EYW, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119: 3888–3900 [DOI] [PubMed] [Google Scholar]
- Zhang F, Yu X (2011) WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 41: 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11: 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.