Abstract
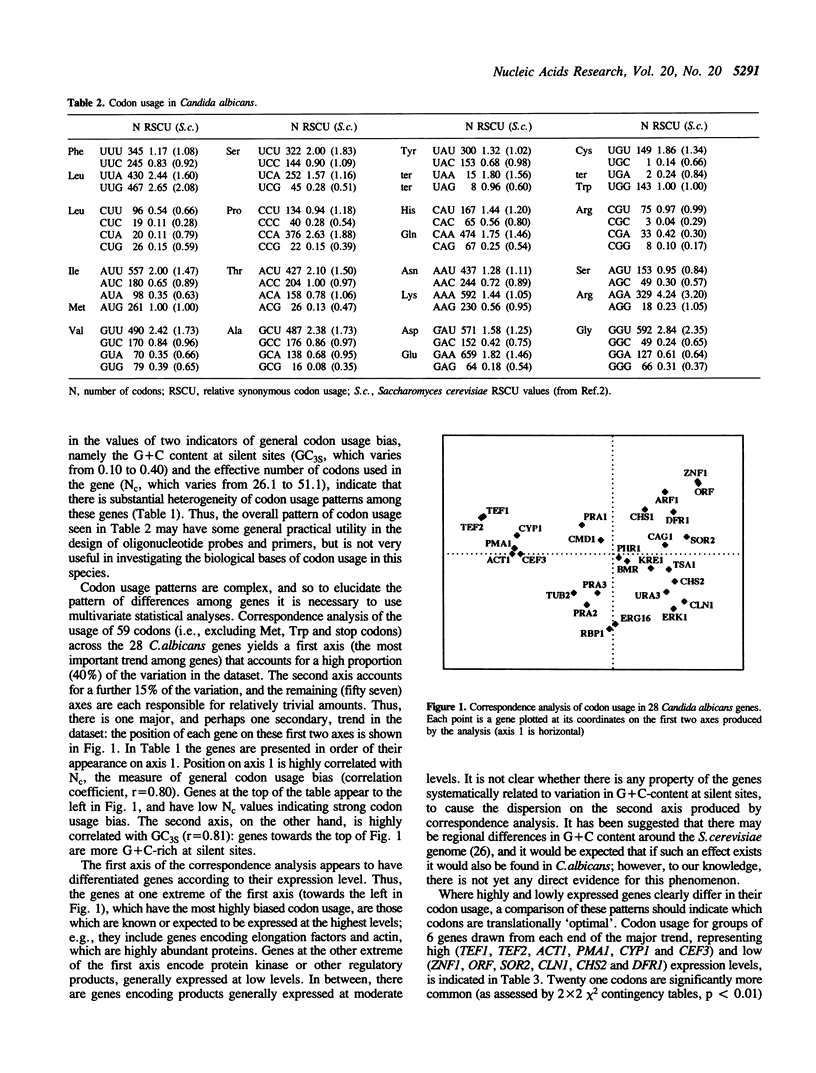
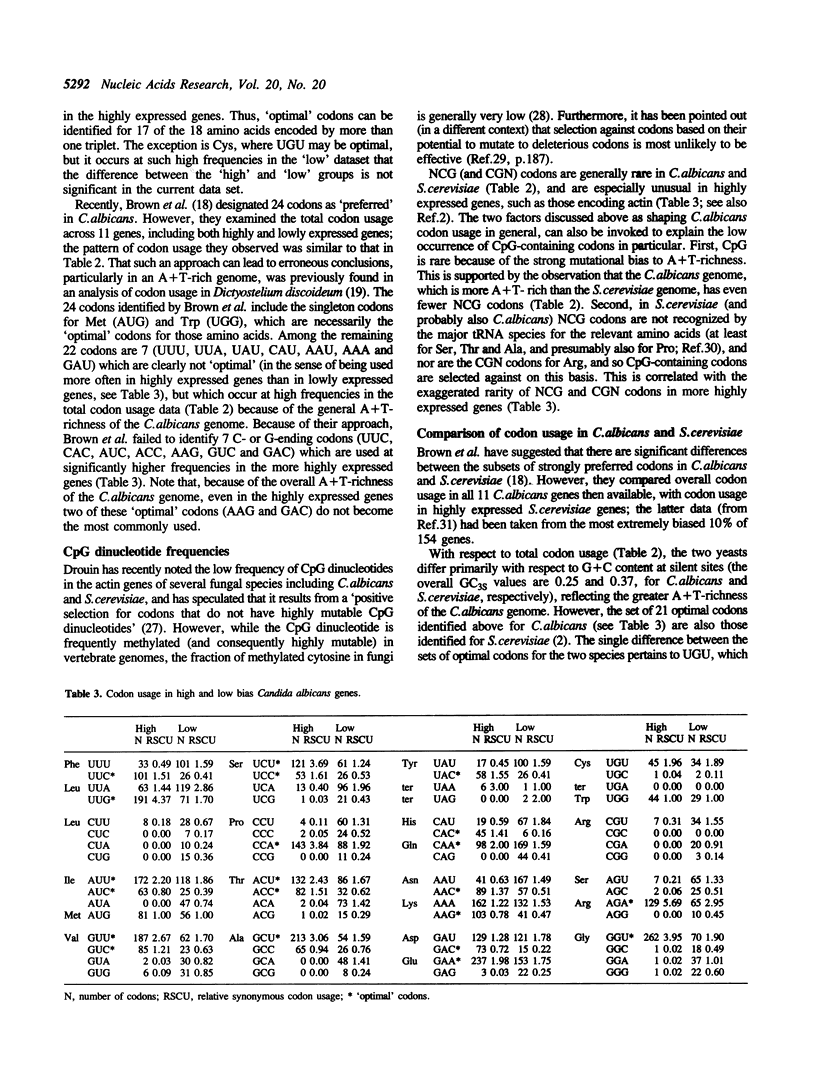
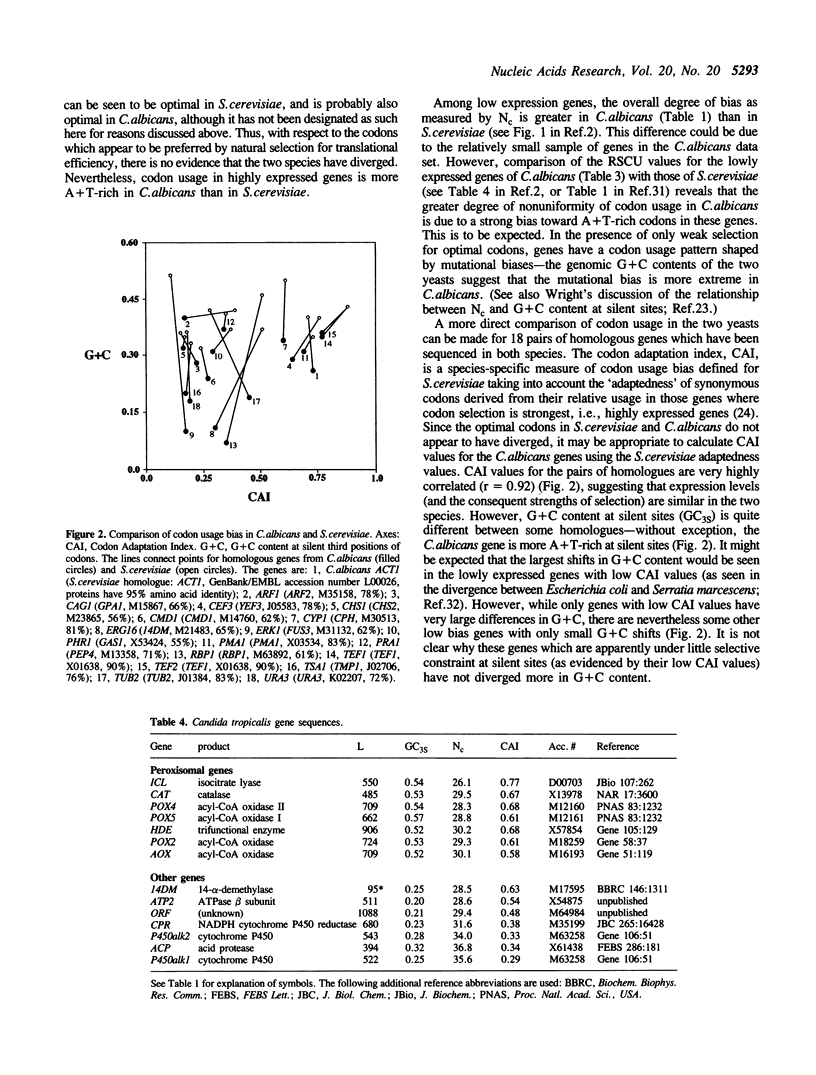
Codon usage in a sample of 28 genes from the pathogenic yeast Candida albicans has been analysed using multivariate statistical analysis. A major trend among genes, correlated with gene expression level, was identified. We have focussed on the extent and nature of divergence between C.albicans and the closely related yeast Saccharomyces cerevisiae. It was recently suggested that significant differences exist between the subsets of preferred codons in these two species [Brown et al. (1991) Nucleic Acids Res. 19, 4293]. Overall, the genes of C.albicans are more A + T-rich, reflecting the lower genomic G + C content of that species, and presumably resulting from a different pattern of mutational bias. However, in both species highly expressed genes preferentially use the same subset of 'optimal' codons. A suggestion that the low frequency of NCG codons in both yeast species results from selection against the presence of codons that are potentially highly mutable is discounted. Codon usage in C.albicans, as in other unicellular species, can be interpreted as the result of a balance between the processes of mutational bias and translational selection. Codon usage in two related Candida species, C.maltosa and C.tropicalis, is briefly discussed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Tamame M., Villanueva J. R., Santos T. DNA methylation in the fungi. J Biol Chem. 1984 Jul 10;259(13):8033–8036. [PubMed] [Google Scholar]
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atomi H., Ueda M., Hikida M., Hishida T., Teranishi Y., Tanaka A. Peroxisomal isocitrate lyase of the n-alkane-assimilating yeast Candida tropicalis: gene analysis and characterization. J Biochem. 1990 Feb;107(2):262–266. doi: 10.1093/oxfordjournals.jbchem.a123036. [DOI] [PubMed] [Google Scholar]
- Au-Young J., Robbins P. W. Isolation of a chitin synthase gene (CHS1) from Candida albicans by expression in Saccharomyces cerevisiae. Mol Microbiol. 1990 Feb;4(2):197–207. doi: 10.1111/j.1365-2958.1990.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Brown A. J., Bertram G., Feldmann P. J., Peggie M. W., Swoboda R. K. Codon utilisation in the pathogenic yeast, Candida albicans. Nucleic Acids Res. 1991 Aug 11;19(15):4298–4298. doi: 10.1093/nar/19.15.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987 Feb 19;325(6106):728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Bulmer M. The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res. 1990 May 25;18(10):2869–2873. doi: 10.1093/nar/18.10.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. W., Anné J., Volckaert G., Huysmans E., Vandenberghe A., De Wachter R. The nucleotide sequences of the 5 S rRNAs of seven molds and a yeast and their use in studying ascomycete phylogeny. Nucleic Acids Res. 1984 Jun 25;12(12):4881–4892. doi: 10.1093/nar/12.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G. Nonrandom CpG mutations affect the synonymous codon usage of moderately GC-rich single copy actin genes. J Mol Evol. 1991 Sep;33(3):237–240. doi: 10.1007/BF02100674. [DOI] [PubMed] [Google Scholar]
- Fling M. E., Kopf J., Tamarkin A., Gorman J. A., Smith H. A., Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991 Jun;227(2):318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Attimonelli M., Lanave C., di Paola G. ACNUC--a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput Appl Biosci. 1985 Sep;1(3):167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L., Goris A., Van de Peer Y., Neefs J. M., Vancanneyt M., Kersters K., Hennebert G. L., De Wachter R. Phylogenetic analysis of five medically important Candida species as deduced on the basis of small ribosomal subunit RNA sequences. J Gen Microbiol. 1991 May;137(5):1223–1230. doi: 10.1099/00221287-137-5-1223. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. CODONS: a microcomputer program for codon usage analysis. J Hered. 1992 May-Jun;83(3):239–240. doi: 10.1093/oxfordjournals.jhered.a111205. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. Codon usage in Aspergillus nidulans. Mol Gen Genet. 1991 Nov;230(1-2):288–294. doi: 10.1007/BF00290679. [DOI] [PubMed] [Google Scholar]
- Murray W. W., Rachubinski R. A. The nucleotide sequence of complementary DNA and the deduced amino acid sequence of peroxisomal catalase of the yeast Candida tropicalis pK233. Gene. 1987;61(3):401–413. doi: 10.1016/0378-1119(87)90202-2. [DOI] [PubMed] [Google Scholar]
- Saporito S. M., Sypherd P. S. The isolation and characterization of a calmodulin-encoding gene (CMD1) from the dimorphic fungus Candida albicans. Gene. 1991 Sep 30;106(1):43–49. doi: 10.1016/0378-1119(91)90564-r. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991 Oct;7(7):657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Devine K. M. Codon usage and gene expression level in Dictyostelium discoideum: highly expressed genes do 'prefer' optimal codons. Nucleic Acids Res. 1989 Jul 11;17(13):5029–5039. doi: 10.1093/nar/17.13.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C. Switches in species-specific codon preferences: the influence of mutation biases. J Mol Evol. 1990 Aug;31(2):71–80. doi: 10.1007/BF02109476. [DOI] [PubMed] [Google Scholar]
- Smith H. A., Allaudeen H. S., Whitman M. H., Koltin Y., Gorman J. A. Isolation and characterization of a beta-tubulin gene from Candida albicans. Gene. 1988;63(1):53–63. doi: 10.1016/0378-1119(88)90545-8. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure, selective constraints, and genetic equilibria. J Mol Evol. 1992 Feb;34(2):95–114. doi: 10.1007/BF00182387. [DOI] [PubMed] [Google Scholar]
- Wada K., Wada Y., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992 May 11;20 (Suppl):2111–2118. doi: 10.1093/nar/20.suppl.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The 'effective number of codons' used in a gene. Gene. 1990 Mar 1;87(1):23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]



