Abstract
Ebola and Marburg filoviruses cause deadly outbreaks of haemorrhagic fever. Despite considerable efforts, no essential cellular receptors for filovirus entry have been identified. We showed previously that Niemann-Pick C1 (NPC1), a lysosomal cholesterol transporter, is required for filovirus entry. Here, we demonstrate that NPC1 is a critical filovirus receptor. Human NPC1 fulfills a cardinal property of viral receptors: it confers susceptibility to filovirus infection when expressed in non-permissive reptilian cells. The second luminal domain of NPC1 binds directly and specifically to the viral glycoprotein, GP, and a synthetic single-pass membrane protein containing this domain has viral receptor activity. Purified NPC1 binds only to a cleaved form of GP that is generated within cells during entry, and only viruses containing cleaved GP can utilize a receptor retargeted to the cell surface. Our findings support a model in which GP cleavage by endosomal cysteine proteases unmasks the binding site for NPC1, and GP–NPC1 engagement within lysosomes promotes a late step in entry proximal to viral escape into the host cytoplasm. NPC1 is the first known viral receptor that recognizes its ligand within an intracellular compartment and not at the plasma membrane.
Keywords: Ebola virus, Marburg virus, Niemann-Pick C1, viral entry, viral receptor
Introduction
Ebola virus (EBOV) and Marburg virus (MARV) are members of the family Filoviridae of enveloped viruses with non-segmented negative-strand RNA genomes (Kuhn et al, 2010). Filovirus entry is mediated by the membrane glycoprotein, GP, which is organized into trimeric spikes at the viral surface (Lee et al, 2008; White et al, 2008). GP consists of a receptor-binding subunit, GP1, and a membrane fusion subunit, GP2. Following attachment to host cells (Becker et al, 1995; Alvarez et al, 2002; Kondratowicz et al, 2011), viral particles are internalized and delivered to late endosomes (Nanbo et al, 2010; Saeed et al, 2010). Endosomal cysteine proteases then cleave GP1 to remove heavily glycosylated C-terminal sequences, generating an entry intermediate comprising an N-terminal GP1 fragment and GP2 (Supplementary Figure S1A; Chandran et al, 2005; Schornberg et al, 2006; Lee et al, 2008; Hood et al, 2010). An unknown trigger signal acts on this ‘primed’ GP, inducing GP1–GP2 dissociation and driving GP2-mediated membrane fusion (Chandran et al, 2005; Schornberg et al, 2006; Lee et al, 2008; Dube et al, 2009; Wong et al, 2010; Brecher et al, 2012).
Current evidence indicates that an interaction between GP and a cellular protein receptor is required for filovirus entry (Takada et al, 1997; Wool-Lewis and Bates, 1998; Yang et al, 1998; Manicassamy et al, 2005; Kuhn et al, 2006; Brindley et al, 2007; Kaletsky et al, 2007; Dube et al, 2009; Ou et al, 2010). Furthermore, the near-universal tropism of filoviruses for mammalian cell types strongly suggests that this host molecule is widely distributed (Van den Groen et al, 1978; Takada et al, 1997; Wool-Lewis and Bates, 1998). However, none of the candidate receptors proposed to date are required in all cell types susceptible to viral infection, or allow filoviruses to overcome species barriers to infection, suggesting that a critical filovirus receptor remains to be identified (Becker et al, 1995; Chan et al, 2001; Alvarez et al, 2002; Shimojima et al, 2006; Kondratowicz et al, 2011).
We and others recently identified Niemann-Pick C1 (NPC1) to be an essential host factor for filovirus entry and infection in all studied cell types (Carette et al, 2011; Côté et al, 2011). Moreover, we showed that NPC1 is required for pathogenesis in mouse models of filovirus infection (Carette et al, 2011). NPC1 is a large polytopic membrane protein that resides in the late endosomes and lysosomes of all cells and is involved in transport of lysosomal cholesterol to the endoplasmic reticulum and other cellular sites (Supplementary Figure S1B; Carstea et al, 1997; Cruz et al, 2000; Davies and Ioannou, 2000). Mutation of NPC1 in humans causes Niemann-Pick type C1 disease, a rare but fatal disorder associated with lysosomal storage of cholesterol and sphingolipids in the brain and other tissues (Patterson et al, 2001; Walkley and Suzuki, 2004). Analysis of NPC1 mutations that cause Niemann-Pick type C1 disease has revealed key roles for the three large luminal ‘loop’ domains, A, C, and I, and for the ‘sterol-sensing domain’, comprising transmembrane domains 3–7, in lysosomal cholesterol transport by NPC1 (Supplementary Figure S1B; Ioannou, 2000; Ory, 2004; Infante et al, 2008a). While a substantial body of information about the housekeeping functions of NPC1 is available, its specific role in filovirus entry remains unknown. Previous findings suggest that the cholesterol transport function of NPC1 is dispensable for its viral host factor function, and that GP can bind to cellular membranes by associating directly or indirectly with full-length NPC1 (Carette et al, 2011; Côté et al, 2011). However, they do not fully distinguish among three mechanisms of action: NPC1 might (i) play an indirect role in viral entry by regulating endosomal/lysosomal morphology or membrane composition; (ii) participate in trafficking of viral particles to the sites of membrane fusion; or (iii) act directly as a filovirus receptor.
In this study, we demonstrate that filovirus entry does not require the full-length NPC1 protein. Instead, we provide multiple lines of evidence that a single luminal domain of NPC1 mediates filovirus entry by binding specifically and directly to the viral glycoprotein. Our work reveals that the NPC1-binding site within the GP1 subunit is recessed beneath the heavily glycosylated mucin and glycan cap GP1 subdomains, and explains our observation that proteolytic removal of these sequences is a prerequisite for direct GP–NPC1 interaction. We exploit our new findings to engineer a synthetic NPC1 analogue that is targeted to the cell surface and that mediates cell attachment and entry only by cleaved viruses containing exposed NPC1-binding sites. Furthermore, we provide evidence that human NPC1 can render cells from a non-permissive species susceptible to filovirus entry and infection. Cumulatively, our results indicate that NPC1 is an essential intracellular receptor for EBOV and MARV that promotes a late step in viral entry by binding to a proteolytically primed form of the viral glycoprotein within the host endosomal/lysosomal pathway.
Results
Human NPC1 renders non-permissive reptilian cells susceptible to filovirus entry and infection
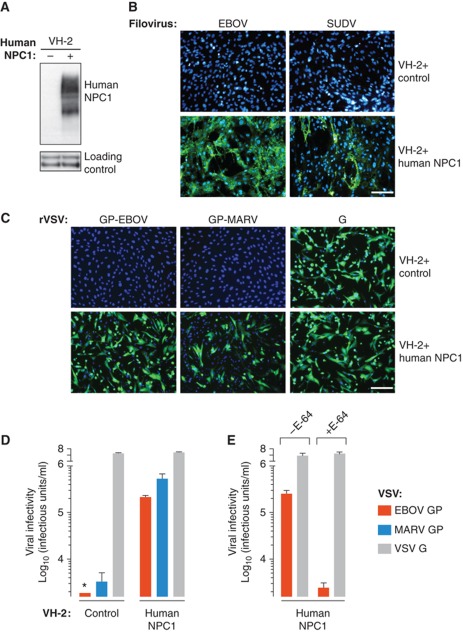
Mammalian cells are broadly susceptible to filoviruses, but reptilian and amphibian cells are reported to be refractory to infection (Van den Groen et al, 1978; Takada et al, 1997). Consistent with these previous findings, wild-type (WT) EBOV and Sudan virus (SUDV) did not infect VH-2 cells derived from the Russell's viper (Daboia russellii). The recent discovery of NPC1 as a critical host factor for filovirus entry and infection (Carette et al, 2011; Côté et al, 2011) led us to speculate that this protein could determine the species tropism of filoviruses. Accordingly, we engineered VH-2 cells to stably express human NPC1 (Figure 1A), and challenged them with WT EBOV and SUDV (Figure 1B). Remarkably, human NPC1 rendered these cells highly susceptible to filovirus infection. The block to infection in VH-2 cells and its rescue by ectopic expression of human NPC1 were recapitulated both with recombinant vesicular stomatitis viruses bearing EBOV or MARV GP (rVSV-GP-EBOV/MARV) (Figure 1C) and with VSV pseudotypes (Figure 1D), showing that VH-2 cells resist filovirus infection at the level of viral entry. Human NPC1 was dispensable for viral attachment and internalization, since fluorescently labelled rVSV-GP particles were delivered to perinuclear sites in both VH-2 cells lacking or expressing human NPC1 (Supplementary Figure S2A). Furthermore, VH-2 cells were replete with endosomal cysteine protease activities that could mediate viral entry upon provision of human NPC1 (Figure 1E), strongly suggesting that the entry block in these cells does not arise from the failure to generate a proteolytically primed GP intermediate within endosomes and/or lysosomes. Instead, ectopic expression of human NPC1 was associated with a reduction in virus-positive intracellular puncta (Supplementary Figure S2B and C), suggesting that this protein facilitates viral escape from the endosomal/lysosomal pathway of Russell's viper VH-2 cells, as observed previously in human and rodent cells (Carette et al, 2011). Finally, the NPC1-dependent entry block in VH-2 cells was not a consequence of defective lysosomal cholesterol transport, since the aberrant accumulation of cholesterol in lysosomes was only observed when these cells were exposed to U18666A, a small molecule that mimics the cellular effects of NPC1 deficiency (Supplementary Figure S2D; Liscum and Faust, 1989). These results demonstrate that the transmembrane protein NPC1 possesses a cardinal property of viral receptors: it allows filoviruses to overcome a species barrier to cell entry and infection.
Figure 1.
Human NPC1 confers susceptibility to filovirus entry and infection. (A) Viper VH-2 cells were engineered to express empty vector or human NPC1. Expression of human NPC1 was determined by immunoblotting (IB) with an anti-NPC1 antibody. Proteins non-specifically detected by this antibody were used as a loading control. Samples for IB of NPC1 were resolved on one gel and loading controls were resolved on another. (B, C) Infection of control and NPC1-expressing VH-2 cells by wild-type filoviruses (B), or recombinant VSVs bearing VSV or filovirus glycoproteins (C). Infected cells (green) and counter-stained nuclei (blue) were visualized by fluorescence microscopy. Scale bars, 20 μm. (D, E) Infectivity of VSV pseudotypes bearing VSV or filovirus glycoproteins in control and NPC1-expressing VH-2 cells. (E) NPC1-expressing VH-2 cells were pretreated with the cysteine protease inhibitor E-64 (300 μM) for 4 h at 37°C and then exposed to virus. (D, E) Results (n=3) are representative of two independent experiments. Error bars indicate s.d. Asterisk indicates values below the limit of detection. Figure source data can be found in Supplementary data.
Filovirus entry requires the luminal domain C of NPC1, but not the full-length protein
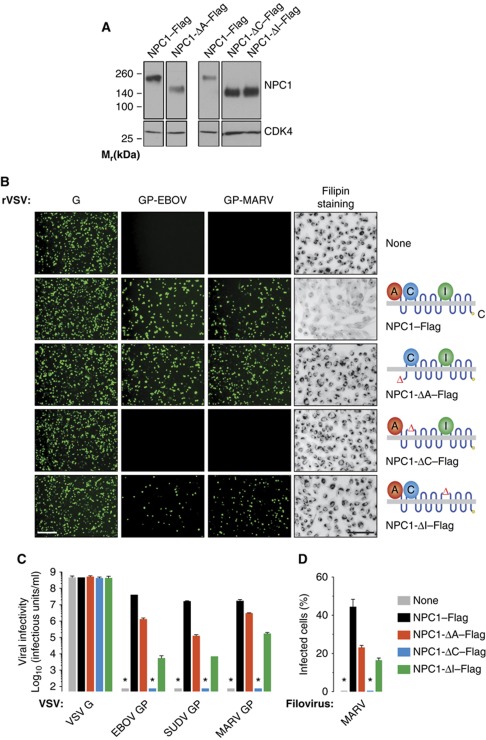
All experiments to date examining the role of NPC1 in filovirus entry have been carried out with the full-length protein. To determine if viral entry requires the entire protein or can instead be attributed to a discrete region within it, we expressed NPC1 deletion mutants individually lacking the large luminal loop domains A, C, and I in an NPC1-null cell line (Chinese hamster ovary (CHO) CT43) (Cruz et al, 2000; Figure 2A), and examined their capacity to mediate lysosomal cholesterol transport and viral infection (Figure 2B–D). CT43 cells accumulated lysosomal cholesterol (Cruz et al, 2000), and they were completely resistant to infection by WT EBOV/MARV and rVSV-GP-EBOV/MARV (Carette et al, 2011). As we showed previously, expression of Flag epitope-tagged WT NPC1 (NPC1–Flag) in these cells not only corrected their cholesterol transport defect but also rendered them highly susceptible to infection by WT filoviruses and rVSVs bearing filovirus GPs (Carette et al, 2011). All three ‘loop-minus’ NPC1 mutants were inactive at lysosomal cholesterol transport (Figure 2B), despite their significant localization to LAMP1-positive late endosomal/lysosomal compartments (Supplementary Figure S3), confirming that this cellular activity of NPC1 requires all three luminal domains A, C, and I. However, the mutants differed in their capacity to support filovirus GP-mediated entry (Figure 2B and C). Both NPC1-ΔA–Flag and NPC-ΔI–Flag could mediate entry, albeit at reduced levels relative to WT NPC1–Flag. In striking contrast, NPC1-ΔC–Flag was unable to rescue viral entry (Figure 2B and C) even though it resembled the other mutants in expression level and intracellular distribution (Figure 2A; Supplementary Figure S3). Similar results were obtained in infection assays with WT MARV (Figure 2D). These findings unequivocally separate NPC1's functions in lysosomal cholesterol transport and filovirus entry. More importantly, they demonstrate that a discrete region within NPC1, the luminal domain C, is essential for EBOV and MARV entry.
Figure 2.
NPC1 luminal loop domain C is required for filovirus entry, but full-length NPC1 is dispensable. (A) NPC1-null CHO CT43 cells were engineered to express mutant forms of human NPC1–Flag lacking domains A, C, or I. NPC1 expression was determined by IB with an anti-Flag antibody. Cyclin-dependent kinase 4 (CDK4) in each sample was detected by IB (Abcam) and provided a loading control. Samples for IB of each NPC1 mutant and its paired WT NPC1 control were resolved on the same gel. NPC1 and CDK4 were detected on separate gels. (B) Capacity of mutant NPC1 proteins to rescue viral entry and transport lysosomal cholesterol. (Left) Infection of NPC1-null CHO CT43 cells expressing mutant NPC1–Flag proteins by recombinant VSVs bearing VSV G or filovirus glycoproteins. Infected cells (green) were visualized by fluorescence microscopy. (Right) Cholesterol clearance by mutant NPC1–Flag proteins in CT43 cells was determined by filipin staining and fluorescence microscopy. Images were inverted for clarity. Scale bars, 20 μm. (C, D) Infectivity of VSV pseudotypes bearing VSV or filovirus glycoproteins (C) and wild-type MARV (D) in CT43 cells expressing mutant NPC–Flag proteins. SUDV, Sudan virus. Results in (C) (n=4–6) are from two independent experiments. Results in (D) (n=3) are from a representative experiment. Error bars indicate s.d. Asterisks indicate values below the limit of detection. Figure source data can be found in Supplementary data.
NPC1 binds specifically and directly to a proteolytically cleaved form of EBOV GP
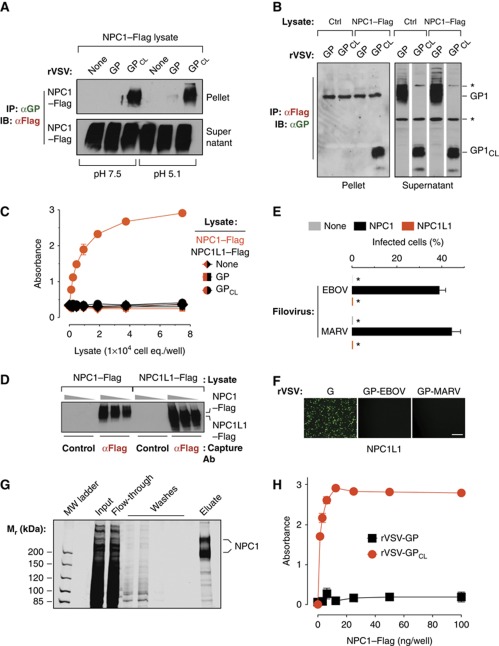
A viral receptor mediates entry by binding specifically and directly to a viral surface protein. Recent work indicated that proteolytically cleaved GP could associate with endosomal membranes derived from WT but not NPC1-deficient cells, suggesting that GP and NPC1 interact (Côté et al, 2011). To examine this hypothesis, we first tested if EBOV GP could bind to NPC1 in a cell- and membrane-free system. Concentrated rVSV-GP-EBOV particles were solubilized in a non-ionic detergent-containing buffer, and GP in these extracts was captured by magnetic beads coated with the GP-specific monoclonal antibody KZ52 (Maruyama et al, 1999; Lee et al, 2008). These GP-decorated beads did not retrieve NPC1–Flag from CT43 detergent extracts in a co-immunoprecipitation (co-IP) assay (Figure 3A). We next incubated rVSV-GP-EBOV with the bacterial metalloprotease thermolysin to generate a GP intermediate (GPCL) that resembles the product of endosomal/lysosomal GP cleavage (Chandran et al, 2005; Schornberg et al, 2006). GPCL could capture NPC1–Flag at both neutral and acid pH (Figure 3A). Similar results were obtained in a reciprocal co-IP experiment: magnetic beads displaying NPC1–Flag captured GPCL but not GP (Figure 3B).
Figure 3.
NPC1 binds specifically and directly to a cleaved form of the Ebola virus glycoprotein. (A) Co-immunoprecipitation (co-IP) of NPC1 by EBOV GP. Magnetic beads coated with GP-specific monoclonal antibody KZ52 were incubated with detergent extracts containing no virus (None), uncleaved rVSV-GP, or cleaved rVSV-GPCL. The resulting control or glycoprotein-decorated beads were mixed with cell lysates containing human NPC1–Flag at pH 7.5 or 5.1 and 4°C. Beads were then retrieved and NPC1–Flag in the immune pellets and supernatants was detected by IB with an anti-Flag antibody. Pellets and supernatants were resolved on separate gels but exposed simultaneously to the same piece of film. (B) Reciprocal co-IP of GP by NPC1. Cell lysates lacking (Ctrl) or containing NPC1–Flag were incubated with anti-Flag antibody-coated magnetic beads. The resulting control or NPC1-decorated beads were mixed with detergent extracts of rVSV-GP or rVSV-GPCL at pH 7.5 and 4°C. Beads were then retrieved and GP or GPCL in the immune pellets and supernatants was detected by IB with an anti-GP antiserum. Pellets and supernatants were resolved on separate gels but exposed simultaneously to the same piece of film. Asterisks indicate bands detected non-specifically by the antiserum. (C) GPCL captures NPC1 but not NPC1-like1 (NPC1L1) in an ELISA. Plates coated with rVSV-GP or rVSV-GPCL were incubated with cell extracts containing NPC1–Flag or NPC1L1–Flag, and bound Flag-tagged proteins were detected with an anti-Flag antibody. Results (n=3) are representative of at least four independent experiments. (D) Cell extracts used in (C) were incubated with plates coated with an anti-Flag antibody or an isotype-matched control, and captured proteins were eluted and detected by IB with the anti-Flag antibody. Samples were resolved on the same gel. (E, F) NPC1L1 cannot support filovirus entry and infection. CT43 cells expressing NPC1L1–Flag were exposed to wild-type EBOV or MARV (E) or to recombinant VSVs (F), and infected cells were visualized and enumerated by fluorescence microscopy. Asterisks in (E) indicate values below the limit of detection. Scale bar, 20 μm. (G, H) GPCL but not GP captures affinity-purified NPC1–Flag in an ELISA. (G) NPC1–Flag was purified from CT43 CHO cell lysates by Flag affinity chromatography and visualized by SDS–PAGE and staining with Krypton infrared protein stain. Samples were resolved on the same gel. (H) ELISA plates coated with rVSV-GP or rVSV-GPCL were incubated with NPC1–Flag purified in (G), and bound Flag-tagged proteins were detected with an anti-Flag antibody. Results (n=3) are representative of at least four independent experiments. Error bars indicate s.d. Figure source data can be found in Supplementary data.
To confirm these findings, we examined the capacity of rVSV-derived GP and GPCL to capture NPC1–Flag from 293T human embryonic kidney cell extracts using an enzyme-linked immunosorbent assay (ELISA). GP and GPCL were captured onto antibody KZ52-coated ELISA plates, and then incubated with CT43 extracts containing NPC1–Flag. We found that NPC1–Flag bound saturably to wells coated with GPCL but not with GP, consistent with our results from the co-IP assay (Figure 3C).
To establish the specificity of GPCL–NPC1 association, we used the ELISA to determine if GPCL could capture NPC1-like1 (NPC1L1), a cholesterol transport protein that resembles NPC1 in topology and sequence (∼50% similarity) (Davies et al, 2000; Wang et al, 2009). NPC1L1–Flag did not detectably bind to wells coated with either GP or GPCL (Figure 3C), despite its greater abundance in cell extracts relative to NPC1–Flag (Figure 3D). Consistent with these binding results, NPC1L1 could not substitute for NPC1 in mediating infection by WT filoviruses (Figure 3E) and rVSVs bearing filovirus GPs (Figure 3F). Therefore, NPC1 associates specifically with GPCL and is specifically required for filovirus entry.
Finally, affinity-purified NPC1–Flag bound saturably to wells coated with GPCL but not with GP in the ELISA, providing evidence that GPCL directly interacts with NPC1 (Figure 3G and H). Cumulatively, these findings demonstrate that the proteolytic priming of EBOV GP creates, or unmasks, a specific and direct binding site for NPC1.
NPC1 luminal domain C is necessary and sufficient for GP–NPC1 binding and viral entry mediated by filovirus GPs
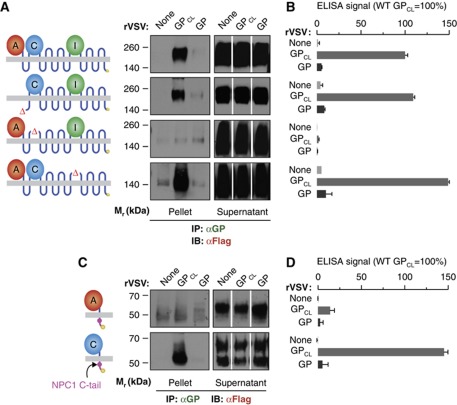
To begin to map the GP binding site within NPC1, we tested the loop-minus NPC1 mutants for GPCL-binding activity in the co-IP and ELISA assays, as described above (Figure 4A and B). NPC1-ΔA–Flag and NPC1-ΔI–Flag in detergent extracts of CT43 cells were fully competent to bind to GPCL, but little or no binding was obtained with NPC1-ΔC–Flag. Therefore, the same region of NPC1, the luminal domain C, is absolutely required for both EBOV GP–NPC1 binding and NPC1-mediated filovirus entry.
Figure 4.
NPC1 luminal domain C is necessary and sufficient for GP–NPC1 binding. (A, B) Binding of NPC1 mutants lacking domains A, C, or I to EBOV GP. (A) Co-IP of mutant NPC1–Flag proteins from CT43 cell extracts by rVSV-GP was carried out as described in Figure 3A. Pellet samples for each NPC1 construct were resolved on the same gel. Pellets and supernatants were resolved on separate gels but exposed simultaneously to the same piece of film. (B) Capture of mutant NPC1–Flag proteins by rVSV-GP in an ELISA was carried out as described in Figure 3C. (C, D) CT43 cells were engineered to express synthetic single-pass membrane proteins comprising individual luminal domains of NPC1 fused to the first transmembrane domain of NPC1, the NPC1 cytoplasmic tail, and a Flag tag. Binding of these NPC1 mutants to EBOV GP was determined by co-IP (C) and ELISA (D) as described above. Results in (B) and (D) (n=3) are representative of at least three independent experiments. Error bars indicate s.d. Figure source data can be found in Supplementary data.
We then used multiple approaches to test if domain C is not only necessary but also sufficient to mediate EBOV GPCL–NPC1 binding. First, we engineered synthetic single-pass membrane proteins comprising each luminal domain fused to the first transmembrane domain of NPC1, the NPC1 cytoplasmic tail (which contains a lysosomal targeting signal; Scott et al, 2004), and a Flag tag, and expressed them in CT43 cells. All three proteins were expressed at similar levels (Supplementary Figure S4A), and domain A-Flag and domain C-Flag localized significantly to late endosomes and/or lysosomes (Supplementary Figure S4B). However, domain I-Flag appeared to be entirely restricted to the endoplasmic reticulum, suggesting that it misfolds (Supplementary Figure S4B); accordingly, we excluded it from further analysis. In both the co-IP and ELISA assays (Figure 4C and D), domain C-Flag bound as well as WT NPC1–Flag to GPCL, and it bound poorly, or not at all, to uncleaved GP. Domain A-Flag did not detectably bind to GP or GPCL (Figure 4C and D).
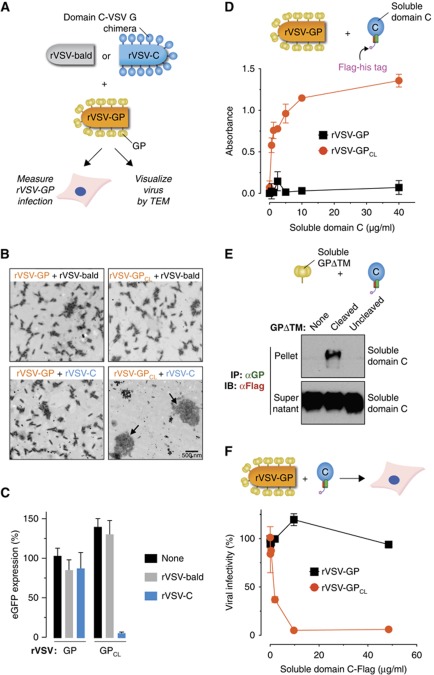
The preceding binding experiments were carried out with detergent-solubilized membrane proteins, either in cell extracts (Figures 3 and 4) or as purified preparations (Figure 3G and H). To examine the GPCL–NPC1 interaction with ‘soluble proteins’ in the absence of detergent, we first engineered recombinant VSV particles to display a chimeric domain C-VSV G glycoprotein at their surface (Figure 5A; Supplementary Figure S5A), and assessed their capacity to bind to rVSV-GPCL. Preincubation of rVSV-GPCL with rVSV-domain C induced the dramatic clustering of viral particles (Figure 5B; Supplementary Figure S5B) and neutralized rVSV-GPCL infection (Figure 5C). By contrast, rVSV-domain C had no discernible effect on uncleaved rVSV-GP. Neither rVSV-GPCL nor rVSV-GP was clustered or neutralized by ‘bald’ rVSV particles lacking any virus-encoded surface glycoproteins (Figure 5B and C; Supplementary Figure S5B).
Figure 5.
Soluble forms of NPC1 domain C bind directly to GP and selectively neutralize infection by viral particles containing cleaved glycoproteins. (A–C) Capacity of an rVSV displaying a domain C-VSV G chimera (rVSV-domain C) to bind to rVSV-GP and neutralize rVSV-GP infection. (A) Schematic of experiments shown in (A–C). ‘rVSV-bald’ particles lack any virus-encoded glycoproteins. (B) rVSV-GP and rVSV-GPCL were preincubated with rVSV-domain C or rVSV-bald, and virus mixtures were stained with phosphotungstic acid and visualized by electron microscopy. Arrows indicate large clusters of viral particles. A larger version of this image is available in Supplementary Figure S5. (C) Virus mixtures were exposed to WT CHO cells, and expression of the rVSV-GP-encoded eGFP gene was quantitated at 7 h post infection (see Materials and methods for details). eGFP signal was normalized to that of uncleaved virus. (D) The capacity of rVSV-GP and rVSV-GPCL to capture a purified, soluble form of domain C containing Flag and hexahistidine tags was determined in an ELISA, as described in Figure 3C. Results (n=3) are representative of at least three independent experiments. (E) The capacity of a purified, soluble form of GP lacking the transmembrane domain (GPΔTM) to associate with purified, soluble domain C was determined by co-IP, as described in Figure 3A. Pellets and supernatants (one gel for each) were resolved on separate gels but exposed simultaneously to the same piece of film. (F) rVSV-GP and rVSV-GPCL were preincubated with soluble domain C, and virus–protein mixtures were exposed to the Vero African grivet monkey kidney cell line. Viral infection was enumerated by fluorescence microscopy. Results are from two independent experiments (n=4). Error bars indicate s.d. Figure source data can be found in Supplementary data.
We next asked if a soluble, secreted, and biologically active form of domain C described recently (Deffieu and Pfeffer, 2011) could bind to GPCL. Cleaved rVSV-GPCL, but not uncleaved rVSV-GP, captured purified domain C in an ELISA (Figure 5D; Supplementary Figure S6). Even more stringently, GPCL derived from a purified, soluble GP lacking the transmembrane domain (GPΔTM) co-precipitated purified domain C, whereas uncleaved GPΔTM did not (Figure 5E; Supplementary Figure S6). Consistent with its capacity to bind directly and stably to GPCL, soluble domain C neutralized infection by rVSV-GPCL but not by rVSV-GP in a dose-dependent manner (Figure 5F).
Finally, we tested the capacity of the synthetic single-domain transmembrane proteins to mediate viral entry in CT43 cells (Figure 6). Remarkably, only domain C-Flag afforded measurable, although incomplete, rescue of filovirus GP-dependent entry, in full agreement with the GPCL-binding activity of domain C (Figure 6A and B). Taken together, these results indicate that sequences essential for both the EBOV GP binding and entry host factor activities of NPC1 reside within domain C, a 248-amino acid domain of this 1278-amino acid protein that protrudes into the endosomal lumen.
Figure 6.
A synthetic single-pass membrane protein containing domain C can mediate filovirus entry. CT43 cells expressing synthetic membrane proteins containing individual NPC1 luminal domains (see Figure 4C) were exposed to rVSVs bearing uncleaved or cleaved filovirus glycoproteins. Infected cells (green) were visualized (A) and enumerated (B) by fluorescence microscopy. Results in (B) (n=3) are representative of at least three independent experiments. Error bars indicate s.d. Asterisks in (B) indicate values below the limit of detection. Scale bar in (A), 20 μm.
The NPC1-binding site in EBOV GP is recessed beneath the mucin and glycan cap subdomains and is unmasked by proteolytic cleavage
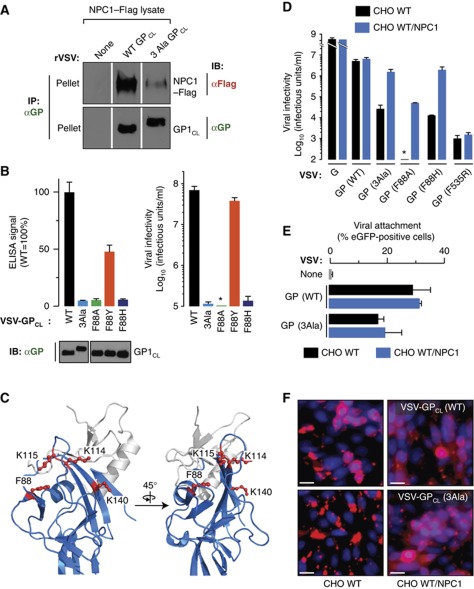
Previous work identified a potential receptor-binding site (RBS) within the EBOV GP1 ‘head’, a structural subdomain composed of N-terminal GP1 sequences (Supplementary Figures S1A and S7C; Kuhn et al, 2006; Lee et al, 2008; Dube et al, 2009, 2010). This site has a key property predicted by our GP–NPC1 binding studies: it is recessed beneath the C-terminal ‘glycan cap’ and mucin GP1 subdomains, and becomes fully exposed only upon their removal during GP → GPCL cleavage (Supplementary Figure S1A; Kaletsky et al, 2007; Lee et al, 2008; Dube et al, 2009). To examine the relationship between the NPC1-binding site and the proposed RBS, we tested the NPC1-binding capacity of a GP containing three mutations that were previously used to define the RBS (K114A+K115A+K140A; ‘3Ala’) (Figure 7A and C; Dube et al, 2009). GPCL(3Ala) bound poorly to purified NPC1–Flag relative to GPCL(WT) in the co-IP and ELISA assays (Figure 7A and B), even though both GPCL proteins were captured by the GP conformation-specific antibody KZ52 in equivalent amounts in both assays (Figure 7A). Similarly, mutation of the nearby highly conserved F88 residue to alanine, which is proposed to remove a key receptor contact or to alter RBS conformation (Manicassamy et al, 2005; Brindley et al, 2007; Lee et al, 2008; Ou et al, 2010), greatly diminished GPCL–NPC1 binding in the ELISA (Figure 7B and C). Therefore, the EBOV GP RBS proposed previously on the basis of mutagenesis and structural modelling contains the binding site for the NPC1 protein.
Figure 7.
Binding of NPC1 to a site within the Ebola virus glycoprotein is required at a late step in entry proximal to viral escape into the cytoplasm. (A) Co-IP of NPC1 by cleaved VSV-GPCL particles containing EBOV GP(WT) or GP(3Ala) carrying three point mutations in the proposed GP receptor-binding site (see text for details). NPC1–Flag and GP in immune pellets were detected by IB with an anti-Flag antibody and an anti-GP antiserum, respectively. Samples for IB of NPC1 and GP were resolved on separate gels (one gel for detection of each protein). (B) (Left) Capture of NPC1–Flag proteins by VSV-GPCL containing WT or mutant GPs in an ELISA. An IB of input cleaved viral particles is shown below the graph. Samples were resolved on the same gel. Equivalent amounts of each GP bound to the prefusion conformation-specific antibody KZ52 were used in the NPC1-capture ELISA. (Right) Infectivity of WT and mutant VSV-GP in WT CHO cells. Results (n=3) are representative of two independent experiments. (C) Cartoon representations of the putative NPC1-binding site in GP1 (PDB id: 3CSY; Lee et al, 2008). Blue, head subdomain; grey, glycan cap subdomain. Mutated residues are shown as red ball-and-sticks. (D) Infectivity of VSV-GP bearing WT or mutant GPs in control and NPC1-overexpressing WT CHO cells. GP(F535R) contains a mutation in the GP2 fusion loop that inhibits membrane insertion (Supplementary Figure S9). Asterisk indicates values below the limit of detection. Results (n=3) are representative of at least two independent experiments. (E) Attachment of eGFP-containing virus-like particles (VLPs) bearing GP(WT) or GP(3Ala) to control and NPC1-overexpressing WT CHO cells. VLPs were exposed to cells at 4°C for 30 min and binding was quantitated by flow cytometry. Percent of eGFP-positive cells was determined by gating on the eGFP signal (see Supplementary Figure S8). Results (n=4–5) are from two independent experiments. (F) Viral escape into the cytoplasm was measured by release of VSV M protein derived from entering VSV-GPCL. Control and NPC1-overexpressing WT CHO cells were exposed to VSV-GPCL containing GP(WT) or GP(3Ala) (multiplicity of infection, MOI=200) for 3 h and processed for VSV M staining. VSV M (red) and counter-stained nuclei (blue) were visualized by fluorescence microscopy. Scale bar, 20 μm. Figure source data can be found in Supplementary data.
Interaction between GP and NPC1 is required for a late step in filovirus entry
We next determined the consequences of these GP mutations for viral entry (Figure 7B). Infection by VSV-GP(3Ala) and VSV-GP(F88A) (Manicassamy et al, 2005; Brindley et al, 2007; Ou et al, 2010) was severely reduced in WT CHO cells. We also tested two additional GP mutations at residue F88 with differential effects on entry. As reported previously, F88Y and F88H were mildly and strongly deleterious for GP-dependent infection, respectively (Ou et al, 2010). The effects of these mutations on viral entry and GP–NPC1 binding were congruent: GPCL(F88Y) displayed substantial, but reduced, binding to NPC1 relative to GPCL(WT), whereas little binding was obtained with GPCL(F88H) (Figure 7B). In sum, this series of experiments defining the molecular basis of the GP–NPC1 interaction strongly supports the hypothesis that GP–NPC1 binding, and not just the presence of the NPC1 protein, is required for filovirus GP-dependent entry.
Finally, we hypothesized that infection by GP mutants attenuated (but not fully deficient) for NPC1 binding may be at least partially rescued by provision of excess NPC1 within late endosomes and lysosomes. Accordingly, we tested the behaviour of VSVs bearing GP(WT), GP(3Ala), GP(F88A), and GP(F88H) in WT CHO cells containing only endogenous NPC1 or additionally expressing high levels of NPC1 (Figure 7D; Supplementary Figure S7). We found that NPC1 overexpression could indeed confer a dramatic enhancement in viral entry mediated by GP. Moreover, this enhancement appeared to be specific for the mutants that showed reduced binding to NPC1; no increase in infectivity was observed with either GP(WT) or GP(F535R), a highly attenuated GP2 mutant (Ito et al, 1999) that bound efficiently to NPC1 in vitro (Supplementary Figure S9). These findings argue that the low binding affinity of the GP(3Ala) and the GP(F88) mutants for NPC1 impairs viral infection, and that increased NPC1 concentrations within endosomal/lysosomal compartments can compensate for this defect, leading to rescue of viral entry and infection.
We then exploited this NPC1-specific rescue assay to investigate the step(s) in viral entry at which GP–NPC1 binding is required. Viral attachment to WT CHO cells mediated by GP(3Ala) was only modestly reduced relative to that mediated by GP(WT), and it was not greatly affected by NPC1 overexpression (Figure 7E; Supplementary Figure S8). Therefore, viral attachment does not require GP–NPC1 binding. We then examined a late entry step: release of the VSV matrix protein, M, into the cytoplasm concomitant with GP-mediated membrane fusion (Figure 7F; Carette et al, 2011). Diffuse M staining indicating productive entry was obtained with in vitro-cleaved VSV-GPCL(WT) particles in both WT and NPC1-overexpressing CHO cells. By striking contrast, only punctate perinuclear M staining was obtained in WT cells exposed to VSV-GPCL(3Ala), indicating a significant reduction in cytoplasmic delivery of viral nucleocapsids. Importantly, NPC1 overexpression substantially overcame this M-release block (Figure 7F). These findings support the conclusion that cleaved viral particles containing GP(3Ala) cannot exit from endosomal/lysosomal compartments, and accumulate within them, because they cannot efficiently bind to NPC1. The corollary to this conclusion is that GP–NPC1 binding is required for a late step in entry, at or near viral escape into the cytoplasm.
Interaction between GP and NPC1 can be retargeted to the cell surface by a synthetic receptor
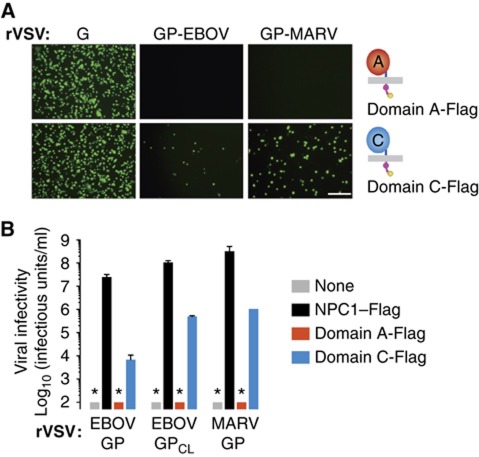
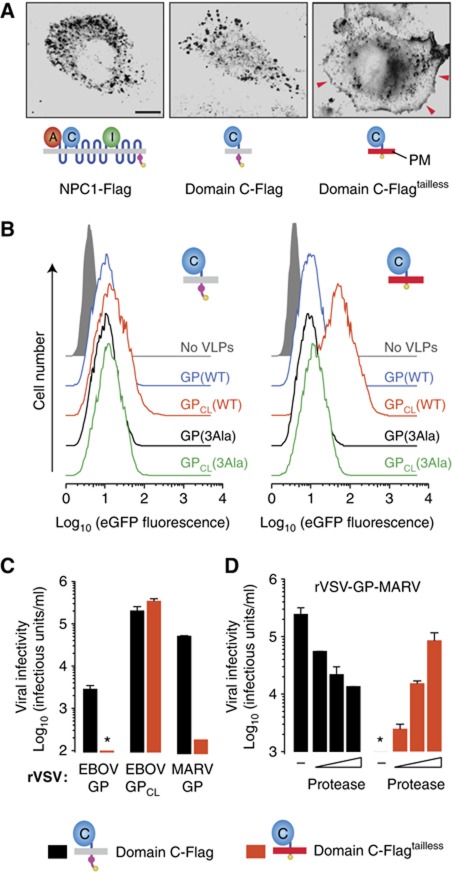
To our knowledge, all known viral receptors and co-receptors engage their viral ligands at the cell surface. However, our findings suggested that the physiological GP–NPC1 interaction occurs only within the endosomal/lysosomal pathway. First, only proteolytically cleaved GPCL could bind specifically and directly to NPC1 in vitro (Figures 3, 4 and 5), and GP cleavage within cells is known to require viral internalization and exposure to endosomal proteases (Chandran et al, 2005; Schornberg et al, 2006). Second, NPC1 is predominantly localized to late endosomes and lysosomes, and its trafficking to these compartments bypasses the plasma membrane (Scott et al, 2004; Berger et al, 2007). To further explore the cellular location of GP–NPC1 interaction, we asked if GP could recognize and exploit domain C if it were made available at the cell surface. To engineer a synthetic cell-surface receptor for filoviruses, we removed the cytoplasmic tail from the minimal NPC1 domain C-Flag protein, which contains a lysosomal targeting signal (Scott et al, 2004), to create domain C-Flagtailless. In contrast to NPC1–Flag and domain C-Flag, which have a predominantly intracellular distribution, domain C-Flagtailless localizes extensively to the plasma membrane (Figure 8A; Supplementary Figure S10). We then assessed the capacity of viral particles to attach to CT43 cells expressing domain C-Flag or domain C-Flagtailless (Figure 8B). A large increase in viral attachment to the domain C-Flagtailless cells relative to the domain C-Flag cells was observed with cleaved viral particles bearing GPCL(WT). By contrast, no such increase was noted with the cleaved NPC1-binding mutant GPCL(3Ala), or with uncleaved viruses bearing GP(WT) or GP(3Ala) (Figure 8B). Therefore, EBOV GPCL but not GP could bind to the plasma membrane-targeted NPC1 domain C in these cells.
Figure 8.
Cleaved but not uncleaved filovirus glycoproteins can utilize a synthetic cell surface receptor based on NPC1 domain C for viral entry. (A) Distribution of NPC1 in CT43 cells expressing WT NPC1–Flag, loop C-Flag, and loop C-Flagtailless (lacking the NPC1 cytoplasmic tail). Cells were immunostained with an anti-Flag antibody and visualized by fluorescence microscopy. Red arrowheads indicate signal at the cell surface. Images were inverted for clarity. The capacity of each protein to localize to the plasma membrane (PM) (grey, no localization to PM; red, localization to PM) is indicated in the cartoon below each image. Scale bar, 10 μm. (B) Attachment of VLPs containing uncleaved or cleaved GP(WT) or GP(3Ala) to CT43 cells expressing domain C-Flag or domain C-Flagtailless. VLPs were exposed to cells at 4°C for 30 min and binding was determined by flow cytometry. (C) Infectivity of rVSV-GP-EBOV, EBOV rVSV-GPCL-EBOV, and rVSV-GP-MARV in CT43 cells expressing domain C-Flag or domain C-Flagtailless. Results (n=4) are from two independent experiments. (D) rVSV-GP-MARV was allowed to attach to CT43 cells expressing domain C-Flag or domain C-Flagtailless at 4°C for 45 min, and cells were treated with 0 (−), 50, 100, or 150 μg/ml thermolysin at 37°C for 5 min. Cells were then washed to remove residual protease and unbound virus, and viral infectivity was determined at 16 h post infection. Asterisks indicate values below the limit of detection. Results (n=4) are from two independent experiments. Error bars indicate s.d.
We then examined the capacity of CT43 cells expressing domain C-Flag or domain C-Flagtailless to support filovirus GP-dependent entry. We found that cleaved rVSV-GPCL-EBOV could infect both cell types, but remarkably, uncleaved rVSV-GP-EBOV could infect only the domain C-Flag cells (Figure 8C). Similar results were obtained with uncleaved rVSV-GP-MARV (Figure 8C). However, we were unable to test if precleaved rVSV-GPCL-MARV could infect domain C-Flagtailless cells, because MARV GP is highly unstable to in-vitro proteolysis and viral particles preincubated with protease rapidly lose their infectivity (Wong and Chandran, in preparation). In an attempt to circumvent this obstacle, we first bound rVSV-GP-MARV to cells expressing domain C-Flag or domain C-Flagtailless at 4°C, and then briefly subjected the cells to protease at 37°C (Figure 8D). This in-situ protease treatment reduced MARV GP-dependent infection in cells expressing domain C-Flag; however, it mediated a substantial, dose-dependent increase in infection of cells expressing domain C-Flagtailless (Figure 8D). Therefore, cleaved forms of EBOV and MARV GP, whether stably generated by in-vitro preincubation with protease or transiently generated by in-situ protease treatment, can utilize a synthetic cell-surface receptor for entry, whereas uncleaved EBOV and MARV GP cannot. These observations provide direct evidence for a physiological GP–NPC1 interaction and demonstrate that this interaction normally occurs within the endosomal/lysosomal pathway after GP cleavage (Figure 9).
Figure 9.
A model for the role of NPC1 in Ebola virus and Marburg virus entry. Virus particles adhere to the plasma membrane by binding to one or more cell attachment factors. They are then trafficked to early/late endosomal (EE/LE) compartments where host cysteine proteases, including cathepsin B (Chandran et al, 2005), cleave GP to remove the mucin and glycan cap domains and expose the NPC1-binding site. This cleaved GP species, which resembles in vitro-cleaved GPCL, can then bind to NPC1's domain C, presumably on the limiting membrane of an LE or lysosomal (Lys) compartment. GP–NPC1 interaction may play a direct role in triggering GP conformational changes that bring about viral membrane fusion and cytoplasmic release of the viral nucleocapsid payload. The molecular requirements for, and intracellular location of, this final step in filovirus entry remain to be defined.
Discussion
In this study, we show that the NPC1 protein is a critical receptor for cell entry by filoviruses. Despite the large size, complexity, and hydrophobicity of NPC1, we have demonstrated that this ∼1300 residue, 13-pass transmembrane protein binds specifically and directly to its viral GP ligand. Unlike NPC1's housekeeping function in lysosomal cholesterol transport, which requires all three of its major luminal loop domains, we show that NPC1's function as a filovirus receptor absolutely requires only its second luminal loop domain (C), which directly engages filovirus GP. Remarkably, a synthetic membrane protein comprising NPC1 domain C appended to a single transmembrane domain constitutes a receptor for filovirus GP-mediated infection.
While domain C plays a central and minimally sufficient role, our failure to fully rescue infection with only this domain implies indirect or direct contributions from other NPC1 sequences. These sequences may facilitate the optimal folding and endosomal/lysosomal delivery of domain C (indirect), contact GP during entry (direct), or participate in putative NPC1-dependent steps in entry downstream of GP–NPC1 binding (direct).
Genetic evidence indicates that cholesterol efflux from endosomal/lysosomal compartments requires both NPC1 and a small lysosomal cholesterol-binding protein, NPC2 (Naureckiene et al, 2000; Sleat et al, 2004). Previous biochemical studies suggested that transfer of cholesterol from NPC2 to NPC1 is a crucial part of the efflux mechanism, but direct evidence for an interaction between the proteins was lacking (Infante et al, 2008b; Kwon et al, 2009). Recently, however, two of us showed that a purified, soluble form of domain C could bind directly to cholesterol-loaded NPC2 in vitro at acid pH but not at neutral pH (Deffieu and Pfeffer, 2011). These results lend support to a model in which docking of NPC2 to NPC1 domain C facilitates transfer of cholesterol from NPC2 to NPC1's cholesterol-binding domain A. Here we show that, in contrast to its interaction with its cellular ligand NPC2, NPC1 domain C binds to its viral ligand, GP, in a manner that requires neither acid pH nor cholesterol. Whether NPC2 and GP contact the same or different surfaces of domain C remains to be seen.
Using a receptor engineering approach, we demonstrate that the use of NPC1 by filovirus GP exemplifies a novel type of virus–receptor interaction: while productive GP–NPC1 interaction can be contrived to take place at the plasma membrane, NPC1 normally binds to viral particles within the endosomal/lysosomal pathway and not at the cell surface (Figure 9). Our in vitro binding experiments provide an explanation for this result. Purified forms of full-length NPC1 and soluble domain C directly bind not to intact GP but to GPCL, which resembles a cleaved entry intermediate generated by the action of host proteases within endosomes (Chandran et al, 2005; Schornberg et al, 2006). These findings account for and extend recent observations that only EBOV GPCL could bind to endosomal membranes containing NPC1 and directly or indirectly co-precipitate NPC1 from membrane extracts (Côté et al, 2011). We delineate the NPC1-binding site within EBOV GP, and show why only cleaved GP can recognize NPC1: the binding site is occluded by heavily glycosylated C-terminal GP1 sequences and becomes fully unmasked only after these sequences have been proteolytically removed. Finally, we provide the first evidence that MARV GP, despite its instability to proteolysis, must nevertheless undergo intracellular cleavage to a species that can engage NPC1 within the endosomal/lysosomal pathway. We speculate that programmed exposure of the NPC1-binding site within intracellular compartments inaccessible to antibodies represents a strategy by which filoviruses evade the humoral immune response in their natural hosts.
Our findings imply that filoviruses must use host molecules other than NPC1 to attach to the cell surface and internalize into endosomes, and indeed, several have been implicated in previous work. Multiple C-type lectins (Alvarez et al, 2002; Simmons et al, 2003) and the T-cell immunoglobulin and mucin-1 protein (Kondratowicz et al, 2011) bind to uncleaved GP and enhance virus-cell attachment in antigen-presenting and epithelial cells, respectively. The Tyro3 receptor tyrosine kinases enhance viral internalization in some cells (Shimojima et al, 2006; Hunt et al, 2011). Because mutations within the GP RBS tested in this study drastically diminish NPC1 binding but have only a modest effect on virus-cell attachment, it appears likely that other, more surface-exposed, sequences in uncleaved GP are primarily responsible for the initial events at the plasma membrane. Regardless of which cell-surface factors mediate viral attachment and internalization, our findings argue that GP in internalized virus must first undergo cleavage by endosomal cysteine proteases and then engage NPC1 where it resides, at the limiting membrane of a late endosomal/lysosomal compartment (Figure 9).
Our results indicate that GP–NPC1 interaction is required at a late step in entry proximal to viral release into the cytoplasm, and they are fully consistent with our previous and current findings that viral particles accumulate in the lysosomes of cells lacking a suitable NPC1 protein (Carette et al, 2011; Supplementary Figure S2). Interestingly, GP1 residues that are necessary for NPC1 binding and that contact GP2 in the trimeric viral spike are closely apposed (Supplementary Figure S9). We thus propose that NPC1 plays a role in triggering GP1–GP2 dissociation, setting in motion the GP2 conformational rearrangements that bring about viral membrane fusion and cytoplasmic escape (Figure 9; Supplementary Figure S1). However, we found that an acid-buffer wash did not allow infection of rVSV-GPCL at the plasma membrane of cells expressing the synthetic domain C-Flagtailless receptor, suggesting that GP–NPC1 binding and acid pH are insufficient for productive membrane fusion and implicating one or more additional endosomal/lysosomal host factors. The requirement for an intracellular receptor revealed in this study, and the possible requirement for additional endosomal trigger factor(s) (Brecher et al, 2012; Schornberg et al, 2006; Wong et al, 2010; this study), at least partially explain the longstanding failure to reconstitute high levels of filovirus GP-catalysed membrane fusion in vitro or at the cell surface (Takada et al, 1997; Bar et al, 2006).
The capacity of human NPC1 to extend the species tropism of filoviruses to reptilian cells strongly supports our claim that it is a filovirus receptor. Because NPC1 is highly conserved among animals (Loftus et al, 1997), and is indispensable for filovirus infection of diverse vertebrate cell types (Carette et al, 2011; Côté et al, 2011; this study) and for viral pathogenesis in mouse models (Carette et al, 2011), it is likely that NPC1 is broadly, if not universally, required in this role. Moreover, a phylogenetically diverse group of filoviruses requires NPC1 for entry and infection (Carette et al, 2011; Côté et al, 2011), and GP1 residues we have shown to be important for NPC1 interaction are highly conserved among filoviruses (Kuhn et al, 2006), suggesting that all known members of the family Filoviridae utilize NPC1 as a receptor. It will therefore be of considerable interest to determine if incompatibilities between GPs and specific NPC1 orthologues during viral entry limit filovirus host range in a natural setting and impose barriers to viral interspecies transmission and zoonotic disease.
Materials and methods
Viruses and infections
Recombinant VSVs (serotype Indiana) expressing eGFP, and EBOV or MARV GP in place of VSV G (rVSV-GP-EBOV/MARV) were recovered and amplified as described previously (Whelan et al, 1995; Wong et al, 2010). The EBOV and MARV GP genes encoded by these viruses were derived from the Mayinga and Musoke isolates, respectively (Genbank accession numbers NP_066246 and YP_001531156). VSV pseudotypes bearing GPs derived from VSV, EBOV, SUDV, and MARV were generated as described previously (Takada et al, 1997). The WT filoviruses EBOV-Zaire 1995 and MARV-Ci67 used in this study have been described previously (Jahrling et al, 1999; Swenson et al, 2008). VSV particles containing GPCL were generated by incubating rVSV-GP-EBOV with thermolysin (200 μg/ml) for 1 h at 37°C. The protease was inactivated by addition of phosphoramidon (1 mM), and reaction mixtures were used immediately. Infectivities of VSV pseudotypes were measured by manual counting of eGFP-positive cells using fluorescence microscopy at 16–24 h post infection, as described (Chandran et al, 2005). Infectivities of rVSVs were measured by fluorescent-focus assay, as described (Wong et al, 2010). Alternatively, NH4Cl (20 mM) was added to infected cell cultures at 2 h post infection to block viral spread, and individual eGFP-positive cells were manually counted at 12–14 h post infection. Neutralization of uncleaved and cleaved rVSV-GP-EBOV by rVSV-domain C in Figure 3C was measured by quantitation of virus-encoded eGFP expression in cell monolayers with a Typhoon 9400 imager (GE Healthcare).
GP–NPC1 co-IP assays
Protein G-coated magnetic beads (20 μl/reaction; Spherotech) were incubated with the GP-specific monoclonal antibody KZ52 (5 μg) (Maruyama et al, 1999) for 1 h, washed to remove unbound antibody, and then added to uncleaved or in vitro-cleaved rVSV-GP-EBOV or VSV-GP-EBOV particles (5 μl concentrated virus; 107–108 infectious units), or to purified EBOV GPΔTM (9 μg) in NTE-CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) buffer (10 mM Tris-Cl (pH 7.5), 140 mM NaCl, 1 mM EDTA, 0.5% vol/vol CHAPS). Bead-virus mixtures were incubated for 2 h at room temperature, and then added to crude detergent extracts of CHO CT43 cells expressing a Flag-tagged NPC1 protein (NPC1–Flag) (2 × 106 cell equivalents in 150 μl; prepared as described in Supplementary Materials and methods), or to purified, soluble NPC1 domain C (5 μg/ml). After overnight incubation with mixing at 4°C, beads were retrieved with a magnet, extensively washed with NTE-CHAPS, and heated in Laemmli sample buffer to elute bound proteins. Solubilized proteins were subjected to SDS–polyacrylamide gel electrophoresis, and NPC1 and GP were detected by immunoblotting with anti-Flag (Sigma-Aldrich) and anti-GP1 (Wong et al, 2010) antibodies, respectively. Typically, 50–100% of each pellet sample and 5–10% of each supernatant sample were loaded on gels. Reciprocal co-IPs were carried out essentially as above, except that Flag-tagged proteins in CT43 cell extracts were first captured onto Flag antibody-coated magnetic beads (20 μl/reaction; Sigma), and the beads were then incubated with uncleaved or cleaved rVSV-GP-EBOV particles in NTE-CHAPS buffer.
GP–NPC1 capture ELISA
Ninety-six-well high-binding ELISA plates (Corning) were coated with the GP-specific monoclonal antibody KZ52 (2 μg/ml in PBS), and then blocked with PBS containing 3% bovine serum albumin and 0.5% CHAPS (PBSA-CHAPS). Uncleaved or in vitro-cleaved rVSV-GP or VSV-GP particles solubilized in PBSA-CHAPS buffer were added to the blocked plates, and GP capture was allowed to proceed for 1 h at 37°C. After washing to remove unbound GP, serial dilutions of NPC1–Flag partially purified from CT43 cells (0–100 ng/well), crude detergent extracts of 293T cells expressing Flag-tagged NPC1 or NPC1L1 proteins (0–2 × 105 cell equivalents), or purified, soluble domain C (0–40 μg/ml) were added to the wells. After an overnight incubation at 4°C, plates were extensively washed, and bound Flag-tagged proteins were detected with an anti-Flag antibody-horseradish peroxidase conjugate and Ultra-TMB substrate (Thermo).
Protease-mediated infection by rVSV-GP-MARV
rVSV-GP-MARV was centrifuged onto monolayers of CT43 cells expressing domain C-Flag or domain C-Flagtailless cells at 150 g at 4°C for 45 min. Cells were removed onto ice and the growth medium was replaced with prewarmed PBS containing increasing concentrations of thermolysin (0–150 μg/ml) at 37°C for 5 min. Protease and unbound virus were removed by washing with growth medium, and cells were then incubated at 28°C. Viral infectivity was scored 16–18 h later by enumerating eGFP-positive cells.
Supplementary Material
Acknowledgments
We thank Margaret C Kielian, Max L Nibert, Vinayaka R Prasad, Steven U Walkley, and Anthony C Wong for critical reading of the manuscript and valuable advice; and T-Y Chang, DR Burton, and T Kirchhausen for their generous gifts of the WT and NPC1-null CHO cells, KZ52 antibody, and a plasmid encoding LAMP1-eGFP, respectively. This research was supported by NIH grants R01 AI088027 (to KC), AI081842 and U54 AI057159 (NERCE-BEID) (to SPW), and R21 HG004938 (to TRB); by the DTRA project, #CBM.VAXPLAT.05.10.RD.005 (to JMD); and by the Ara Parseghian Medical Research Foundation (to SRP). EHM was additionally supported by NIH-funded training program T32 GM007288 at the Albert Einstein College of Medicine. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Army.
Author contributions: EHM, KC, TRB, SPW, and JD conceived the study and wrote the paper. EHM, EN, and KC devised and implemented the GP–NPC1 binding assays. EHM, GO, JEC, and KC designed mutant forms of NPC1 and generated stable cell lines expressing them. EHM, MR, and KC carried out the filovirus entry and infection studies with VSV pseudotypes and recombinants. GO and TRB generated the viper cell lines and NPC1 loop-deletion mutants. AK generated the cell line expressing NPC1L1. MR created and tested the rVSV-loop C virus, and carried out studies of viral entry and protein localization using confocal fluorescence and electron microscopy. MD and SRP designed and produced purified soluble domain C. ASH and JMD generated purified GPΔTM. ASH, AIK, GR, and JMD performed experiments with the wild-type agents.
Footnotes
Brummelkamp, Carette, Chandran, Raaben, and Whelan are co-inventors on a patent application that describes NPC1 as a target for development of antiviral therapies against filoviruses.
References
- Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R (2002) C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 76: 6841–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar S, Takada A, Kawaoka Y, Alizon M (2006) Detection of cell-cell fusion mediated by Ebola virus glycoproteins. J Virol 80: 2815–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Spiess M, Klenk HD (1995) The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J Gen Virol 76(Part 2): 393–399 [DOI] [PubMed] [Google Scholar]
- Berger AC, Salazar G, Styers ML, Newell-Litwa KA, Werner E, Maue RA, Corbett AH, Faundez V (2007) The subcellular localization of the Niemann-Pick Type C proteins depends on the adaptor complex AP-3. J Cell Sci 120: 3640–3652 [DOI] [PubMed] [Google Scholar]
- Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM (2012) Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion relevant conformational change. J Virol 86: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley MA, Hughes L, Ruiz A, McCray PB Jr, Sanchez A, Sanders DA, Maury W (2007) Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J Virol 81: 7702–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477: 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie EJ et al. (1997) Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277: 228–231 [DOI] [PubMed] [Google Scholar]
- Chan SY, Empig CJ, Welte FJ, Speck RF, Schmaljohn A, Kreisberg JF, Goldsmith MA (2001) Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 106: 117–126 [DOI] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM (2005) Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308: 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté M, Misasi J, Ren T, Bruchez A, Lee K, Chandran K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J (2011) Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477: 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Sugii S, Yu C, Chang TY (2000) Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J Biol Chem 275: 4013–4021 [DOI] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA (2000) Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem 275: 24367–24374 [DOI] [PubMed] [Google Scholar]
- Davies JP, Levy B, Ioannou YA (2000) Evidence for a Niemann-pick C (NPC) gene family: identification and characterization of NPC1L1. Genomics 65: 137–145 [DOI] [PubMed] [Google Scholar]
- Deffieu M, Pfeffer SR (2011) Niemann-Pick type C1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA 108: 18932–18936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM (2009) The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol 83: 2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D, Schornberg KL, Shoemaker CJ, Delos SE, Stantchev TS, Clouse KA, Broder CC, White JM (2010) Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci USA 107: 16637–16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ (2010) Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol 84: 2972–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CL, Kolokoltsov AA, Davey RA, Maury W (2011) The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J Virol 85: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante RE, Radhakrishnan A, Abi-Mosleh L, Kinch LN, Wang ML, Grishin NV, Goldstein JL, Brown MS (2008a) Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem 283: 1064–1075 [DOI] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL (2008b) NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA 105: 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou YA (2000) The structure and function of the Niemann-Pick C1 protein. Mol Genet Metab 71: 175–181 [DOI] [PubMed] [Google Scholar]
- Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y (1999) Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol 73: 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling PB, Geisbert TW, Geisbert JB, Swearengen JR, Bray M, Jaax NK, Huggins JW, LeDuc JW, Peters CJ (1999) Evaluation of immune globulin and recombinant interferon-alpha2b for treatment of experimental Ebola virus infections. J Infect Dis 179(Suppl 1): S224–S234 [DOI] [PubMed] [Google Scholar]
- Kaletsky RL, Simmons G, Bates P (2007) Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol 81: 13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB Jr, Chiorini J, Maury W. (2011) From the cover: T-cell immunoglobulin, mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA 108: 8426–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB (2010) Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol 155: 2083–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M (2006) Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem 281: 15951–15958 [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE (2009) Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454: 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L, Faust JR (1989) The intracellular transport of low density lipoprotein-derived cholesterol is inhibited in Chinese hamster ovary cells cultured with 3-beta-[2-(diethylamino)ethoxy]androst-5-en-17-one. J Biol Chem 264: 11796–11806 [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, Pentchev PG, Pavan WJ (1997) Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277: 232–235 [DOI] [PubMed] [Google Scholar]
- Manicassamy B, Wang J, Jiang H, Rong L (2005) Comprehensive analysis of ebola virus GP1 in viral entry. J Virol 79: 4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR (1999) Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol 73: 6024–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y (2010) Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 6: e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P (2000) Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290: 2298–2301 [DOI] [PubMed] [Google Scholar]
- Ory DS (2004) The Niemann-Pick disease genes; regulators of cellular cholesterol homeostasis. Trends Cardiovasc Med 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Ou W, King H, Delisle J, Shi D, Wilson CA (2010) Phenylalanines at positions 88 and 159 of Ebolavirus envelope glycoprotein differentially impact envelope function. Virology 396: 135–142 [DOI] [PubMed] [Google Scholar]
- Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea E, Neufeld EB, Branchette-Mackie JE, Pentchev P (2001) Niemann-Pick disease type C: a lipid trafficking disorder. In The Metabolic and Molecular Bases of Inherited Disease, , Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds), pp 3611–3634. New York: McGraw-Hill [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA (2010) Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 6: e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J (2006) Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80: 4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C, Higgins ME, Davies JP, Ioannou YA (2004) Targeting of NPC1 to late endosomes involves multiple signals, including one residing within the putative sterol-sensing domain. J Biol Chem 279: 48214–48223 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y (2006) Tyro3 family-mediated cell entry of ebola and marburg viruses. J Virol 80: 10109–10116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pöhlmann S (2003) DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305: 115–123 [DOI] [PubMed] [Google Scholar]
- Sleat DE, Wiseman JA, El-Banna M, Price SM, Verot L, Shen MM, Tint GS, Vanier MT, Walkley SU, Lobel P (2004) Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc Natl Acad Sci USA 101: 5886–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson DL, Warfield KL, Larsen T, Alves DA, Coberley SS, Bavari S (2008) Monovalent virus-like particle vaccine protects guinea pigs and nonhuman primates against infection with multiple Marburg viruses. Expert Rev Vaccines 7: 417–429 [DOI] [PubMed] [Google Scholar]
- Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y (1997) A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA 94: 14764–14769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Groen G, Webb P, Johnson K, Lange JV, Lindsay H, Eliott L (1978) Growth of Lassa and Ebola viruses in different cell lines. In Ebola Virus Haemorrhagic Fever, Pattyn SR (ed), pp 172–176. Amsterdam: Elsevier/North-Holland Biomedical Press [Google Scholar]
- Walkley SU, Suzuki K (2004) Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim Biophys Acta 1685: 48–62 [DOI] [PubMed] [Google Scholar]
- Wang J, Chu BB, Ge L, Li BL, Yan Y, Song BL (2009) Membrane topology of human NPC1L1, a key protein in enterohepatic cholesterol absorption. J Lipid Res 50: 1653–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN, Wertz GT (1995) Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA 92: 8388–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K (2008) Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43: 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K (2010) A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J Virol 84: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool-Lewis RJ, Bates P (1998) Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol 72: 3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Delgado R, Xu L, Todd RF, Nabel EG, Sanchez A, Nabel GJ (1998) Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279: 1034–1037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.