Abstract
Invariant Natural Killer T (iNKT) cells use highly restricted αβ T cell receptors (TCRs) to probe the repertoire of lipids presented by CD1d molecules. Here, we describe our studies of lysophosphatidylcholine (LPC) presentation by human CD1d and its recognition by a native, LPC-specific iNKT TCR. Human CD1d presenting LPC adopts an altered conformation from that of CD1d presenting glycolipid antigens, with a shifted α1 helix resulting in an open A’ pocket. Binding of the iNKT TCR requires a 7-Å displacement of the LPC headgroup but stabilizes the CD1d–LPC complex in a closed conformation. The iNKT TCR CDR loop footprint on CD1d–LPC is anchored by the conserved positioning of the CDR3α loop, whereas the remaining CDR loops are shifted, due in part to amino-acid differences in the CDR3β and Jβ segment used by this iNKT TCR. These findings provide insight into how lysophospholipids are presented by human CD1d molecules and how this complex is recognized by some, but not all, human iNKT cells.
Keywords: CD1d, iNKT TCR, lysophospholipid, structure
Introduction
Invariant Natural Killer T (iNKT) cells are a specialized lineage of αβ T cells that recognize the major histocompatibility complex (MHC) class I-like molecule, CD1d (Bendelac et al, 2007). Unlike conventional αβ T cells restricted to classical class I and II MHC molecules presenting peptides, iNKT cells have an innate-like phenotype, which allows them to respond quickly and potently to an appropriate antigenic stimulus. iNKT cells are known to be able to secrete a variety of different cytokines, including both Th1 and Th2 types (Venkataswamy and Porcelli, 2010) and can respond to CD1d presenting lipids from both exogenous and endogenous sources. The strength of the antigenic signal delivered through the iNKT TCR and the presence of other cytokines (i.e., IL-12; Brigl et al, 2003) during iNKT cell stimulation have been shown to be key factors influencing the cytokines produced by iNKT cells (Wang et al, 2008; Bricard et al, 2010; Brigl et al, 2011). Much of the work on iNKT cell recognition has been focussed on bacterial glycolipids, however, there is increasing evidence that recognition of endogenous antigens, or iNKT cell autoreactivity, contributes to their function in the immune response and some of those selfantigens have been characterized (Zhou et al, 2004b; Paget et al, 2007; Salio et al, 2007; Brennan et al, 2011). As iNKT cells are implicated in a range of infectious and cancerous diseases (Bendelac et al, 2007), understanding how these cells are tuned by endogenous antigenic stimuli is becoming an important focus of the NKT cell field.
iNKT cells generally express a restricted repertoire of T-cell receptors (TCRs), in humans using the Vα24 and Vβ11 subunits while mice use Vα14 and Vβ8, Vβ7 or Vβ2 in rearranging their TCRs. The consistent use of the Jα18 segment in rearrangement of the α chain in both species is one of the defining aspects of the iNKT TCR, as the residues encoded within this segment are critical for iNKT cell recognition of CD1d (Scott-Browne et al, 2007; Wun et al, 2008). The only variable components of iNKT TCRs is the CDR3β loop, encoded by the V-D-J gene segment junction, and the Jβ segment used in this rearrangement. This loop does not appear to have a conserved sequence motif in the iNKT cell population, indeed variation in this loop has been shown to modulate iNKT cell reactivity for certain lipid antigens (Mallevaey et al, 2009; Matulis et al, 2010). iNKT TCRs engineered for high affinity for CD1d show dependence on residues in the CDR3β (Mallevaey et al, 2009, 2011) and mutagenic data suggest that the CDR3β contributes to ligand specificity in the mouse (Scott-Browne et al, 2007; Florence et al, 2009).
Most of the lipids identified as antigens for iNKT cells are glycolipids and these derive from both exogenous and endogenous sources. In particular, iNKT cells characteristically share an ability to recognize an unusual foreign glycolipid called α-galactosylceramide (αGalCer or KRN7000) (Kobayashi et al, 1995). Structural efforts have demonstrated why the α-glycosidic linkage is a key feature of αGalCer's potency (Borg et al, 2007; Pellicci et al, 2009), as well as the positioning of the hydroxyl groups in the galactose headgroup (Scott-Browne et al, 2007; Wun et al, 2008). Key contacts with the CDR1α and CDR3α loops of the iNKT TCR also explain the restricted usage of the Vα24 (human) and Vα14 (mouse) variable genes and the Jα18 gene segment in iNKT TCR rearrangement (Borg et al, 2007; Scott-Browne et al, 2007; Wun et al, 2008; Pellicci et al, 2009). Several other reports using native iNKT TCRs or iNKT TCRs engineered for high affinity to study recognition of other glycolipid antigens, both of the α and β linkage and with varying sugar headgroups and lipid chains, have shown that despite the variation in the chemistry and structure of these ligands, these iNKT TCRs adopt a highly conserved docking orientation on the CD1d molecule, requiring the lipid antigen to bend or shift to accommodate this conserved architecture (Li et al, 2010; Sullivan et al, 2010; Wang et al, 2010; Aspeslagh et al, 2011; Kinjo et al, 2011; Pellicci et al, 2011; Wun et al, 2011; Yu et al, 2011).
Recently, we have discovered that some, but not all, human iNKT cells are reactive to a specific lysophospholipid, lysophosphatidylcholine (LPC) (Fox et al, 2009), one of the endogenous lipid ligands eluted from human CD1d molecules (Cox et al, 2009; Yuan et al, 2009). Generated through enzymatic cleavage via phospholipase A2 (PLA2) (Schmitz and Ruebsaamen, 2009) or through oxidation of diacylated lipid species such as plasmalogens, LPC is present constitutively within cells and in many extracellular environments. Notably, LPC levels increase markedly during inflammation, and are present at high concentrations in oxidized Low Density Lipoproteins (oxLDLs; Croset et al, 2000), elevated in the serum of multiple myeloma patients (Sasagawa et al, 1999) and have also been found to be generated at high quantities during atherosclerotic plaque formation (Portman and Alexander, 1969; Thukkani et al, 2003). Due to the potential immunological importance of lysophospholipids and because structural information is lacking for how lysophospholipids are presented by CD1 molecules and limited in general for human iNKT TCRs/CD1d complexes (Borg et al, 2007; Pellicci et al, 2009), we sought to investigate the molecular basis for lysophospholipid presentation by CD1d and its recognition by iNKT TCRs. To this end, we present structures of CD1d complexed with LPC, the unliganded LPC-reactive iNKT TCR and the complex of this LPC-reactive NKT TCR with CD1d–LPC. Our structures provide molecular evidence that human iNKT TCR recognition of LPC requires substantial structural shifts in CD1d and the positioning of the LPC headgroup. We also find that LPC specificity of this iNKT TCR is due to subtle shifts in docking of the iNKT TCR established through contributions of the amino-acid diverse CDR3β and use of a particular Jβ segment that shifts the positioning of the Vβ in relation to the Vα, Cα and Cβ of the TCR.
Results
LPC presentation by CD1d
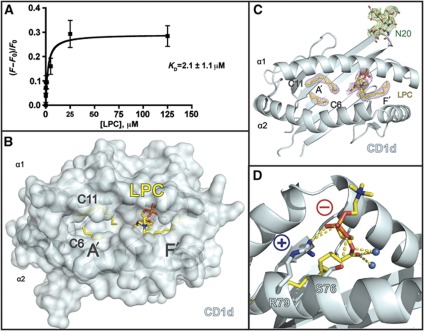
We have shown previously via lipid competition assays that the lysophospholipids LPC and LPE (lysophosphatidylethanolamine) are bound by recombinant CD1d molecules (Fox et al, 2009). In order to quantitate LPC's interaction with CD1d, we performed tryptophan fluorescence titration spectroscopy, a technique that does not require alteration of the protein or lipid ligand to measure association. Recombinant, fully glycosylated CD1d expressed in insect cells was titrated with solubilized LPC and the intrinsic tryptophan fluorescence change was measured and used to calculate the apparent equilibrium binding constant (KD) (Figure 1A), a value dependent on the solubility of the lipid (the critical micellar concentration or CMC), its ability to displace lipids already bound in the insect expressed CD1d, and finally its interaction with CD1d. From our measurements, LPC has an apparent KD of ∼2.1 μM, which is on the lower end of lipid ligand affinities for CD1 molecules (Ernst et al, 1998; Naidenko et al, 1999; Cantu et al, 2003; Scharf et al, 2010). Because LPC is highly soluble, this KD is likely more representative of the low affinity of LPC for CD1d due to its single acyl chain anchor. For comparison, we have also used this technique to calculate the apparent affinity of αGalCer (Supplementary Figure S1A), producing an affinity for CD1d of ∼0.11 μM, similar to the affinity of αGalCer for mouse CD1d calculated by SPR (∼0.34 μM) (Naidenko et al, 1999).
Figure 1.
LPC presentation by human CD1d. (A) The change in intrinsic tryptophan fluorescence of CD1d as a function of titrated amounts of LPC. The increase of tryptophan fluorescence with increasing amounts of LPC was expressed as a ratio (F−Fo)/Fo, the change in fluorescence divided by the control fluorescence in the absence of added LPC. The data were fit to a one-site binding model to determine the apparent KD of this interaction. (B) Semitransparent surface and ribbon diagram of the structure of human CD1d (cyan) presenting LPC (yellow). Two spacer lipids identified in the electron density, C11 and C6 and the location of the A’ and F’ pockets are also shown. (C) Electron density of lysophosphatidylcholine (LPC) and the N-linked glycosylation in the CD1d–LPC structure. CD1d is shown from above as a ribbon diagram in cyan and 2Fo−Fc electron density maps contoured at 1α around the LPC ligand, C11 and C6 is coloured in pink, and around the glycosylation at N20 is coloured in green. (D) Phosphorylcholine headgroup coordination of LPC by CD1d. Main chain of CD1d is shown as cyan ribbon, whereas side chains mediating the coordination are shown as stick. Atoms are coloured as follows: blue=nitrogen, red=oxygen, orange=phosphorus and yellow=carbon. Two hydrogen bonded water molecules are shown as spheres coloured in marine. The positive and negative charges of the Arg79 and the phosphate moiety are shown as ‘+’ and ‘−’, respectively. Hydrogen bonds are shown as dashed yellow lines: highly likely H-bonds (>3.3 Å) are shown with heavy dashed lines, probable H-bonds (4 Å> but >3.3 Å) are shown as light dashed lines.
The three-dimensional structure of CD1d presenting LPC
To first understand how human CD1d molecules present LPC, we used glycosylation minimization strategies on insect expressed, human CD1d to generate crystals of CD1d/LPC that diffracted to 2.8 Å resolution (see Table I for crystallization and refinement statistics). There were three molecules in the asymmetric unit that reflected the same topology with small root mean squared deviations (RMSDs) from molecule 1 (0.755 for molecule 2, chains C and D, and 0.802 for molecule 3, chains E and F). For simplicity, molecule 1 (chains A and B) will be discussed for the remainder of the manuscript. A semitransparent surface representation of this structure of CD1d with a ribbon representation showing the location of the α1 and α2 helices is shown in Figure 1B. LPC is shown in yellow, as are two spacer hydrocarbon chains which were identified in the A’ pocket, named C11 and C6. Electron density for LPC was unambiguous (Figure 1C; Supplementary Figure S1B) and positioned the sn1 chain of LPC in the F’ pocket. The phosphorylcholine headgroup of LPC is positioned high and outside of the lipid binding groove of CD1d, coordinated by a salt bridge between the positively charged Arg79 of the CD1d α1 helix and the negative charge of the phosphate moiety (Figure 1D). Hydrogen bonds are also evident between the Oγ of the adjacent Ser76 CD1d residue and a phosphate oxygen and the Nη2 of Arg79's guanidinium group and two of the phosphate oxygens. Two water molecules, hydrogen bonded by the ester oxygens of LPC are also evident in our electron density and are represented as blue spheres in Figure 1D.
Table 1. Data collection and refinement statistics (molecular replacement).
| J24.L17 | CD1d–LPC | J24.L17–CD1d–LPC | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | I222 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 64.60, 65.76, 66.99 | 108.61, 127.19, 332.64 | 66.59, 117.61, 190.66 |
| α, β, γ (deg) | 103.89, 106.32, 100.84 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 20–2.8 (2.85–2.80) | 46–2.8 (2.85–2.80) | 20.02–3.06 (3.10–3.05) |
| Rsym | 0.073 (0.272) | 0.124 (0.748) | 0.108 (0.732) |
| I/σI | 10.7 (2.5) | 13.2 (1.1) | 10.9 (1.7) |
| Completeness (%) | 97.4 (89.4) | 97.5 (78.1) | 93.7 (92.0) |
| Redundancy | 1.1 (1.1) | 2.1 (1.5) | 10.9 (1.7) |
| Refinement | |||
| Resolution (Å) | 2.80 | 2.80 | 3.05 |
| No. of reflections | 42 643 | 52 389 | 25 892 |
| Rwork/Rfree | 0.22 (0.32)/0.27 (0.43) | 0.22 (0.34)/0.26 (0.41) | 0.22 (0.30)/0.29 (0.39) |
| No. of atoms | |||
| Protein | 6920 | 8571 | 9438 |
| Ligand/ion | 6 | 353 | 83 |
| Water | 36 | 123 | 15 |
| B factors | |||
| Protein | 58.9 | 67.6 | 62.6 |
| Ligand/ion | 48.7 | 93.0 | 90.6 |
| Water | 33.8 | 49.5 | 41.8 |
| RMSDs | |||
| Bond lengths (Å) | 0.003 | 0.0116 | 0.010 |
| Bond angles (deg) | 0.776 | 1.174 | 0.884 |
| Values in parentheses are for highest resolution shell. | |||
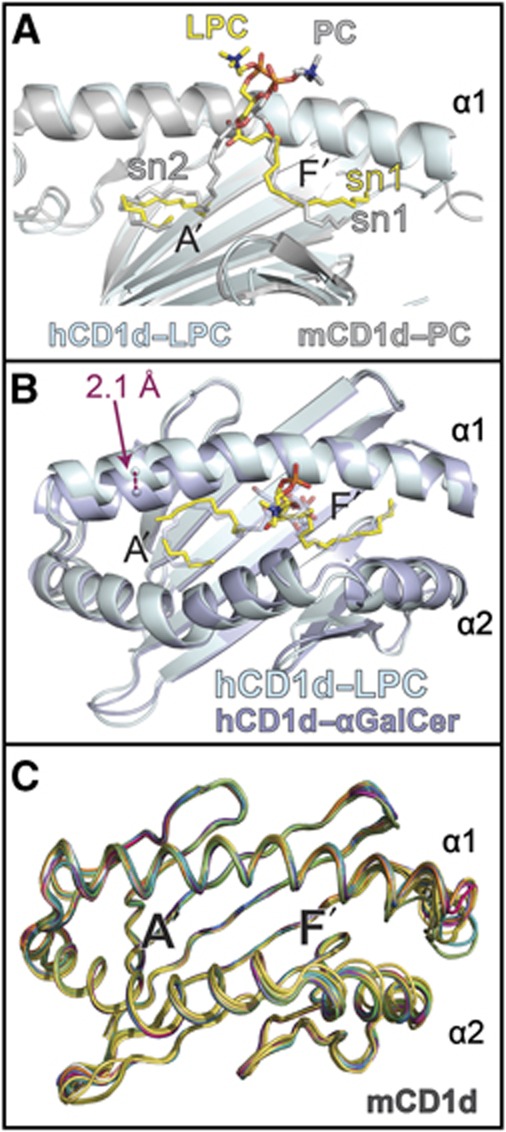
This coordination of the phosphorylcholine headgroup is similar to that seen in the structure of murine CD1d presenting PC (Giabbai et al, 2005), indeed, superposition of our human CD1d–LPC structure with that of mouse CD1d–PC shows the orientation of the sn1 acyl chain and phosphate moiety are similarly positioned between the two structures (Figure 2A). Also, both Ser 76 and Arg79 of mouse CD1d play prominent roles in coordination of the phosphate moiety of PC, forming both hydrogen-bonding and electrostatic interactions (Giabbai et al, 2005). Comparison of our human CD1d–LPC structure with that of the only other human CD1d structure, that with αGalCer (Koch et al, 2005), shows a distinct conformational change in the α1 helix (Figure 2B). When presenting LPC, the CD1d α1 helix shifts ∼2.1 Å from the α1 helical position of the CD1d–αGalCer structure, disrupting the A’ roof by increasing the distance between Leu66 and T165 by 2.1 Å and Ile69 and Leu161 by 1.8 Å (as measured from the Cδ1- and Cδ2-terminal carbons) and opening up the A’ pocket to solvent. A shift similar in nature was also noted in the unloaded human CD1d structure co-crystallized with the CD1d–αGalCer structure (Koch et al, 2005). Such an extreme shift has not be noted in any of the mouse CD1d structures solved thus far, as shown through superposition of the 16 published, unliganded, mCD1d heavy chains presenting diverse lipid antigens (Figure 2C). However, flexibility of the α1 and α2 helical distances composing the A’ roof has been noted in some mouse iNKT TCR liganded structures as a result of ligand accommodation (Aspeslagh et al, 2011). Variation in the α-helical distances forming the F’ pocket are very apparent in mouse CD1d (Figure 2C).
Figure 2.
Comparison of human CD1d-LPC with other CD1d structures. (A) Human CD1d–LPC is shown in cyan compared with mouse CD1d presenting PC (Giabbai et al, 2005) (PDB ID: 1ZHN), shown in grey. Structures were aligned via the main-chain CA carbons of the heavy chains without ligand included. LPC in yellow and PC from the mouse structure in grey; the α2 helix has been omitted for clarity are shown. The sn1 and sn2 chains of the lipids are shown where present. Atoms are coloured as in Figure 1. (B) Human CD1d–LPC (cyan–yellow) is aligned with human CD1d–αGalCer shown in lavender (PDB ID: 1ZT4) via their main-chain CA carbons of the heavy chains. The ∼2.1-Å shift in the CD1d–LPC α1 helix main chain relative to CD1d–αGalCer structure as well as the relative positioning of the bound lipid ligands is shown. (C) Superposition of published mouse CD1d α1 and α2 platform domains aligned through the graphics program Pymol. CD1d structures are shown as cartoon loop format and are derived from the following PDB IDs: 3MA7, EILP, 3ILQ, 3GO8, 3GML, 3GMN, 3GMO, 3GMP, 3GMQ, 3GMR, 2Q7Y, 2GAZ, 2FIK, 2AKR, 1Z5L and 1ZHN.
Conformational shift of CD1d and the phosphorylcholine headgroup upon TCR binding
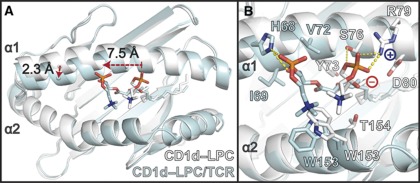
Upon binding of the iNKT TCR to the CD1d–LPC surface, two major shifts occur in both the heavy chain of CD1d and the positioning of the LPC ligand. The outward-shifted α1 helix observed in the unliganded CD1d presenting LPC is shifted 2.3 Å inward, closing the A’ pocket and adopting a conformation close to that observed in the CD1d–αGalCer structures (Figure 3A). In addition, the phosphorylcholine headgroup is shifted ∼7.5 Å towards the A’ pocket, making room to accommodate the CDR loops of the TCR that occupy the position of the unliganded LPC headgroup (Figure 3A), which is discussed in more detail later. This movement includes breaking of hydrogen bonds and a salt bridge between the negatively charged phosphate group and the positively charged Arg79 of CD1d in the unliganded state (Figure 3B). In the liganded state, LPC is stabilized by hydrogen bonds with His68 of CD1d, which moves into hydrogen-bonding distance through the inward shift of the α1 helix (Figure 3B) and a salt bridge with K71 from iNKT TCR's HV4α loop (see Figure 6A and B). Overall, the contacts made between CD1d and the headgroup of LPC are almost completely unique between the liganded and unliganded states, with only a van der Waals (VDW) interaction with Trp153 shared between the two positions (Figure 3B). Electron density of the phosphorylcholine headgroup of LPC in our liganded structure is apparent, however, it is discontinuous with the electron density observed for the acyl tail, consistent with the acyl tail occupying either the A’ or F’ pockets (see Figure 6B). We have positioned the acyl tail in the F’ pocket for simplicity, but our data are consistent with occupancy of either pocket, suggesting that either LPC occupies both pockets in our structure and we are seeing an average of this in our electron density, or that the electron density we observe is due to positioning of the acyl tail in one pocket and spacer fatty acid chains that are typically seen filling CD1 molecules in the other pocket.
Figure 3.
Conformational shift of the phosphorylcholine headgroup of LPC and the α1 helix of CD1d between the unliganded and TCR-liganded states. (A) Structures of CD1d–LPC in the unliganded (white) and TCR-liganded (cyan) states are shown aligned via the main-chain CA carbons of their heavy chains. Two major shifts are seen: (1) the phosphorylcholine headgroup of LPC shifts 7.5 Å towards the A’ pocket and (2) the α1 helix moves inward ∼2.3 Å to adopt a conformation similar to that of CD1d–αGalCer. (B) Side-chain residues of the α1 and α2 helices of CD1d that coordinate the phosphorylcholine headgroup in the unliganded (white) and TCR-liganded (cyan) state. Side-chain atoms are shown as sticks, and are coloured as described in Figure 1A. The amino acids coordinating the two states are unique, save Trp153, which adopts different side-chain conformations in each state. The positive and negative charge of Arg79 and the phosphate moiety are shown as blue ‘+’ and red ‘−’, respectively. Hydrogen bonds are shown as dashed yellow lines: highly likely H-bonds (>3.3 Å) are shown with heavy dashed lines, probable H-bonds (4 Å> but >3.3 Å) are shown as light dashed lines.
Docking orientation of the iNKT TCR on CD1d–LPC
Our initial characterization of 10 human CD1d-restricted clonal T-cell lines demonstrated that some, but not all, responded specifically to LPC presented by recombinant human CD1d molecules (Fox et al, 2009). One of these clones, J24.L17, which expresses an iNKT TCR (Vα24-Jα18 and Vβ11) and demonstrated nearly 100-fold over background responses to LPC, was used for the structural and biochemical studies presented here. We expressed the TCR chains of J24.L17 in E. coli as inclusion bodies and used an optimized refolding strategy described in Materials and methods to produce the full-length ectodomain of the TCR. While multiple attempts at measuring an interaction between recombinant J24.L17 and CD1d–LPC failed, we did succeed in crystallizing this TCR with our glycosylation minimized CD1d loaded with LPC. We obtained weakly diffracting crystals that, through iterative rounds of optimization, generated a data set that resulted in the three-dimensional structure presented here. This data set was processed and refined to 3.0 Å resolution (see Table I for crystallization and refinement statistics). Within the asymmetric unit was one CD1d–LPC–iNKT TCR complex and one unliganded iNKT TCR.
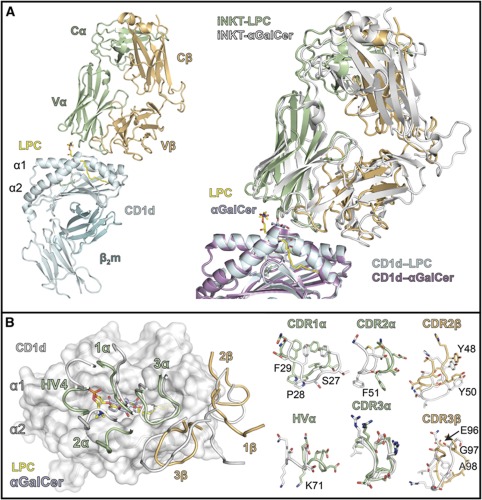
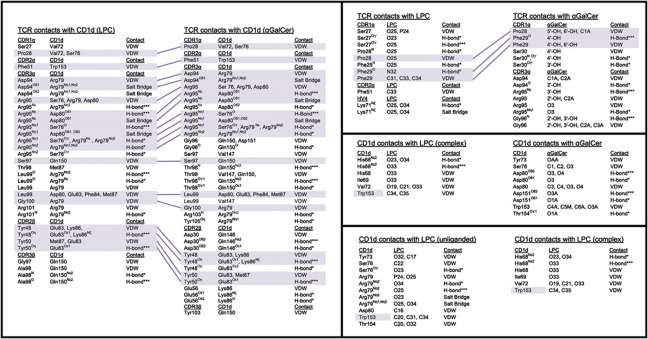
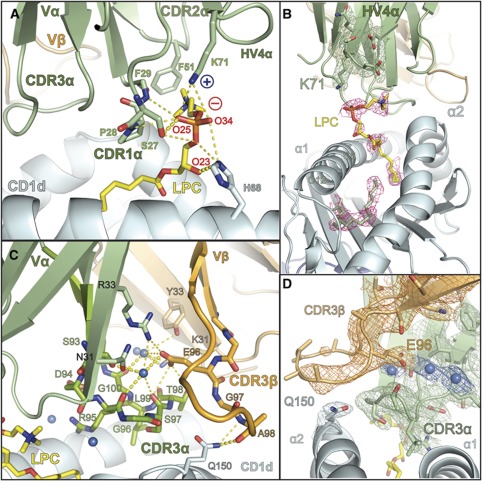
A ribbon diagram of the CD1d–LPC–iNKT TCR complex is shown in Figure 4A (left panel). The overall docking orientation was similar, but not identical, to that of the CD1d–αGalCer–iNKT TCR structure previously reported (Borg et al, 2007; Pellicci et al, 2009) as shown by a superposition of these complexes through alignment of the CD1d heavy chains (Figure 4A, right panel). The interface area between the iNKT TCR and the CD1d–LPC surface was calculated to be 850 Å2 using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), only slightly smaller than that of the iNKT TCR–CD1d–αGalCer structure (900 Å2). The distribution of interface surface varies considerably between the two structures, however, where the α chain contributes 660 Å2 (∼78%) to the interface with CD1d–LPC and only 570 Å2 (63%) to CD1d–αGalCer. This is in part due to the higher contact surface between the α chain and LPC (190 Å2 versus 120 Å2 for αGalCer), mediated by unique CDR1α, CDR2α and HV4α contacts to this antigen (discussed further below).
Figure 4.
Complex structure of the J24.L17 iNKT TCR with CD1d–LPC. (A) Ribbon diagram of the human iNKT TCR J24.L17 (α chain in light green and β chain in light orange) in complex with human CD1d (cyan) and LPC (yellow). β2 microglobulin (β2m) is shown in teal. To the right is a superposition of the human iNKT TCR–CD1d–αGalCer complex (Pellicci et al, 2009) (PDB ID: 3HUJ; TCR in white, CD1d in mauve and αGalCer in lavender) with that of the iNKT TCR–CD1d–LPC complex structure. Both are shown as ribbons with ligands in stick representation. Complex structures were aligned via the main-chain CA carbons of the CD1d heavy chain. (B) Positioning of the iNKT TCR CDR loops on the CD1d-ligand surface. Left: CD1d is shown as semitransparent surface in white with ribbons showing the location of the α1 and α2 helices. LPC is shown in yellow and αGalCer in lavender. The CDR loops are coloured according to their TCR chain colouring in (A). Right panel shows the conformation of the side-chain residues (shown as sticks) of the CDR loops involved in CD1d–LPC recognition; main chains are shown as ribbon loops.
The LPC-reactive iNKT TCR (iNKT-LPC) has a counter-clockwise rotation in relation to the iNKT TCR of the αGalCer complex, resulting in shifts of all CDR loops except the CDR3α (Figure 4B). The positioning of the CDR3α loop, as well as many of the contacts established with the CD1d surface, is conserved across the two complexes (Figures 4B and 5). Of the 31 contacts that the iNKT-LPC CDR3α loop makes with CD1d, 20 (∼65%) of these are identical to contacts made by the CDR3α loop of the iNKT TCR in the CD1d/αGalCer structure (Figure 5). A notable difference in our structure is that the CDR3α loop does not contact the LPC ligand. CDR3α loop contact, either directly or via water-mediated bridges, is a feature seen in all previously reported iNKT complex structures. Here, the CDR1α, CDR2α and HV4α loops establish all contacts with the LPC ligand and the CDR1α and CDR2α essentially block contact by the CDR3α (Figure 6). Serine at position 27 of the CDR1α establishes both VDW and hydrogen bond interactions with LPC's phosphate moiety, with minor contacts contributed by Pro28 and Phe29 with the phosphorylcholine headgroup. In the iNKT TCR complex structure with αGalCer, Pro28 and Phe29 play a more prominent role in contacting αGalCer, establishing contacts with the 4′OH, 5′OH and 6′OH groups of the galactose headgroup. Serine 27 does not make contacts with αGalCer. Furthermore, the positioning of the CDR2α in our complex structure allows for a VDW interaction of Phe51 with a methyl group of the quaternary ammonium component of the phosphorylcholine headgroup.
Figure 5.
Atomic contact comparison of iNKT TCRs, CD1d and lipid ligands. The contacts for (1) the TCR and CD1d heavy chain of the LPC and αGalCer complex structures (left panel); (2) the TCR contacts with LPC and αGalCer (right top panel); (3) the CD1d heavy chain contacts between the liganded LPC and αGalCer structures; (4) the CD1d heavy chain and the phosphorylcholine headgroup of the unliganded CD1d–LPC and complexed CD1d–LPC structures (right lower panel) are shown. VDW, van der Waals interactions, based on a distance ⩽4 Å; salt bridge, based on electrostatic interactions of 4 Å or less; H-bonds*=probable hydrogen bonds based on a distance between 3.3 Å and 4 Å; H-bonds***=highly likely hydrogen bonds based on a distance of 3.3 Å or less. Conserved contacts are shaded in purple.
Figure 6.
Specificity of the iNKT TCR for CD1d–LPC is mediated by contacts with the CDR loops of the Vα domain and intricate coordination of the CDR3β loop. (A) The contacts of the iNKT TCR with LPC are mediated exclusively through the CDR1α, CDR2α and HV4α loops shown in green. Side- and main-chain atoms are shown where they establish contacts with the LPC phosphorylcholine headgroup. The positive and negative charge of Lys71 and the phosphate moiety are shown as blue ‘+’ and red ‘−’, respectively. (B) Electron density of the LPC ligand in the CD1d–LPC–iNKT TCR complex. Electron density corresponds to a composite omit map (2Fo−Fc) contoured at 1α around the LPC ligand (pink) and C12 and C6 fatty acids shown in green. Electron density for the HV4α loop and the K71 side chain are shown in green, contoured as with LPC. (C) Hydrogen bonding network of the water-mediated coordination of the CDR3β loop. The CDR3β loop and Vβ domain are shown in light orange, the CDR3α loop and the Vα domain are shown in light green. Stick representation is used to show the side- and main-chain atom contacts between these loops and their coordination by three water molecules, shown as marine coloured spheres. CDR3β's contact with CD1d (coloured cyan), mediated by the Gln150 side chain, is shown as stick representation. LPC is coloured in yellow. In both panels, hydrogen bonds are shown as dashed yellow lines: highly likely H-bonds (>3.3 Å) are shown with heavy dashed lines, probable H-bonds (4 Å> but >3.3 Å) are shown as light dashed lines. (D) Electron density of the CDR3β–CDR3α–CD1d interface. A 2Fo-Fc map is shown, contoured at 1σ around the CDR3β (orange) and the Glu96β side chain, CDR3α (green) and the Gln150 residue of the CD1d α2 helix (cyan). Electron density for three water molecules located in the CDR3α-CDR3β interface is also shown, the waters are represented by spheres coloured in marine.
Another key interaction of the iNKT TCR with CD1d–LPC is found in the HV4α loop. This loop is typically not involved in ligand recognition in conventional αβTCR and MHCp interactions, although an equivalent lysine (K69) is noted to establish a ligand contacts in murine iNKT TCR/CD1d complex structures (Pellicci et al, 2011; Yu et al, 2011). Here, the HV4α loop appears to play a pivotal role in LPC recognition with the establishment of a salt bridge between the HV4α's K71 and the phosphate of the phosphorylcholine headgroup (Figure 6A and B). This interaction likely compensates for the breaking of the salt bridge with CD1d's Arg79 coordinating LPC in the unliganded CD1d–LPC structure.
While unique contacts are made between the iNKT TCR and the CD1d–LPC surface, there are contacts that are conserved despite main-chain movement of the CDR loops. Tyr48 and Tyr50 of the CDR2β loop, shown to be critical in human iNKT TCR engagement of CD1d–αGalCer (Wun et al, 2008), establish almost identical contacts to those seen in the αGalCer complex (Figure 5). The Tyr48 and Try50 contacts to CD1d are preserved through rotation of their tyrosyl side chains despite shifts of their main-chain CAs by 3.4 Å and 2.8 Å, respectively (Figure 4B, right panel). Overall, 32 of the 47 (68%) contacts between CD1d–LPC and the iNKT TCR are observed in the complex structure with αGalCer. However, these shared contacts only make up 32 out of the 53 total contacts observed in the iNKT TCR–CD1d–αGalCer structure (60%). Therefore, a considerable number of the intermolecular contacts established (32% for the LPC complex and 40% for the αGalCer complex) are unique to each complex.
The molecular basis for LPC specificity: role of the CDR3β and Jβ segment
iNKT TCRs have been described as ‘pattern-recognition receptors’ due to the invariant docking mode and conserved contacts seen in mouse iNKT TCR structures with different endogenous and exogenous glycolipid antigens presented by CD1d (Scott-Browne et al, 2007; Li et al, 2010; Sullivan et al, 2010; Wang et al, 2010; Aspeslagh et al, 2011; Kinjo et al, 2011; Pellicci et al, 2011; Wun et al, 2011; Yu et al, 2011). Our findings that some, but not all, human iNKT T cells are reactive to LPC and the ability to recognize LPC does not correlate with the strength of the response to αGalCer (Fox et al, 2009) suggest there are inherent yet subtle differences in the human iNKT TCR population that confer specificity to particular antigens. Our complex structure provides molecular evidence for the role of the CDR loops of the Vα chain in LPC specificity, but also contributions from the variable CDR3β, as well as conformational variability of the TCR structure caused by use of particular Jβ segments during rearrangement.
In our complex structure, the CDR3β is locked into a semi-rigid state by an intricate water-mediated hydrogen-bonding network involving the CDR3α loop and several β-strands from both the Vα and Vβ domains (Figure 6C). The involvement of the CDR3β in this network is mediated primarily through Glu96, a residue encoded by the variable component of the CDR3β. This positions the N-terminal portion of the CDR3β, including Gly97 and Ala98, in close proximity to Gln150 of the CD1d molecule, where two main-chain hydrogen bonds are established (Figure 6C). The electron density for this network is shown in Figure 6D. Electron density for the C-terminal portion of the CDR3β is weak and for some residues is absent, suggesting that this part of the loop is disordered in our crystal structure. The amino-acid sequence of the CDR3β in the J24.L17 iNKT TCR thus produces a specific, semirigid conformation that establishes a main chain-mediated contact with CD1d and likely contributes to the specificity of this iNKT TCR for LPC.
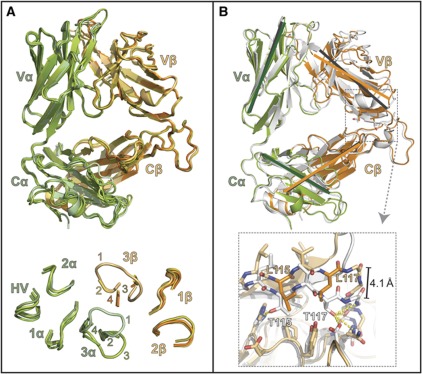
The unconventional involvement of the CDR1α, CDR2α and HV4α loops linked to the counter-clockwise rotation of the CDR loops around the CDR3α loop anchor led us to investigate whether there were additional factors that contributed to these structural features. First, to establish whether CDR loop conformational change played a role in CD1d–LPC recognition, we used three independent unliganded structures of our LPC-reactive iNKT TCR for comparison with our liganded TCR. One of these TCR structures was derived from the complex data set and two derived from an independent crystallization of the J24.L17 iNKT TCR alone. This data set was refined to 2.8 Å resolution (Table I for crystallization and refinement statistics), with two iNKT TCRs in the asymmetric unit. Superposition of the unliganded TCRs with that from our complex structure showed neither major conformational movement between the V and C domains of the heterodimer (Figure 7A), nor changes in the structure of the CDR1 and CDR2 loops from both chains (Figure 7A). However, in the unliganded TCRs, poor electron density of the CDR3α and CDR3β loops made modelling of these regions impossible; clear main-chain electron density was only observed in the CDR3α of TCR #1 in the TCR-only data set, and this conformation was unlike that of the liganded CDR3α (Figure 7A).
Figure 7.
Comparison of the iNKT TCR structure between J24.L17 unliganded and liganded, and that of the iNKT TCR co-crystallized with CD1d–αGalCer. (A) Alignment of the complexed J24.L17 iNKT TCR with that of the three uncomplexed J24.L17 iNKT TCRs derived from our studies. The complexed iNKT TCR is coloured in light green (α chain) and light orange (β chain) as in other figures; the uncomplexed iNKTs are coloured in related shades. No major domain shifts are noted between the complexed and uncomplexed TCRs. The CDR loop conformations; no major shifts are seen between the CDR1, CDR2 loops of the α and β chains, or for the HV4α loop are shown at the bottom. Electron density was poor for all uncomplexed CDR3β loops; these loops are shown truncated. Electron density was also poor for two of the three unliganded CDR3α loops; they are shown truncated as well. The loops are labelled according to the TCRs to which they correspond: 1=complexed iNKT TCR; 2=uncomplexed iNKT TCR from the complexed data set; 3=iNKT TCR (1) from the uncomplexed data set; and 4=iNKT TCR (2) from the uncomplexed data set. Electron density for the main-chain atoms of the CDR3α loop of #3 was continuous and positioned this loop in an alternate conformation from that of the liganded CDR3α loop. (B) Alignment of the LPC-specific iNKT TCR with that of iNKT TCR from the αGalCer complex structure (Pellicci et al, 2009) (PDB#3HUJ). αGalCer iNKT TCR is shown in white, LPC iNKT TCR is shown in green (α chain) and orange (β chain); these were aligned based on the Cα and Cβ main-chain CA carbons. Grey lines represent the orientation of the Ig domains that make up the αGalCer iNKT TCR core structure and green and orange lines represent the Ig domains from the LPC iNKT TCR; lines were drawn from identical atom locations on each TCR. Black lines were drawn similarly and shown the pivot point of the Vβ domain in relation to the Vα, Cα and Cβ domains. At bottom is a magnified view of the C-terminal residues of the Jβ segment, side- and main-chain atoms contributing to the shift are shown as stick representation, LPC β-chain residues as orange, αGalCer β-chain residues shown in white. Hydrogen bonds contributing to this interaction are shown as dashed yellow lines: highly likely H-bonds (>3.3 Å) are shown with heavy dashed lines, probable H-bonds (4 Å> but >3.3 Å) are shown as light dashed lines. A 4.1-Å shift is apparent in the main-chain CA carbons between the two TCRs within this region.
When we compared our iNKT TCR structure with that of another human iNKT TCR structure, we noted a shift in the orientation of the Vβ domain in relation to the Vα, Cα and Cβ domains (Figure 7B). Upon closer inspection, we discovered this shift correlated with two C-terminal residues in the Jβ segment that differ between these two structures. In our structure, positions 115 and 117 of the Vβ domain encode non-polar leucine residues due to the use of the TRGJ2-5 segment (…QETQYFGPGTRLLVL), whereas in the other iNKT TCR, these residues are polar threonines due to the use of the TRBJ2-7 segment (…SYEQYFGPGTRLTVT). Threonine at these positions establishes a hydrogen-bond network in the Vβ–Cβ elbow, locking the Vβ domain to the Cβ domain at an acute angle. Thr117Vβ establishes main-chain hydrogen bond contacts with Glu118Cβ and Asp119Cβ and side-chain hydrogen bond contacts with Asn122Cβ and Asp180Cβ; Thr115Vβ hydrogen bonds with His157Cβ (Figure 7B). This hydrogen-bonding network is disrupted when leucines are at positions 115 and 117, and the repulsion between the polar and non-polar residues results in an ∼4.1 Å shift between the main chains of this region in these two TCRs. This shift translates down the Vβ domain, pivoting through the Vα–Vβ interface resulting in a similar shift in the CDRβ loops (∼4.2 Å shift as measured between CA carbons of the CDR1β Lys31). We postulate the role of the Jβ segment in establishing the core structure of the TCR contributes to the observed shift in the CDR loops in our iNKT TCR–CD1d–LPC complex and contributes to the unique associations we observe in our structure, such as the involvement of the CDR1α, CDR2α and HV4α loops in LPC specificity. This variation in CD1d docking raises the question as to whether it is due to recognition of LPC, or whether it is intrinsic to this TCR. Structural information of this TCR with other CD1d/lipid antigen complexes is lacking; however, the binding affinity of this TCR for CD1d/αGalCer (∼3.3 μM) (Supplementary Figure S2) is similar to that of certain other iNKT TCRs (e.g., 4–6 μM in Gadola et al, 2006), suggesting that any docking variation that may exist does not profoundly affect the affinity of binding.
Discussion
Our investigation into LPC presentation by CD1d molecules revealed a low relative KD of binding (∼2 μM), suggesting that the overall half-life of this lipid in CD1d may be lower than that of other lipids. For CD1d–LPC, our structural data suggest that LPC is predominantly bound in the F’ pocket, with a surprising shift in CD1d's α1 helix resulting in an A’ pocket open to solvent. This feature, similarly to that noted in the ‘empty’ human CD1d structure (Koch et al, 2005), suggests that human CD1d has intrinsic flexibility in the association of its α1 and α2 helices, and our structure likely represents one of many conformations that exist during the process of lipid loading. In this model, the interaction between the α helices is dynamic, allowing for the insertion of lipid molecules directly, without need for their threading into the A’ and/or F’ pockets. Lipid transfer proteins (LTPs) such as saposins (Zhou et al, 2004a) and microsomal triglyceride transport protein (MTP) (Brozovic et al, 2004) have been demonstrated to play an important role in lipid transfer to CD1d molecules; it is intriguing to speculate that they may facilitate the lipid transfer process through a temporary stabilization of these open structures, however, this has not yet been explored experimentally. This conformational shift observed in our structure may also explain, in part, why the apparent affinity of LPC for CD1d is low (∼2 μM) and why recycling of CD1d molecules into the endocytic pathway may rapidly remove bound LPC molecules (Fox et al, 2009). While our structural data suggest that LPC is predominantly bound in the F’ pocket, we cannot rule out that this conformation was favoured during crystallization and that LPC can also be presented in the A’ pocket.
Our complex structure provides several new perspectives on human iNKT TCR recognition of antigens. First, comparisons between the unliganded and liganded TCRs suggest that our iNKT TCR engagement of CD1d is not a classic ‘lock and key’ type recognition, with little conformational changes occurring in the CDR loops between the unliganded state and after engagement of CD1d. Instead, one of our unliganded TCR structures reveals a CDR3α loop structure unlike that of it in the liganded state, suggesting that, at a minimum, this loop must undergo some sort of conformational transition upon binding. This contrasts with findings from other groups whereby the unliganded TCR maintained a similar CDR3α loop conformations as those in the liganded state (Gadola et al, 2006; Borg et al, 2007). While crystal structures are only snap shots of a likely range of dynamic conformations, our data confirm that the CDR3α loop of this iNKT TCR can adopt a different conformation from that of the liganded state. It is tempting to speculate that the lack of electron density in the CDR3 loop structures of the remaining unliganded TCRs is due to backbone flexibility, however, because there are other factors that can produce poor electron density in these regions this cannot be used as a conclusive argument for conformational plasticity. The contacts made upon complex formation for the CDR3α and CDR2β are highly similar between our structure and that of previously published iNKT TCR/CD1d complexes, consistent with previous work demonstrating these loops provide an important adhesive force for iNKT TCR binding (Scott-Browne et al, 2007; Wun et al, 2008). However, it is only the CDR3α loop that adopts a similar docking position in our structure, contacts established by the CDR2β are due to alternate conformations of the Tyr48 and Tyr50 tyrosyl headgroups with Glu83, Lys86 and Met87 of CD1d, as the CDR2β main chains deviate up to ∼3 Å between the LPC and αGalCer complex structures. These particular tyrosine residues have also been shown to be part of the ‘codon’ recognition model of conventional αβTCR recognition of MHCp, establishing critical contacts with MHC helical surfaces of varying amino-acid composition through movement of the tyrosyl headgroups (Feng et al, 2007; Dai et al, 2008). Shifts in CDRβ loop positioning have been noted for mouse NKT TCRs in complex with CD1d when alternate Vβ domains are used (Pellicci et al, 2009), however, this is due to accommodation of the alternative amino-acid motifs that make up these CDR loops (CDR2β loop in particular).
Since all of the iNKT cell clones we have analysed share the same CDR3α and CDR2β sequences, interactions of the CDR3α and CDR2β loops with the CD1d molecule do not explain why only some, but not all, of the iNKT cell clones respond to CD1d-mediated presentation of LPC. Our structures provide evidence that specificity for particular antigens results from contributions from additional elements of the TCR, in particular variable amino acids of the CDR3β and use of different Jβ segments that affect Vβ–Cβ association and translate into shifts in positioning of the CDR loops relative to each other. This shift results in the alternative docking we see in our structure, with contacts between CD1d–LPC by all the Vα CDR loops (including the HV4α) and the CDR2 and CDR3 loops of the β chain. The contribution of the HV4α loop to ligand discrimination in the form of a salt bridge between K71 and LPC's phosphate is unconventional among classical αβ TCR recognition; however, an analogous lysine is conserved in the mouse Vα14 domain, and has been demonstrated in these mouse NKT TCRs to contribute to ligand discrimination as well.
Our structure shows a highly coordinated CDR3β loop primarily anchored by a water-mediated hydrogen-bonding network with the CDR3α. This structure positions the main-chain structure of the CDR3β to provide main-chain interactions with CD1d at Q150. Sequence analysis of TCRs from six other LPC-reactive NKT cell clones has revealed that these do not have identical sequences in their CDR3β, and none have a glutamate at position 96. Only one of the other LPC-reactive clones uses the TRBJ2-5 segment (clone GG1.2). One of the LPC-reactive clones (J24N.22) uses Vα2.3 joined to Jα18 instead of the Vα24-Jα18 rearrangement; and therefore, this LPC-reactive clone is not an iNKT cell in its true sense. Because there is no clear motif shared between these clones, it is likely that each may have a different solution to CD1d–LPC recognition. J24.L17, the clone used in these studies, thus likely combines a semicoordinated CDR3β loop sequence with shifts in the Vα/Vβ domain interface due to the Jβ segment to recognize LPC in the context of CD1d. Whether the TCR from this T-cell clone would maintain the same docking footprint when recognizing other CD1d–lipid complexes (i.e., αGalCer) is unclear; however, its affinity for αGalCer is similar to that of other human iNKT TCR/CD1d–αGalCer measurements.
The search for physiological lipids in humans that can directly stimulate the NKT cell population has revealed few compelling candidates that can be studied at the molecular level. LPC represents a novel class of ligands for NKT cells and has potential applications to human disease due to the rapid generation of LPC in states such as cancer (Umezu-Goto et al, 2002) and atherosclerosis (Portman and Alexander, 1969; Thukkani et al, 2003), and the suggested role of NKT cells in these diseases (Swann et al, 2004). LPC can be generated in many tissues via PLA2 activity and can be found at significant levels in human serum, with levels as high as mM concentrations seen in hyperlipidemic subjects (Chen et al, 1997). Our work provides a molecular mechanism for iNKT cell recognition of CD1d presenting the endogenous ligand, LPC and provides a model by which this system can be tested for its relevance for human inflammatory diseases such as atherosclerosis and cancers such as multiple myeloma.
Materials and methods
Human wild-type CD1d, glycosylation-minimized human CD1d and LPC-reactive iNKT TCR expression
The cDNAs corresponding to the ectodomain region of human CD1d and human β2 microglobulin (β2m) were separately cloned into the pAcGP67A vector (BD Biosciences); the CD1d heavy chain was engineered with a C-terminal hexahistidine tag. Both proteins were co-expressed in Hi5 cells via baculovirus transduction and co-purified with Ni2+ beads and anion exchange chromatography. For crystallographic purposes, three of the four potential N-glycosylation sites (N42, N108 and N163) of CD1d were mutated to Q to reduce protein heterogeneity. This protein was expressed as described above for wild type. The hexa-His tag was removed by Carboxypeptidase A (Sigma) digestion at a 1:100 enzyme to protein ratio for 16 h. The protein sample was then buffer exchanged to 20 mM Tris, pH 8.7 and further purified under anion exchange chromatography. The protein was additionally treated with endoglycosidase F3 in 50 mM sodium acetate, pH 5.5, 5 mM CaCl2 buffer at 37°C for 2 h to further minimize heterogeneity present by the one remaining potential N-linked glycosylation site (N20). The glycosylation-minimized CD1d was purified by one round of anion exchange followed by size-exclusion chromatography.
The ectodomain of the LPC-reactive invariant TCR clone J24.L.17 was modified to favour proper heterodimeric refolding as described (Boulter et al, 2003) by inserting T48C and S57C mutations at the α and β constant domains, respectively, and eliminating the wild-type interchain disulphide cysteines. These constructs were cloned in the pET28a vector (Novagen) in frame with a C-terminal hexahistidine tag. Both α and β chains were expressed separately as inclusion bodies in Rosetta™ (DE3) pLysS cells (Novagen). For TCR refolding, 100 mg of each chain was dissolved in 6 M Guanidine hydrochloride, 50 mM Tris pH 8.1 and 10 mM β-mercaptoethanol, heated at 50°C for 10 min, spun and mixed together. The protein mixture was then injected drop wise at 4°C under continuous stirring in a 200-ml buffer containing 50 mM Tris pH 8.1, 5 M Urea, 0.4 M L-arginine, 0.2 mM PMSF, 5 mM reduced L-glutathione and 0.5 mM oxidized L-glutathione. A same amount of inclusion bodies were added 2 h later. Four hours later, a six-fold dilution process was initiated with 1 L of 50 mM Tris pH 8.1 that was added drop wise along a 24-h period of time under continuous stirring at 4°C. The diluted protein sample was kept at 4°C for an extra 24 h of continuous stirring. Then, the sample was filtered and the his-tagged protein captured with Ni2+ beads and digested with Carboxypeptidase A as described previously for the CD1d mutant. Further TCR purification was accomplished by buffer exchanging the protein sample to 20 mM Tris pH 8.1 and subjecting it to a monoQ column (GE Healthcare) under an NaCl gradient from 0 to 0.5 M in 40 column volumes. This later step allowed the isolation of heterodimeric TCR that was further purified by size-exclusion chromatography with an S200 column (GE Healthcare), yielding a single monodispersed peak at the expected elution volume.
CD1d loading with LPC
LPC (18:1) was obtained from Avanti Polar Lipids (Catalogue #845875C). The lipid in chloroform was dried under N2 stream and the resultant lipid film resuspended in HBS buffer and incubated at 37°C for 20 min, which rendered a clear lipid solution. To load wild-type and glycosylation-minimized CD1d with LPC a protein–lipid mixture was prepared at a 1:10 protein to lipid ratio and incubated overnight at room temperature. Finally, the excess of the lipid was removed by extensive dilution and concentration cycles in a 30-kDa molecular weight cutoff centrifugal device (Millipore).
Analysis of CD1d–LPC interaction with fluorescence spectroscopy
Human soluble CD1d was incubated with increasing concentrations of LPC (18:1) in HBS for 2 h at RT and the intrinsic fluorescence was monitored using a 10-mm light path Quartz cell (Starna Cells Inc, Atascadero, CA, USA) in a PTI (model 810) spectrofluorimeter. Excitation wavelength was set to 280 nm and the emission fluorescence recorded at 340 nm. The excitation and emission slit widths were 2 and 4 nm, respectively. Readings of samples in the absence of protein were subtracted from the matched samples containing soluble CD1d and lipids. The changes in the intrinsic fluorescence of soluble CD1d upon LPC incubation were calculated using the equation: (F−F0)/F0, where F0 is the intrinsic fluorescence of soluble CD1d and F its fluorescence in the presence of the lipid. The signal ratio was plotted against LPC concentration and fit using non-linear regression in GraphPad Prism™ software to a one-site binding model.
CD1d loading with αGalCer and measurement of molecular interactions by SPR
A human CD1d construct bearing a 3C site-his tag at the C-terminus was expressed in Hi5 cells and purified as described above. The processed sample was then treated with 3C protease for 16 h at 4°C to remove the his tag and further purified by anion exchange chromatography in a MonoQ column (GE Healthcare) before loading it onto a nickel agarose column. CD1d-enriched flow-through fraction was used for loading with αGalCer at room temperature with a three molar excess of lipid for 16 h. The excess of lipid was then removed with a Superdex 200 (10/30) column (GE Healthcare). All interaction experiments were performed using a BIAcore 3000 Instrument (GE Healthcare). J24.L17 NKT TCR from E. coli inclusion bodies was refolded and purified as mentioned above and captured to a density of 2000 RU in the flow channel 2 of an NTA sensor chip (GE Healthcare) previously treated with NiCl2. Insect cell-derived recombinant FC was captured in the flow channel 1 to account for non-specific binding events. Increasing concentrations (0.111, 0.333, 1, 3 and 9 μM) of CD1d–αGalCer were injected at a flow rate of 30 μl/min in 10 mM Hepes pH 7.4, 150 mM NaCl and 0.005% Tween-20. Kinetic and equilibrium parameters were calculated using BIAevaluation software 3.2RC1 (GE Healthcare) and GraphPad Prism.
Protein crystallization
Original crystallization attempts with LPC-loaded human wild-type CD1d yielded large, sharp-edge crystals that diffracted to 4 Å. In order to improve the resolution, the glycosylation minimized CD1d was used for crystallization and optimized crystals of LPC-loaded, glycosylation minimized human CD1d were obtained in sitting drop plates by mixing 0.5 μl of protein sample at 10 mg/ml and 0.5 μl of mother liquor (0.1 M Tris–HCl pH 8.5, 2.0 M Ammonium Sulphate and 0.03 M glycyl-glycyl-glycine) at 18°C. Crystals were cryo-cooled in mother liquor supplemented with 20% glycerol prior to data collection and diffracted to 2.8 Å. Crystals of the J24.L17 iNKT TCR grew in 0.1 M Hepes pH 7.0, 17% PEG 4000 and 0.1 M BaCl2 at a protein concentration of 10 mg/ml. The crystals were equilibrated in the precipitant solution containing 20% glycerol and diffracted to 2.8 Å. Data sets for both crystal forms were collected at 100° K at a wavelength of 1.033 Å. For LPC-loaded human CD1d–TCR complex crystallization, both CD1d and TCR protein samples were mixed in HBS at 1:1 molar ratio and concentrated in Nanosep Centrifugal Devices (Pall Life Sciences). Initial trials with wild-type human CD1d yielded plate-like crystals diffracting to 4 Å. The ternary complex with glycosylation minimized CD1d crystallized at 20 mg/ml in 0.1 M Hepes pH 7.0, 17% PEG 4000 and 0.1 MgCl2 and after flash freezing in the presence of 20% glycerol, diffracted to 3.0 Å. X-ray diffraction data were collected at 100° K, at a wavelength of 0.979 Å. All data sets reported in this manuscript were collected on a MAR300 CCD at beamline 23 ID-D at the Advanced Photon Source (APS) at Argonne National Laboratory and processed with HKL2000 (Otwinowski and Minor, 1997).
Structure determination and refinement
Structures were solved by molecular replacement with the program Phaser (McCoy et al, 2007). For the LPC-loaded human CD1d, the search model was the human CD1d–β2m structure (Protein Data Bank (PDB) accession number 1ZT4) with all non-protein coordinates removed to reduce model bias. The LPC-reactive TCR structure was solved using the invariant Vα24 TCR (PDB accession number 2EYS) as search model with the coordinates corresponding to the CDR3β loop omitted. Finally, the crystal structure of the LPC-loaded CD1d in complex with the iNKT TCR was solved using search models CD1d–β2m–αGalCer (PDB accession number 3HUJ) and the same TCR model mentioned above.
Refinement was performed with the Phenix software suite (Adams et al, 2010). Initial steps of refinement were accomplished through rigid body, individual sites and B-factor refinement in the presence of non-crystallographic symmetry (NCS) restraints. Next, subsequent cycles of manual building in Coot (Emsley and Cowtan, 2004) and refinement were carried out and ligands such as lipids or covalently bound sugar moiety in the case of the CD1d molecules were introduced guided by Fo−Fc positive electron density. Ligands structures and chemical parameters were defined with C.C.P.4.'s Sketcher (Collaborative Computational Project, Number 4, 1994) and included in subsequent refinement and manual building steps. Translation/libration/screw (TLS) partitions were calculated at later refinement stages with the TLSMD server. Structure stereochemistry was further optimized by real space refinement with a two-step algorithm developed by EJ Haddadian, KF Freed and TR Sosnick (Haddadian et al, 2011). All the refinement procedures were performed taking a random 5% of reflections and excluding them for statistical validation purposes (Rfree). Ramachandran statistics were as follows: CD1d/LPC structure: 96.4% preferred, 3.3% allowed; J24.L17 TCR: 96.0% preferred, 3.9% allowed; CD1d/LPC/J24.L17: 96.0% preferred, 3.6% allowed.
Structure analysis
Intermolecular contacts and distances were calculated using the program Contacts from the CCP4 software package (Collaborative Computational Project, Number 4, 1994), interface surface areas were calculated using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) and all structural figures were generated using the program Pymol (DeLano, 2006).
Accession numbers
Coordinates and structure factors for the CD1d–LPC, J24.L17 TCR and iNKT TCR–CD1–LPC complex have been deposited in the Protein Data Bank under the accession codes 3U0P, 3TYF and 3TZV, respectively.
Supplementary Material
Acknowledgments
We thank the staff of the Advanced Proton Source at GM/CA-CAT (23ID) and LS-CAT (21ID) for their use and assistance with X-ray beamlines; Ruslan Sanishvili and Joseph Brunzelle in particular for help and advice during data collection. This study was supported by National Institutes of Health grant R01AI073922 and the Kinship Foundation Searle Scholars Award to Erin J Adams. We also acknowledge the Ministry of Education of the Government of Spain for supporting Jacinto López-Sagaseta with a post-doctoral fellowship (Programa Nacional de Movilidad de Recursos Humanos del Plan Nacional de I-D+i 2008-2011).
Author contributions: JL-S, LVS and JEK performed protein purification; JL-S and LVS performed crystallization; JL-S performed fluorescence experiments, structure determination and refinement; JL-S, JG and EJA intellectual input; JG and EJA project conception and EJA investigation leadership and manuscript preparation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspeslagh S, Li Y, Yu ED, Pauwels N, Trappeniers M, Girardi E, Decruy T, Van Beneden K, Venken K, Drennan M, Leybaert L, Wang J, Franck RW, Van Calenbergh S, Zajonc DM, Elewaut D (2011) Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J 30: 2294–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L (2007) The biology of NKT cells. Annu Rev Immunol 25: 297–336 [DOI] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J (2007) CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448: 44–49 [DOI] [PubMed] [Google Scholar]
- Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK (2003) Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng 16: 707–711 [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, Gadola SD, Hsu FF, Besra GS, Brenner MB (2011) Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol 12: 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricard G, Venkataswamy MM, Yu KO, Im JS, Ndonye RM, Howell AR, Veerapen N, Illarionov PA, Besra GS, Li Q, Chang YT, Porcelli SA (2010) Alpha-galactosylceramide analogs with weak agonist activity for human iNKT cells define new candidate anti-inflammatory agents. PLoS One 5: e14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB (2003) Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol 4: 1230–1237 [DOI] [PubMed] [Google Scholar]
- Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu FF, Besra GS, Brenner MB (2011) Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med 208: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, Chen D, Kaser A, Glickman J, Kuo T, Little A, Morrison J, Corazza N, Kim JY, Colgan SP, Young SG, Exley M, Blumberg RS (2004) CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med 10: 535–539 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cantu C III, Benlagha K, Savage PB, Bendelac A, Teyton L (2003) The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J Immunol 170: 4673–4682 [DOI] [PubMed] [Google Scholar]
- Chen L, Liang B, Froese DE, Liu S, Wong JT, Tran K, Hatch GM, Mymin D, Kroeger EA, Man RY, Choy PC (1997) Oxidative modification of low density lipoprotein in normal and hyperlipidemic patients: effect of lysophosphatidylcholine composition on vascular relaxation. J Lipid Res 38: 546–553 [PubMed] [Google Scholar]
- Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D, Gumperz J, Hildebrand W (2009) Determination of cellular lipids bound to human CD1d molecules. PLoS One 4: e5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset M, Brossard N, Polette A, Lagarde M (2000) Characterization of plasma unsaturated lysophosphatidylcholines in human and rat. Biochem J 345 (Pt 1): 61–67 [PMC free article] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW (2008) Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity 28: 324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2006) MacPyMOL. DeLano Scientific: San Carlos, CA, USA [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, Kronenberg M, Modlin RL (1998) Molecular interaction of CD1b with lipoglycan antigens. Immunity 8: 331–340 [DOI] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC (2007) Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol 8: 975–983 [DOI] [PubMed] [Google Scholar]
- Florence WC, Xia C, Gordy LE, Chen W, Zhang Y, Scott-Browne J, Kinjo Y, Yu KO, Keshipeddy S, Pellicci DG, Patel O, Kjer-Nielsen L, McCluskey J, Godfrey DI, Rossjohn J, Richardson SK, Porcelli SA, Howell AR, Hayakawa K, Gapin L et al. (2009) Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J 28: 3579–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR, Cook ME, Adams EJ, Hildebrand WH, Gumperz JE (2009) Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol 7: e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadola SD, Koch M, Marles-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, Cerundolo V, Jones EY (2006) Structure and binding kinetics of three different human CD1d-alpha-galactosylceramide-specific T cell receptors. J Exp Med 203: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M (2005) Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol 175: 977–984 [DOI] [PubMed] [Google Scholar]
- Haddadian EJ, Gong H, Jha AK, Yang X, Debartolo J, Hinshaw JR, Rice PA, Sosnick TR, Freed KF (2011) Automated real-space refinement of protein structures using a realistic backbone move set. Biophys J 101: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH et al. (2011) Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol 12: 966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi E, Motoki K, Uchida T, Fukushima H, Koezuka Y (1995) KRN7000, a novel immunomodulator, and its antitumor activities. Oncol Res 7: 529–534 [PubMed] [Google Scholar]
- Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V (2005) The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol 6: 819–826 [DOI] [PubMed] [Google Scholar]
- Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM (2010) The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med 207: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI, Marrack P, Rossjohn J, Gapin L (2011) A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, Howell AR, McCluskey J, Godfrey DI, Rossjohn J, Marrack P, Gapin L (2009) T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity 31: 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schumperli D, Schmidt RR, Villiger PM, Jakobsen BK, Gadola SD (2010) Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol 8: e1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidenko OV, Maher JK, Ernst WA, Sakai T, Modlin RL, Kronenberg M (1999) Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J Exp Med 190: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC, Capron M, Ryffel B, Faveeuw C, Leite de Moraes M, Platt F, Trottein F (2007) Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity 27: 597–609 [DOI] [PubMed] [Google Scholar]
- Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J (2011) Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol 12: 827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, Rossjohn J (2009) Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity 31: 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman OW, Alexander M (1969) Lysophosphatidylcholine concentrations and metabolism in aortic intima plus inner media: effect of nutritionally induced atherosclerosis. J Lipid Res 10: 158–165 [PubMed] [Google Scholar]
- Salio M, Speak AO, Shepherd D, Polzella P, Illarionov PA, Veerapen N, Besra GS, Platt FM, Cerundolo V (2007) Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci USA 104: 20490–20495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa T, Okita M, Murakami J, Kato T, Watanabe A (1999) Abnormal serum lysophospholipids in multiple myeloma patients. Lipids 34: 17–21 [DOI] [PubMed] [Google Scholar]
- Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ (2010) The 2.5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 33: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Ruebsaamen K (2009) Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis 208: 10–18 [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L (2007) Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol 8: 1105–1113 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M (2010) Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J Immunol 184: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann J, Crowe NY, Hayakawa Y, Godfrey DI, Smyth MJ (2004) Regulation of antitumour immunity by CD1d-restricted NKT cells. Immunol Cell Biol 82: 323–331 [DOI] [PubMed] [Google Scholar]
- Thukkani AK, McHowat J, Hsu FF, Brennan ML, Hazen SL, Ford DA (2003) Identification of alpha-chloro fatty aldehydes and unsaturated lysophosphatidylcholine molecular species in human atherosclerotic lesions. Circulation 108: 3128–3133 [DOI] [PubMed] [Google Scholar]
- Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H (2002) Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol 158: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataswamy MM, Porcelli SA (2010) Lipid and glycolipid antigens of CD1d-restricted natural killer T cells. Semin Immunol 22: 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM (2010) Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA 107: 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen X, Rodenkirch L, Simonson W, Wernimont S, Ndonye RM, Veerapen N, Gibson D, Howell AR, Besra GS, Painter GF, Huttenlocher A, Gumperz JE (2008) Natural killer T-cell autoreactivity leads to a specialized activation state. Blood 112: 4128–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, Godfrey DI, McCluskey J, Rossjohn J (2008) A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med 205: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J (2011) A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity 34: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ED, Girardi E, Wang J, Zajonc DM (2011) Cutting edge: structural basis for the recognition of {beta}-linked glycolipid antigens by invariant NKT Cells. J Immunol 187: 2079–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Kang SJ, Evans JE, Cresswell P (2009) Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol 182: 4784–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Cantu C III, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L (2004a) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A (2004b) Lysosomal glycosphingolipid recognition by NKT cells. Science 306: 1786–1789 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.