Abstract
Eukaryotic mRNA decapping proteins are essential for normal turnover of mRNA. Yet, the mechanism of bulk mRNA turnover during stress responses and its importance to stress tolerance are poorly understood. Here, we showed that dehydration stress activated MPK6 to phosphorylate serine 237 of Arabidopsis DCP1 and phospho-DCP1 preferentially associated with DCP5 to promote mRNA decapping in vivo. This process was essential for stress adaption as dcp5-1 and DCP1-S237A plants were hypersensitive to stress compared with wild-type (WT) plants. Microarray analysis revealed that dehydration-induced expression of many stress responsive genes was compromised in dcp5-1, whereas a subset of transcripts was over-represented in this mutant. Further analysis revealed that this subset of transcripts was likely the direct targets of stress-triggered mRNA decapping in WT. Our results suggest that mRNA decapping through MPK6-DCP1–DCP5 pathway serves as a rapid response to dehydration stress in Arabidopsis.
Keywords: arabidopsis, dehydration stress, mitogen-activated protein kinase, mRNA decapping, phosphorylation
Introduction
Eukaryotes respond to various environmental stresses by regulating transcriptional and post-transcriptional processes to bring about changes in their protein repertoire. An immediate response to stress onset is the cessation of translation of a subset of mRNAs. The translationally repressed mRNAs are aggregated into processing bodies (P-bodies) where they are stored and/or degraded (Brengues et al, 2005). In yeast, decapping enzyme DCP2 is phosphorylated during stress, suggesting regulation of the decapping process in vivo (Yoon et al, 2010). However, the signalling pathways and mechanisms involved in mRNA decapping and degradation during stress response in multicellular eukaryotes remain unclear.
Plant P-bodies contain DCP1, DCP2 (TDT) and VCS (Xu et al, 2006). Arabidopsis mutants deficient in any one of these three proteins are post-embryonic lethal, indicating that each of them is required for mRNA decapping in vivo (Xu et al, 2006; Goeres et al, 2007; Iwasaki et al, 2007; Brodersen et al, 2008). Plant DCP1s share with other DCP1s a similar EVH1 domain at the N-terminus. In contrast to yeast DCP1, they also contain an extended C-terminal region with a proline-rich unstructured motif and a trimerization domain, which is required for efficient mRNA decapping in vivo (Tritschler et al, 2009). In addition to stimulating DCP2 decapping activity, DCP1 may serve as a regulatory platform for an mRNA decapping network. Several lines of evidence support this notion. First, yeast DCP1 has been identified as a phospho-protein (LaGrandeur and Parker, 1998) and stress induces phosphorylation of human DCP1 (Blumenthal et al, 2009) although the responsible human kinase has not yet been identified. The post-translational modification of DCP1 suggests a connection between stress signalling pathway and the mRNA decapping network. Second, in addition to DCP2, DCP1 was found to associate with other proteins including eIF4G, Pab1, Hedls/VCS, Edc3, Rck/DHH1 and, RAP55/DCP5 (Vilela et al, 2000; Kshirsagar and Parker, 2004; Fenger-Gron et al, 2005; Xu et al, 2006; Tritschler et al, 2007, 2008; Xu and Chua, 2009). Notably, all these proteins are factors involved in translation and translational repression, which precede decapping.
DCP5 is another Arabidopsis P-body component sharing sequence homology with human RAP55. Similar to DCP1, DCP5 does not have any decapping activity in vitro; however, DCP5 is required for translational repression, P-body formation and mRNA decapping in vivo (Xu and Chua, 2009). Both DCP5 and DCP1 protein levels displayed similar dynamic changes during embryonic and post-embryonic development. DCP5 is known to physically interact with DCP1, but how the association is regulated is unknown. Here, we identified a phosphorylation site of DCP1. Plants expressing non-phosphorylated DCP1 mimicking mutation and dcp5-1 were hypersensitive to osmotic stress. Global transcript changes in response to dehydration were altered in dcp5-1 and subsets of mRNAs that were up- or down-regulated were identified by microarray analysis. Moreover, we found that mitogen-activated protein kinase (MPK) pathway signals DCP1 phosphorylation by MPK6 during stress, thus connecting the MPK signalling pathway to the machinery involved in regulation of mRNA degradation.
Results
DCP1 is phosphorylated at S237 upon dehydration
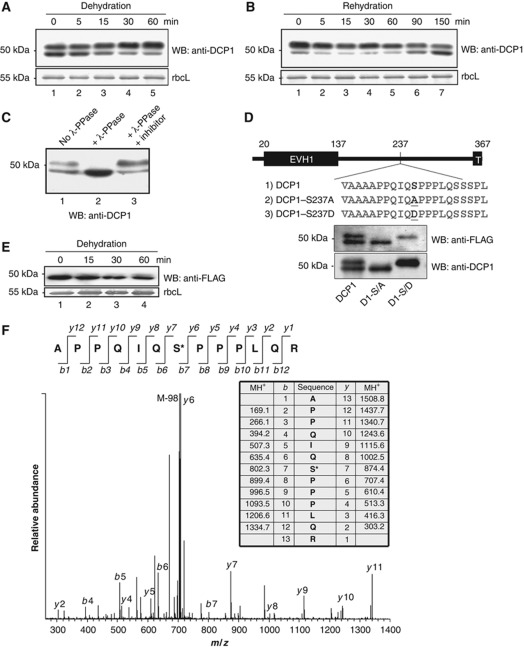
We previously reported that in Arabidopsis, decapping of seed storage protein (SSP) mRNAs required P-body components including DCP1, DCP2 and DCP5 (Xu and Chua, 2009). Translational repression of these mRNAs was also impaired in single mutant deficient in any one of these three components such that SSPs accumulated in the mutants, resulting in post-embryonic lethality. Interestingly, DCP1 and DCP5 levels increased during seed maturation but decreased upon germination (Xu and Chua, 2009). Because dehydration occurs during seed maturation, we reasoned that changes in DCP1 and DCP5 levels might be signalled by stress and environmental cues. To explore this possibility, we investigated DCP1 levels in Arabidopsis seedlings subjected to dehydration. Figure 1A shows that control seedlings accumulated two DCP1-related bands around 50 kDa. Fifteen minutes after dehydration stress, there was a shift in abundance from the lower to the upper band, and at 60 min, the upper band represented about 90% of total DCP1. This shift in apparent molecular size was reversed by rehydration (Figure 1B). Phosphatase treatment (Peck, 2006) suggested that the upper band was likely a phosphorylated form of DCP1 (Figure 1C). This was confirmed by mass spectrometry, which also identified serine 237 (S237) as the phosphorylation site (Figure 1F). Based on this result, we engineered constructs to express two DCP1 mutants using the DCP1 native promoter, with DCP1-S237D mimicking the phosphorylated DCP1 and DCP1-S237A mimicking the non-phosphorylated form. The complementation lines were obtained and each DCP1 mutant was analysed by western blot. Different DCP1 mutant derivatives expressed in transgenic lines migrated differently, mimicking the modified and non-modified form of DCP1 (Figure 1D). These results confirmed S237 as the phosphorylation site in vivo. To rule out the existence of other phosphorylation sites, we analysed transgenic lines expressing DCP1-S237A subject to dehydration treatment. Figure 1E shows that S237 was likely the only site being modified since there was no change in the molecular size of DCP1-S237A upon dehydration.
Figure 1.
Characterization of DCP1 phosphorylation site. (A) Western blot analysis of DCP1 from seedlings subjected to dehydration stress. Samples were taken at different times after dehydration as indicated. Coomassie blue-stained rbcL (large subunit of ribulose-1,5-bisphosphate carboxylase) was used as a loading control. (B) DCP1 from dehydrated seedlings subjected to rehydration. (C) Western blot analysis of DCP1. Samples were either untreated or treated with λ phosphatase (λ-PPase) or with λ-PPase plus inhibitor mix. (D) Schematic presentation and western blot analysis of DCP1 and its S237 mutants in complementation lines. The phosphorylation site and mutated sites were highlighted. EVH1, Enabled/VASP homology 1. T, Trimerization domain. (E) Western blot analysis of transgenic plants expressing FLAG–DCP1-S237A subjected to dehydration stress. (F) MS–MS spectrum of the phosphorylated DCP1 peptide containing the phosphorylation site at S237. Phosphorylated DCP1 was digested with trypsin followed by LC-MS–MS analysis. The monoisotopic mass of the phosphopeptide ion was calculated at 1508.8 (MH1+). The fragment ion at 705.89 (MH22+) displays a strong ion signal intensity corresponding to the loss of phosphoric acid (98) from the precursor ion. Top panel shows the b- and y-fragment ions of the peptide. Both b- series and y-ion series confirmed S237 to be the phosphorylation site.
Phospho-DCP1 promotes mRNA decapping
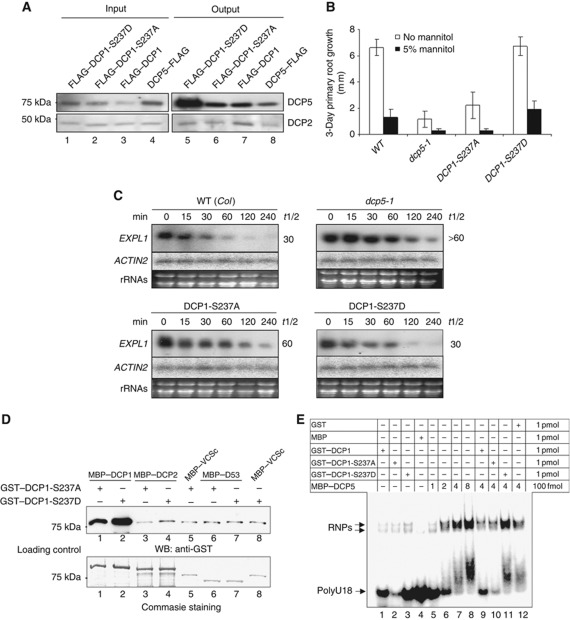
During seed development, phospho-DCP1 gradually accumulates likely resulting from the stresses imposed when the developing embryos become gradually dehydrated (Xu and Chua, 2009). Both phospho-DCP1 and DCP5 accumulate to high levels in dried seed, and consistently, DCP5 preferentially associates with phospho-DCP1 in vivo (Xu and Chua, 2009). Using transgenic plants expressing a transgene encoding either FLAG–DCP1-S237A or FLAG–DCP1-S237D, we found that DCP5 preferentially associated with FLAG–DCP1-S237D compared with FLAG–DCP1-S237A (Figure 2A). Together, these results suggest that DCP1 phosphorylation promotes its association with DCP5 and the association is not limited to seed development.
Figure 2.
Phospho-DCP1 promotes mRNA decapping in vivo. (A) Phospho-DCP1 preferentially associates with DCP5 in vivo. Co-immunoprecipitation was performed with extracts from 12-day-old seedlings expressing either FLAG–DCP1 or FLAG–DCP1 mutants or DCP5–FLAG. Anti-FLAG (mouse monoclonal) M2 affinity gel was used for immunoprecipitation and rabbit anti-DCP5 and anti-DCP2 antibodies were used to detect the related proteins in the immunoprecipitates. Input sample contained 5% extracts. (B) Three-day primary root growth of different mutants, complementation or overexpression lines. Error bars show standard deviations from 15 individual seedlings. Six-day-old seedlings were used as starting materials. (C) EXPL1 transcript accumulation in WT, dcp5-1, DCP1-S237A and DCP1-S237D. Individual transcript (labelled on the left) levels at different time points after cordycepin treatment were determined by northern blot using specific DNA probes. Estimated half-life (min) of specific mRNAs is given on the right of each panel. Ethidium bromide-stained rRNA bands were used as loading controls. (D) Effects of S237A mutation or S237D mutation on interaction between DCP1 and various proteins, as shown by in vitro pull-down assay. D53, the C-terminus of DCP5 (Xu and Chua, 2009); VCSc, the C-terminus of VCS (Xu et al, 2006). The last two proteins served as negative controls. The 75-kDa band is either GST–DCP1-S237A or GST–DCP1-S237D. Note that the former mutant protein migrated slightly faster than the latter mutant protein because of the amino acid change. (E) Effects of DCP1 mutants on DCP5/RNA RNP formation. Protein amounts used in EMSAs are indicated on top with amounts given on the far right column.
The rapid phosphorylation upon dehydration stress raises the possibility that DCP5 and DCP1 may function in mRNA turnover during stress responses and regulation. Although dcp1-1 mutants are arrested in early seedling development (Xu et al, 2006), we were able to recover dcp1-1 mutant lines complemented with DCP1-S237A or DCP1-S237D expressed from a DCP1 native promoter (Figure 1D). The T-DNA insertion in the DCP1 locus of the dcp1-1 mutant conferred resistance to sulfadiazin whereas the T-DNAs we subsequently introduced into dcp1-1 conferred resistance to glufosinate. We recovered >100 T1 plants resistant to both antibiotics. Since the transgene carried a FLAG tag it was possible to compare the expression levels of native DCP1 and FLAG-tagged DCP1 using appropriate antibodies. Among 15 T1 plants of each complemented line, we identified four putative homozygous dcp1-1 complemented with FLAG–DCP1-S237A and four complemented with FLAG–DCP1-S237D. Growth on double selection medium along with protein detection (a typical western blot was shown in Figure 1D) was used to select plants of the T2 and subsequent generations. Finally, we characterized three individual T4 lines homozygous with respect to both T-DNAs. The DCP1-S237D complementation line was indistinguishable from wild type (WT) with respect to plant morphology at various growth stages. By contrast, DCP1-S237A seed germinated and grew poorly, similar to dcp5-1, a knockdown mutant of DCP5 (Figure 2B). Both DCP1-S237A and dcp5-1 were also sensitive to osmotic stress (Figure 2B). The hypersensitivity was readily visible after 12 days of growth on treatment medium, such that these two lines were unable to produce lateral roots, in contrast to the typical response in WT (Supplementary Figure S1). These results suggest that the DCP5 association with phospho-DCP1 is indeed important for stress responses.
We previously reported that a typical unstable Expansin-Like1 (EXPL1) transcripts accumulated in the capped form in decapping mutant dcp1-1 with an estimated half-life of 85 min (Xu et al, 2006). In dcp5-1, EXPL1 transcripts were also stabilized with an estimated half-life of 60 min (Xu and Chua, 2009). We measured EXPL1 transcript stability in DCP1-S237A and DCP1-S237D using the same method as before. The estimated half-life of EXPL1 transcripts in DCP1-S237A was 60 min compared with the estimated half-life of 30 min in WT. This result suggests a partial complementation of dcp1-1 as EXPL1 transcripts were more stabilized in DCP1-S237A (Figure 2C). By contrast, the estimated half-life of EXPL1 transcripts in DCP1-S237D was 30 min, comparable to that in WT (Figure 2C). Notably, there was an apparent accumulation of EXPL1 transcripts in dcp5-1 and DCP1-S237A in comparison with WT and DCP1-S237D even without cordycepin treatment.
To relate the half-life of a transcript to its 5′end structure, we developed a 5′termini characterization assay (see Materials and methods) employing a 5′ phosphate-dependent exonuclease treatment and qRT–PCR. The estimated ratio of capped to uncapped EXPL1 transcripts in WT was 5.54 (Table I), whereas this ratio was increased to 16.22 in dcp5-1, suggesting an inhibition of the decapping process in the latter. In DCP1-S237A, the ratio was 9.48, which was about two-fold higher compared with WT, whereas in DCP1-S237D the ratio was 3.31 close to 5.54 in WT (Table I). These results suggest that EXPL1 transcript stability in different lines can be largely attributed to the difference in their mRNA decapping activities. Thus, DCP1 phosphorylation enhances mRNA decapping and contributes to, but not all of, mRNA decapping activities in vivo.
Table 1. Estimated ratio of capped to uncapped transcripts by transcript 5′ termini characterization assay.
| Normalized transcript levels (EXPL1) | Standard error of the mean (s.e.m.) | Ratio | ||
|---|---|---|---|---|
| WT | RNA1 | 0.70123 | 0.18555 | 5.54 |
| RNA2 | 0.12659 | 0.01671 | ||
| DCP1-S237A | RNA1 | 0.63824 | 0.21756 | 9.48 |
| RNA2 | 0.06731 | 0.01090 | ||
| DCP1-S237D | RNA1 | 0.46628 | 0.04558 | 3.31 |
| RNA2 | 0.14089 | 0.06345 | ||
| dcp5-1 | RNA1 | 0.60343 | 0.08410 | 16.22 |
| RNA2 | 0.03721 | 0.00547 | ||
| RNA1 refers to total RNA after terminator exonuclease treatment; RNA2 refers to total RNA subjected to apex alkaline phosphatase, tobacco acid pyrophosphatase and terminator exonuclease treatment (see Materials and methods). s.e.m.s were based on three biological replicates. | ||||
We also tested the interaction between GST–DCP1 mutant proteins and MBP–DCP2 in vitro. In these assays, we used amylose resins to retrieve the individual MBP-tagged proteins, which were used as baits. Antibody to GST was then used to detect the presence of GST–DCP1-S237A or GST–DCP1-S237D bound to the MBP-tagged protein. Figure 2D shows a significant increase in interaction between GST–DCP1-S237D and MBP–DCP2, as compared with GST–DCP1-S237A and MBP–DCP2. Moreover, GST–DCP1-S237D displayed a higher affinity for MBP–DCP1 compared with GST–DCP1-S237A (Figure 2D), suggesting that DCP1 trimerization (Tritschler et al, 2009) was also enhanced by S237 phosphorylation. The two GST–DCP1 mutant proteins showed equal binding to negative controls proteins D53 (a C-terminal fragment of DCP5; Xu and Chua, 2009) and VCSc (a C-terminal region of VCS; Xu et al, 2006).
DCP5 homologues CAR-1 and RAP55 have been identified as mRNA-binding proteins (Audhya et al, 2005; Tanaka et al, 2006). We tested RNA binding by electrophoretic mobility shift assay (EMSA) using in vitro purified proteins and a synthetic RNA containing 18 uridines (polyU18). Figure 2E shows that GST–DCP1 and mutants could not bind to RNA effectively whereas MBP–DCP5 alone was capable of binding to RNA. Because the DCP1 proteins carried a GST tag, we investigated the effect of GST on RNA binding. Figure 2E shows that addition of GST reduced formation of RNPs mediated by DCP5 (cf. lane 7 and lane 12). Using GST plus MBP–DCP5 as a control (lane 12), we found that compared with GST–DCP1-S237A (lane 10), addition of GST–DCP1-S237D to the MBP–DCP5/RNA mix (lane 11) significantly increased complex formation between MBP–DCP5 and RNA, suggesting that DCP1 phosphorylation promoted RNA/protein complex formation.
All together, these results suggest phosphorylation of DCP1 promotes mRNA decapping in vivo, presumably through mRNP formation by reinforcing its physical interaction with DCP5, DCP2 and DCP1 itself.
Global transcript level change upon dehydration stress
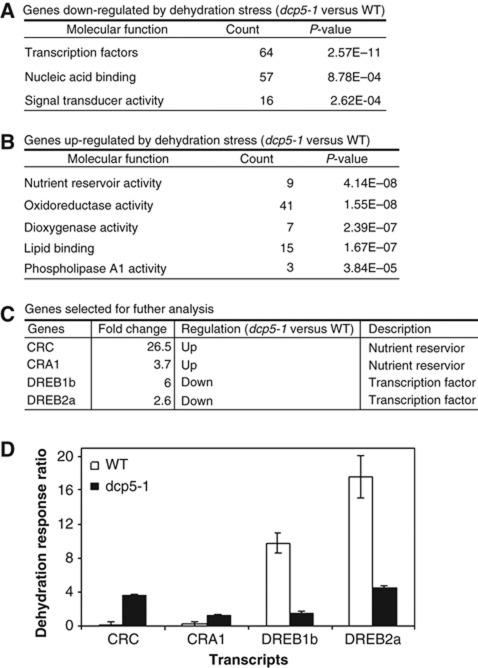
The apparent accumulation of EXPL1 transcripts in dcp5-1 and DCP1-S237A even without cordycepin treatment suggested that DCP1 phosphorylation and subsequent association with DCP5 may contribute to changes in global transcript levels upon dehydration stress. To test this hypothesis, we designed a two-colour expression microarray experiment using Arabidopsis V4 chips (Agilent). We isolated total RNAs from seedlings just before dehydration treatment and at 30 min after dehydration and RNA samples were labelled with different fluorescence dyes (cy3 or cy5). After data processing, we were able to calculate the response ratio (e.g., cy5/cy3) of each transcript, reflecting stress-induced changes in transcript levels in WT or dcp5-1. Six thousand and eighty-four out of 43 663 probes showed significant differences in transcript response ratio between WT and dcp5-1 (Student's t-test, P<0.05). Among them, 710 probes displayed at least a two-fold change in response ratio. Two hundred and eighty-two probes showed a response ratio up-regulated by over two-fold in dcp5-1 compared with WT. The number of probes for the down-regulated response ratio was 428, suggesting that relatively more genes in dcp5-1 could not respond to dehydration stress to the same extent as in WT (Supplementary Table S1). Gene ontology (GO) analysis revealed that genes impaired in their dehydration response in dcp5-1 were significantly enriched in those encoding transcription factors and related proteins (Figure 3A). By contrast, genes over-responsive to stress in dcp5-1 encoded proteins with nutrient reservoir activity or implicated in other metabolic pathways (Figure 3B) (Supplementary Table S2). These results indicate that during stress treatment, DCP5 is required for appropriate response of a subset of mRNAs.
Figure 3.
Microarray analysis of global transcript changes upon dehydration response. (A, B) List of GO terms corresponding to genes whose transcript response ratio is significantly down-regulated (A) or up-regulated (B) in dcp5-1. Details of each probe and their corresponding genes are listed in Supplementary Table S2. (C) List of representative genes selected for further analysis. Fold changes in dehydration response ratio in dcp5-1 compared with WT were shown. (D) Transcript response ratio in WT or dcp5-1 with dehydration treatment (30 min). Transcripts were analysed by qRT–PCR using total RNA as template. Transcript response ratio is defined as the ratio of transcript levels between dehydration stress and control. For details of individual transcript, see Supplementary Figure S2.
To test the reproducibility of the microarray data, we selected CRC (CR; Cruciferin), CRA1, DREB1b (DREB; dehydration responsive element binding) and DREB2a as representative genes (Figure 3C) and measured their transcript levels by qRT–PCR. DREB1b and DREB2a, which are typical dehydration stress responsive genes, encode AP2 domain-containing transcription factors (Oono et al, 2003), whereas CRA1 and CRC encode 12S globulins, which are typical SSPs. Figure 3D shows the estimated response ratio based on qRT–PCR results (Supplementary Figure S2). Compared with in WT, induction of both DREB1b and DREB2a in dcp5-1 was much reduced during dehydration stress, whereas CRC and CRA1 transcripts displayed over-responsiveness (Figure 3D). These results reconfirmed the microarray data.
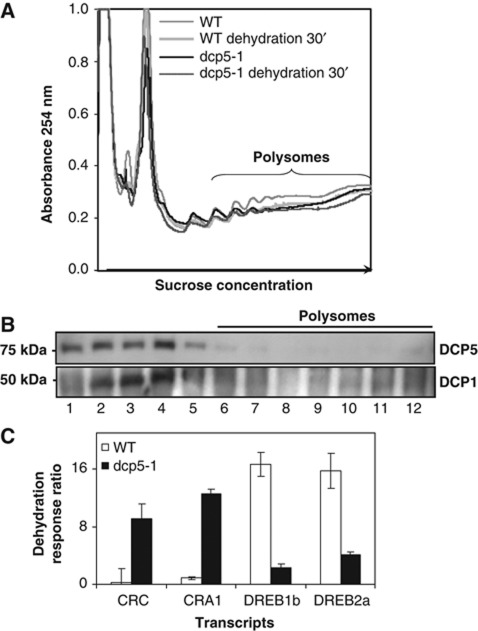
We also isolated polysomal RNAs (Figure 4A) and subsequently performed qRT–PCR (Supplementary Figure S2) using the same materials collected for microarray analysis. Since DCP5 protein was not detected in the polysomal fractions, this protein could not be responsible for the difference in polysomal mRNAs between WT and dcp5-1 (Figure 4B). Both CRC and CRA1 showed a much greater over-responsiveness to dehydration in dcp5-1 polysome-associated mRNAs compared with that in total RNAs (Figure 4C, for details see Supplementary Figure S2). Similar changes were detected in total RNA of WT and dcp5-1 but with a reduced induction ratio (Figure 3D; Supplementary Figure S2). In contrast to CRC and CRA1, the induction ratio of DREB1b and DREB2a transcript levels in polysomal RNAs was decreased in dcp5-1 compared with WT (Figure 4C, for details see Supplementary Figure S2). Response patterns of all four transcripts tested by using total RNAs were reproduced with polysome-associated RNAs, and for CRC/CRA1 the difference between WT and dcp5-1 was even amplified. This result indicates that transcripts with altered response ratio obtained from microarray analysis were largely determined by whether they were engaged in polysomes for active translation where DCP5 apparently played an important role in this process.
Figure 4.
Transcript response ratio in polysomal RNAs in WT or dcp5-1 treated with dehydration stress (30 min). (A) Polysome profiling of WT (Col.) and dcp5-1 with or without dehydration treatment. Polysomal RNAs were extracted from fractions 6–12 as labelled. (B) Proteins were collected for western blots using anti-DCP5 and anti-DCP1. (C) Transcript response ratio in WT or dcp5-1 with dehydration treatment (30 min). Transcripts were analysed by qRT–PCR using polysomal RNA as template. For details of individual transcript, see Supplementary Figure S2.
To test whether the global transcript changes can be attributed to altered mRNA decapping during dehydration, we again used the transcript 5′ termini characterization assay to estimate the ratio of capped to uncapped transcripts. For CRC transcripts this ratio was decreased from 11.65 to 0.01 upon dehydration in WT whereas in dcp5-1 (for unknown reason, the CRC transcript level in dcp5-1 was very low), this decrease was much reduced (Table II, for details see Supplementary Table S4). Considering that dcp5-1 is a knockdown mutant, this result suggested that in WT dehydration stress rapidly enhanced decapping of CRC transcripts via DCP5 in vivo. The similar decrease in WT but not dcp5-1 was also seen with CRA1 transcripts (Table II). However, in both WT and dcp5-1, the estimated ratios for DREB1b and DREB2a transcripts were increased (Table II). This result suggested that (1) DREB1b and DREB2a transcripts were not the direct targets of stress-triggered mRNA decapping and (2) the decreased induction ratio of DREB1b and DREB2a transcript levels in total RNAs and polysomal RNAs in dcp5-1 was not directly related to stress-triggered mRNA decapping.
Table 2. Estimated ratio of capped transcripts to uncapped transcripts during dehydration stress.
| Dehydration | WT |
dcp5-1
|
||
|---|---|---|---|---|
| 0 min/control | 30 min | 0 min/control | 30 min | |
| CRA1 | 3.10 | 1.54 | 3.05 | 3.20 |
| CRC | 11.65 | 0.01 | 0.08 | 0.01 |
| DREB1b | 3.15 | 7.13 | 3.86 | 9.55 |
| DREB2a | 5.40 | 12.99 | 7.06 | 14.59 |
| For details of each transcript, please see Supplementary Table S4. | ||||
MPK6 mediates DCP1 phosphorylation
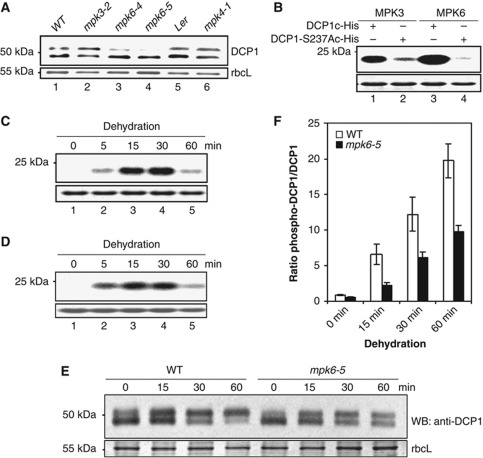
Sequence analysis surrounding S237 revealed a consensus sequence, S/T-P for phosphorylation by MPK (Figure 1D). Among the 20 MPKs in Arabidopsis MPK 3, 4 and 6 may act redundantly in abiotic stress responses (Colcombet and Hirt, 2008). Figure 5A shows that reduced levels of phospho-DCP1 were detected only in seedlings of mpk6-4 and mpk6-5 but not those of mpk3-2 and mpk4-1. We performed in vitro kinase assays using specific immunoprecipitates as enzyme sources (Merkouropoulos et al, 2008) and purified DCP1 C-terminal fragment (DCP1c) containing S237 as a substrate. Both MPK3 and 6 were capable of phosphorylating DCP1c but the activity was significantly compromised with DCP1-S237Ac, which carried the S237A mutation (Figure 5B). More important, we found anti-MPK6 immunoprecipitates from plants without stress treatment were unable to phosphorylate DCP1c but the MPK6 activity was rapidly induced 15 min after dehydration (Figure 5C). The same was seen with anti-MPK3 immunoprecipitates (Figure 5D). The reduced level of phospho-DCP1 in mpk6-5 mutant resulted in an altered ratio between the two bands recognized by DCP1 antibody (Figure 5E). The ratio change was also seen with mpk6 mutants under dehydration stress (Figure 5F). Taken together, these results indicate that MPK6 plays a major role in S237 phosphorylation of DCP1 during dehydration stress, whereas MPK3 plays only a minor role or its activity can be supplanted by those of other MPKs.
Figure 5.
MAPK6 mediates DCP1 phosphorylation in response to dehydration. (A) Western blot analysis of DCP1 protein in WT plants and different mpk mutants. (B) In vitro kinase assays using DCP1c-His and DCP1S237Ac-His as substrates incubated with anti-MPK6 or anti-MPK3 immunoprecipitates. DCP1c is the C-terminal region (192–367 aa) of DCP1 containing ser237. Extracts from seedlings after 30 min dehydration stress were used for immunoprecipitation. (C, D) In vitro kinase assay using DCP1c-His as a substrate incubated with anti-MPK6 (C) or anti-MPK3 (D) immunoprecipitates. Extracts used for immunoprecipitation were from seedlings subjected to dehydration for different periods. (B–D) Lower panels show the substrate band (DCP1c-His or DCP1S237Ac-His; stained with Coomassie brilliant blue), which was used as a loading control. Upper panels show phosphorylated substrates. (E) Western blot analysis of DCP1 in WT (Col.) and mpk6-5 at different time points after dehydration. Bottom panel shows rbcL (55 kDa) as the loading control. (F) Quantitative presentation of the ratio of phospho-DCP1 to DCP1 in WT and mpk6-5. Error bars represent standard deviations (n=3).
Discussion
Abiotic stresses are known to activate several plant MPKs but few stress-related downstream substrates have yet been identified. Here, we show that dehydration activates Arabidopsis MPK6 within 15 min and the activated MPK6 phosphorylates DCP1. As an adaptive response phospho-DCP1 promotes its association with DCP5, DCP2, and itself, and enhances decapping activity in vivo. The MPK6-DCP1–DCP5 pathway is important to global transcript changes during stress response, and stress-triggered decapping is essential for proper stress adaption. The evolutionary conservation of eukaryotic stress-related MPKs suggests that the signalling pathway we have uncovered, from MPK activation to DCP1 phosphorylation and RNP formation, may operate in other eukaryotes as well. Recently, the yeast DCP2 was found to be phosphorylated by Ste20 kinase (MAPKKKK) (Yoon et al, 2010), suggesting additional connections of diverse signalling pathways to P-bodies than previously envisioned.
DCP1 was first characterized in yeast as an essential component of a decapping complex required for cytoplasmic mRNA turnover (Beelman et al, 1996). Although DCP1 does not possess enzyme activity in vitro it interacts directly with DCP2 through an EVH1 domain to activate DCP2 decapping activity (Dunckley and Parker, 1999; Tharun and Parker, 1999; van Dijk et al, 2002; Sakuno et al, 2004; She et al, 2004). Moreover, DCP1 stimulates DCP2 activity by promoting and/or stabilizing DCP2/RNA complex (Deshmukh et al, 2008; She et al, 2008). In contrast to yeast DCP1, Arabidopsis DCP1 shares with human DCP1a a similar long unstructured region and a trimerization domain at the C-terminus (Tritschler et al, 2009). The single phosphorylation (S237) site we identified in plant DCP1 is located between the EVH1 domain and the C-terminal trimerization site, suggesting important function of this unstructured region previously unrecognized. Although S237 is not conserved in human DCP1a, the sites reported for DCP1a phosphorylation (Blumenthal et al, 2009) reside in a similar unstructured region containing S/T-P features (Supplementary Figure S3). In view of our results here it would be not surprising if the human kinase responsible for the human DCP1a phosphorylation is part of the MPK pathway during stress response.
Both transcriptional and post-transcriptional processes contribute to the global changes in transcript levels upon stress. By comparing the response ratio between WT and dcp5-1 in stress response, we were able to focus on a subset of transcripts specifically requiring DCP5 protein for their response. A subset of transcripts, for example, CRC and CRA1, was over-represented in dcp5-1 total RNA and polysomal RNAs, suggesting that in WT, these transcripts were direct targets of DCP5 for decapping. This was consistent with the results from 5′-termini characterization assay and the finding that phosphorylation of DCP1 signalled by MPK6 promoted association with DCP5 and enhanced mRNA decapping. The role of this subset of genes, including genes encoding SSPs, in the abiotic stress pathway, has not yet reported. Here, we provided evidence that decapping of their mRNAs could contribute to dehydration responses in plants. However, not all transcripts show over-responsiveness to dehydration stress in dcp5-1. Another subset of transcripts, including those of DREB1b and DREB2a, which are typical dehydration stress responsive genes (Oono et al, 2003), displayed a significantly reduced response in dcp5-1. In dcp5-1, transcripts in the under-represented category may not be able to effectively compete with those in the over-represented category for polysome occupancy. For example, DCP5 may specifically recognize a subset of mRNAs to form degrading RNPs and consequently allow newly synthesized mRNAs to enter polysome for translation. Our results hence provide evidence that decapping process is employed in dehydration stress response to globally regulate gene expression.
Many mRNAs including unstable mRNAs are stabilized in decapping-deficient mutants suggest a major role of decapping in cytoplasmic RNA decay (Xu et al, 2006). However, at a specific developmental stage, for example, post-embryonic growth, SSP mRNAs accumulate in decapping mutants at a much higher level as compared with other transcripts, suggesting that there is substrate specificity for decapping (Xu and Chua, 2009). Interestingly, many SSP genes belong to the group of genes encoding proteins with nutrition reservoir activity. It is not clear how the lifetime and fate of these mRNAs are determined in P-bodies. A future challenge is to identify sequence or structural specificity of these mRNAs.
Materials and methods
Plant material and stress treatment
Mutants dcp1-1 and dcp5-1 were previously characterized (Xu et al, 2006; Xu and Chua, 2009), mpk 6-5, mpk 6-4, mpk3-2 were from Kazuo Shinozaki (Takahashi et al, 2007), and mpk4-1 from John Mundy (Petersen et al, 2000).
Plants were grown on sterile solid half-strength Murashige-Skoog (MS) medium for 18 days before being transferred to a greenhouse under similar conditions (22°C, 16/8 h photoperiod cycles). For dehydration stress, 12-day-old seedlings were transferred onto 3 MM papers (Whatman) at 22°C exposed to 60% humidity at room temperature under dim light. After 30 min, the fresh weight of dehydrated seedlings was reduced to about 60%. Seedlings after 1 h of dehydration were returned to the original MS medium for rehydration. Osmotic stress treatment was carried out with the same MS medium but containing mannitol (5%).
λ Phosphatase treatment
λ Phosphatase treatment was performed according to Peck (2006).
LC-MS/MS to characterize phosphorylation site
The two bands corresponding to unphosphorylated and phosphorylated proteins were excised from gels, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide and then digested with Glu(C) followed by trypsin digestion at 37°C overnight. Digestion products were analysed by MALDI-TOF to identify phosphopeptides with a PerSeptive MALDI-TOF DE-STR mass spectrometer. Two ions were observed at 1508.8 and 1579.8 Da corresponding to the mass (m/z) of phosphorylated peptides APPQIQSPPPLQR and AAPPQIQSPPPLQR, respectively. These peptides were only found in the upper band. The cleavage at the Ala residue may be due to an unknown proteolytic activity from the Glu (C) enzyme. To determine the phosphorylation site(s), the tryptic peptides were subjected to LC-MS/MS analysis with a LTQ mass spectrometer.
Preparation of recombinant proteins
MBP–DCP1, MBP–DCP2, GST–DCP1, DCP1c-His were purified from Escherichia coli extracts (Xu et al, 2006). Quickchange site-directed mutagenesis kit (Stratagene) was used for mutagenesis to generate GST–DCP1-S237A, GST–DCP1-S237D, and DCP1-S237Ac-His. Freshly purified proteins are dialysed and stored at −80°C in storage buffer (20 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 1 mM DTT and 10% glycerol).
Constructs and transformation
DCP5–FLAG was constructed as described (Xu and Chua, 2009). A similar vector design was used for complementation using DNA sequences encoding FLAG–DCP1, FLAG–DCP1-S237A and FLAG–DCP1-S237D. A genomic fragment containing 6148 bp of the DCP1 locus was used, including 3044 bp upstream of the first ATG and 1059 bp downstream of the stop codon. The DNA fragment encoding the FLAG tag was inserted 3′ to the first ATG to produce FLAG–DCP1. Codon TCA, encoding S237 was changed to GCA (A237) and GAC (D237). The mutagenesis was performed using a Quickchange site-directed mutagenesis kit (Stratagene). Primer sequences are presented in Supplementary Table S3. The expression constructs were used to transform dcp1-1/DCP1-1 heterozygous plants and transformants were selected by resistance to both 10 mg/l glufosinate ammonium (T-DNA co-expressing DCP1s) and 52.5 mg/l sulfadiazin (T-DNA insertion in dcp1-1).
Transcript 5′ termini characterization assay
Three batches of RNA were prepared from total RNA. In all, 5 μg total RNA was treated with terminator 5′-phosphate-dependent exonuclease (Epicentre). The treated RNA was purified, precipitated and collected as RNA1 (representing capped transcripts). The second batch of 5 μg total RNA was treated sequentially with Apex alkaline phosphatase (Epicentre) and tobacco acid pyrophophatase (Epicentre), and followed by the terminator exonulease (Epicentre). RNA was purified and precipitated between each treatment step and finally collected as RNA 2 (representing uncapped transcripts). qRT–PCR was performed to measure transcript levels in each RNA pools. The relative transcript levels normalized to ACTIN2 levels were used to account for possible losses in ethanol precipitation steps. Tables I and II show estimated ratios of RNA1 (capped transcripts) to RNA2 (uncapped transcripts).
MAPK immunoprecipitation and in vitro kinase assays
MAPK immunoprecipitation and in vitro kinase assays were performed according to Merkouropoulos et al (2008) using 12-day-old seedlings subjected to dehydration. Briefly, 3 μl of each antibody and 30 μl Protein A-Sepharose (Santa Cruz Biotechnology) were incubated with protein extracts from 300 mg stress-treated seedlings for 2 h in 1 ml buffer (100 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5% glycerol, 0.1% Tween 20, 0.5% polyvinylpyrrolidine, 1 mM phenyl methyl sulfonyl fluoride). PhosSTOP (Roche) and proteinase inhibitor (Roche) were added to the incubation mix to block dephosphorylation and protein degradation. Immunoprecipitates (10 μl each) were incubated with substrates (10 μg) for 10 min at 30°C in a total volume of 20 μl in kinase buffer (50 mM Tris–HCl, pH 7.5, 10% glycerol, 150 mM NaCl, 5 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 0.2 mM ATP and 1 μCi [γ-32P] ATP). Reactions were stopped by adding SDS loading buffer and heating for 15 min at 65°C.
Immunoprecipitation and western blot
Immunoprecipitations and western blots were performed as described (Xu et al, 2006). Polyclonal antibodies against DCP1, DCP2 and DCP5 were generated in rabbits using recombinant proteins purified from E. coli. For immunoprecipitation of FLAG-tagged proteins, EZview red ANTI-FLAG M2 affinity gel (Sigma) was used.
Cordycepin treatments and northern blot analysis
Cordycepin treatments and northern blot analysis were performed according to Xu and Chua (2009).
Microarray analysis
The microarray data from this publication have been submitted to the Gene expression omnibusdatabase (http://www.ncbi.nlm.nih.gov/geo) and assigned the identifier GSE28493.
Polysomal RNA isolation
Polysomal RNA was isolated by differential centrifugation as described (Mustroph et al, 2009), except with the following changes: gradient (15–45% sucrose w/v) preparation and polysome fractionation were performed using a Biocomp gradient station and fractionation system following the manufacturer's instructions. UV absorbance data were obtained by Bio-Rad EM-1 Econo UV Monitor and were recorded digitally using a DI-154RS (DATAQ Instrument). Polysome fraction was collected starting from the absorbance peaks representing small polysomes (Figure 4A).
qRT–PCR
qRT–PCR was performed as described (Xu and Chua, 2009) using the CFX96 Real-Time PCR Detection System (Bio-Rad). Data were processed using CFX Manager Software (version 1.5). The comparative CT method was used to quantify the relative amounts of target gene transcripts. All qRT–PCR reactions were performed with an annealing temperature of 60°C and a total of 40 cycles of amplification. Primer sequences are presented in Supplementary Table S3.
Electrophoretic mobility shift assay
5′-End labelling was performed with T4 polynucleotide kinase and [γ-32P] ATP. Labelled poly U18 RNAs (20 000 c.p.m., 5 fmol) were incubated with proteins for 30 min at 4°C in 10 μl binding buffer (20 mM HEPES, pH 7.5, 100 mM KAc, 1 mM DTT, 0.1 mg/ml BSA, 2 U/μl RNaseout, 100 nM yeast tRNA). After addition of 2 μl glycerol, the mixture was loaded on 6% DNA retardation gel (Invitrogen) and electrophoresis was performed for 1 h at 100 V at room temperature. Dried gels were subjected to phosphorimaging.
Supplementary Material
Acknowledgments
We thank Haiteng Deng from the Rockefeller University Proteomics Resource Centre for help in characterizing the DCP1 phosphorylation site, Scott C Peck for providing anti-MPK3 and anti-MPK6 anti-bodies, Kazuo Shinozaki for mpk6-5, mpk6-4 and mpk3-2 mutants, John Mundy for mpk4-1 mutant and Manuel Ascano in Thomas Tuschl laboratory for advice on polysomal RNA isolation. This work was supported by NIH GM44640 to N-HC.
Author contributions: JX and N-HC conceived and designed the experiments, analysed the data and wrote the manuscript. JX performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR III, Desai A, Oegema K (2005) A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol 171: 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Blumenthal J, Behar L, Elliott E, Ginzburg I (2009) Dcp1a phosphorylation along neuronal development and stress. FEBS Lett 583: 197–201 [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R (2005) Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang D-U, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD (2008) mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell 29: 324–336 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Parker R (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Takeda A, Motose H, Watanabe Y (2007) Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett 581: 2455–2459 [DOI] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R (2004) Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur TE, Parker R (1998) Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J 17: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Andreasson E, Hess D, Boller T, Peck SC (2008) An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, Is a substrate of MAP kinases 3 and 6. J Biol Chem 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J (2009) Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109–126 [DOI] [PubMed] [Google Scholar]
- Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, Satou M, Sakurai T, Ishida J, Akiyama K, Iida K, Maruyama K, Satoh S, Yamaguchi-Shinozaki K, Shinozaki K (2003) Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J 34: 868–887 [DOI] [PubMed] [Google Scholar]
- Peck SC (2006) Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J 45: 512–522 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Sakuno T, Araki Y, Ohya Y, Kofuji S, Takahashi S, Hoshino S, Katada T (2004) Decapping reaction of mRNA requires Dcp1 in fission yeast: its characterization in different species from yeast to human. J Biochem 136: 805–812 [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H (2004) Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol 11: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H (2008) Structural basis of Dcp2 recognition and activation by Dcp1. Mol Cell 29: 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M (2006) RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem 281: 40096–40106 [DOI] [PubMed] [Google Scholar]
- Tharun S, Parker R (1999) Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151: 1273–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, Weichenrieder O (2009) DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc Natl Acad Sci USA 106: 21591–21596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V (2008) Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol 28: 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O (2007) A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol 27: 8600–8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C, Velasco C, Ptushkina M, McCarthy JE (2000) The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J 19: 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2009) Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Choi EJ, Parker R (2010) Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol 189: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.