Abstract
The functional output of the genome is closely dependent on its organization within the nucleus, which ranges from the 10 nm chromatin fiber to the three-dimensional arrangement of this fiber in the nuclear space. Recent observations suggest that intra-and inter-chromosomal interactions between distant sequences underlie several aspects of transcription regulatory processes. These contacts can bring enhancers close to their target genes, or prevent inappropriate interactions between regulatory sequences via insulators. In addition, intra- and inter-chromosomal interactions can bring co-activated or co-repressed genes to the same nuclear location. Recent technological advances have made it possible to map long-range cis and trans interactions at relatively high resolution. This information is being used to develop three-dimensional maps of the arrangement of the genome in the nucleus and to understand causal relationships between nuclear structure and function.
Keywords: Chromatin, Epigenetics, Transcription, Nucleus
Introduction
The genome of an organism is packed in the nuclei of its cells in a manner that ensures its safe storage, duplication, transmission and expression. This packaging must allow for an easy transition between the relatively uncompacted state during interphase and the highly compacted state during mitosis, while enabling the transmission of genetic and epigenetic information between mother and daughter cells. The question of how DNA is arranged and organized in the eukaryotic nucleus is a critical issue in understanding nuclear processes but one that, until recently, was difficult to address experimentally at an informative level of resolution. The introduction of 3C-related techniques by Job Dekker and collaborators has dramatically changed the landscape of the transcription field by affording the possibility of mapping inter- and intra-chromosomal interactions at high resolution and, therefore, analyzing the relationship between nuclear organization and gene expression (Dekker, Rippe et al. 2002).
Based on our current knowledge of nuclear biology, one could, a priori, speculate on the possible existence of various types of interactions among DNA sequences in the nucleus. It is possible that many interactions that can be measured between different DNA sequences are a consequence of the need for the DNA to be highly folded in order to fit in the nuclear space. If this is the case, these contacts may be random and may not be detected when interactions are measured in a population of cells. It is therefore likely that detectable interactions have biological significance. Some of these intra- and inter-chromosomal interactions may play a structural role in maintaining the genome in an arrangement that facilitates folding of chromosomes during interphase and mitosis. It is possible that these interactions are important in decondensing the genome during anaphase/telophase to ensure rapid expression of genes that need to be transcribed at the M/G1 transition (Strukov et al. 2011). At the other end of the spectrum, other interactions may represent contacts between distal enhancers and promoters in which enhancer-bound transcription factors and the components of Mediator and the transcription complex are directly involved. Finally, a third type of interactions may organize the chromatin to juxtapose very distally-located enhancers and promoters or to ensure that regulatory sequences of a gene do not act on the promoter of a different gene. Included in this class of interactions are those whose function is to bring together groups of co-regulated genes to a specific transcription factory or groups of co-repressed genes to Polycomb bodies (Bantignies and Cavalli 2011; Schoenfelder et al. 2010). It is therefore likely that some intra- and inter-chromosomal interactions are established as a consequence of genome activity i.e. transcription and replication, whereas other interactions may have an organizational role and inform genome function.
Whether inter- and intra-chromosomal interactions have a primary structural role with a secondary functional effect or vice-versa, the ultimate outcome of these interactions is the establishment of a specific three-dimensional arrangement of the genome within the nucleus. The close relationship between organization and function of the genome supports the possibility that this arrangement may be cell-type specific and that it may directly correlate with the functional output of the genome in a particular cell-type. As a consequence, nuclear organization may be a fingerprint of cell identity and a blueprint of the transcriptional output of the cell. This organization should be epigenetically inherited and integrate all other epigenetic information contained in the 10 nm chromatin fiber, such as DNA methylation and histone covalent modifications. Work in the past few years on various aspects of nuclear biology has begun to deconstruct the three-dimensional organization of the DNA in the nucleus by analyzing the role of specific intra- and inter-chromosomal interactions on distinct aspects of transcription regulation.
Regulation of transcription by interactions between the promoter and terminator
Studies carried out in yeast have detected strong interactions between the promoter and termination sites of various genes transcribed by RNA polymerase II (RNAPII) (Ansari and Hampsey 2005; El Kaderi et al. 2009; O'Sullivan et al. 2004; Singh and Hampsey 2007). The presence of these contacts appears to be a general phenomenon that is not restricted to long transcription units but it is also observed in genes as short as 1kb. Promoter-terminator interactions have also been described in mammalian cells at the breast cancer BRCA1 gene (Tan-Wong et al. 2008), at the immunohistological marker CD68 gene (O'Reilly and Greaves 2007) and at proviral HIV-1 integrants (Perkins et al. 2008). One possible explanation for these interactions is that RNAPII and other general transcription factors released after termination of transcription can be efficiently recycled and reused for the initiation of the next round of transcription, since the terminator and promoter are closely positioned as a consequence of these contacts (Figure 1B) (Ansari and Hampsey 2005; Mapendano et al. 2010); nevertheless, there is no evidence at this time to support this hypothesis. While promoter-terminator contacts seem to be dispensable for normal transcription, their presence correlates with rapid reactivation of transcription. This suggests that promoter-terminator interactions are linked to transcription memory in the sense that, when the gene is re-activated following a transient silencing period, such as mitosis, activation of transcription requires a shorter period of stimulation than during the first round. For several inducible genes in yeast, the maintenance of these memory gene loops confers rapid re-activation of transcription involving faster RNAP II recruitment to the gene upon induction following intervening periods of transcriptional repression (Tan-Wong et al. 2009). However, this faster response to induction is not observed for genes lacking memory gene loops. Interestingly, some of these loops have been found interacting with the nuclear pore complex through association with myosin-like protein 1 (Mlp1). This might suggest that memory gene loops readily facilitate reactivation of transcription by efficiently reusing limited transcription machinery proteins when responding to changing environmental conditions (Tan-Wong et al. 2009). However, promoter-terminator loops can be maintained in snf2 mutants in which transcription memory is lost, suggesting that gene looping may not be sufficient for transcription memory under conditions allowing normal transcription (Laine et al. 2009). In mammalian cells, gene looping may not necessarily play a role in transcription or transcription memory. For example, promoter-terminator interactions actually disappear when the BRCA1 gene is highly transcribed (Tan-Wong et al. 2008). The formation of this loop is also not strictly related to transcription activity of the CD68 gene (O'Reilly and Greaves 2007). Thus, promoter-terminator interactions may play limited roles in transcription re-activation and memory in organisms other than yeast, and their contribution to the establishment of a three-dimensional organization of the DNA in the nucleus may not be extensive in higher eukaryotes.
Figure 1.
Summary of insulator function in nuclear organization and gene expression. A. Linear organization of a typical eukaryotic gene. The RNA coding region of the gene is represented by a green arrow, the transcription complex and RNA polymerase II is denoted by an orange oval and the Mediator complex by a green sphere. The cohesion complex is indicated by a red ring; cohesion is also found at some enhancers (not shown). Enhancers in the upstream regulatory region of the gene are indicated by blue ovals of different hues. Insulators are represented by pink spheres. B. Three-dimensional arrangement of the same region represented in panel A. The 3' region of the gene (terminator) contacts the promoter to form a gene loop, a phenomenon that has been observed more frequently in yeast cells. The most proximal enhancer (E1) contacts Mediator and/or the transcription complex; cohesion stabilizes this interaction. Insulator elements, such as CTCF in vertebrates, contact each other to form a loop; this interaction is also mediated by cohesion. As a consequence of the formation of this loop, Enhancer E2 is unable to act on the promoter of the gene while enhancer E3 is brought close to the promoter to activate transcription. C. Many insulator sites come together at one nuclear location to form insulator bodies. This arrangement is similar to that formed by PREs and PcG proteins, which come together at Pc bodies (see Figure 2C).
Enhancer-promoter interactions
The question of how an enhancer activates its target gene over long linear distances has been one of the driving forces behind initial attempts to study whether interactions take place between distant DNA sequences. Out of the various models proposed to explain the mechanisms of enhancer function (Blackwood and Kadonaga 1998; Bulger and Groudine 1999; Dorsett 1999; Ptashne 1986), the looping model has received increasing experimental support from studies using chromosome conformation capture (3C) techniques (Bulger and Groudine 2011; Dekker et al. 2002). It is therefore likely that many of the contacts that contribute to the establishment of a nuclear organization pattern are based on enhancer-promoter interactions (Figure 1A and 1B).
The first attempt to investigate a direct physical interaction between an enhancer and a promoter was carried out nearly two decades ago (Cullen et al. 1993). This report showed a direct interaction between an enhancer and a promoter after estrogen receptor binding at the enhancer region. Since the invention of 3C (Dekker et al. 2002), this and related methodologies have become standard for the investigation of chromatin interactions at different genomic scales, resulting in a wealth of new information demonstrating the requirement of these contacts for activation of transcription (Dostie et al. 2006; Fullwood et al. 2009; Lieberman-Aiden et al. 2009; Simonis et al. 2006; Tiwari et al. 2008; van Steensel and Dekker 2010; Zhao et al. 2006).
One of the earliest applications of 3C in mammalian system was the analysis of interactions in the mouse β-globin locus (Tolhuis et al. 2002). These studies revealed that DNaseI hypersensitive sites within the Locus Control Region (LCR) come into physical proximity with the active globin genes and form an active chromatin hub (ACH) in which DNA containing the inactive β-globin genes loops out (Osborne et al. 2004; Tolhuis et al. 2002). Interestingly, the β-globin locus reorganizes during cell differentiation as the different globin genes are sequentially switched on or off. As interactions of newly activated globin genes with the LCR are established, the loop between the LCR and the globin gene being silenced is disrupted. Thus the ACH dynamically reforms as a new activated gene moves in and a silenced gene moves out (Osborne et al. 2004). This switch happens in part because transcription factors required for globin gene expression are made at different stages during erythroid differentiation.
Studies at the Kit locus have shown that the exchange of transcription factors can effectively reorganize the chromatin to influence enhancer-promoter interactions and affect gene expression during differentiation. GATA-2 and GATA-1 are erythroid specific transcription factors recognizing identical consensus sequences and the expression of GATA-1 follows the silencing of GATA-2 during erythroid differentiation. Prior to GATA-1 expression, an upstream enhancer is bound by GATA-2 and forms a loop with the promoter-proximal region. Conditional activation of GATA-1 abolishes GATA-2 occupancy at the Kit locus and disrupts enhancer-promoter interactions. Interestingly, new loops are also formed among downstream GATA binding sites (Jing et al. 2008). These results suggest that different GATA proteins exert distinct effects on chromosome conformation and that GATA factor exchange at the Kit locus reorganizes physical contacts to form new loops and prevent the interaction of the Kit gene with upstream enhancers.
In the case of the mammalian β-globin locus, multiple protein factors are required for the formation of chromatin loops. GATA-1 and its partners FOG-1, EKLF and Nli/Ldb1 are enriched at both the LCR and the promoter region of the beta globin genes and are necessary for their interaction (Drissen et al. 2004; Song et al. 2007; Vakoc et al. 2005). It is not clear whether simultaneous binding of the same protein factors at separate sites is required for the establishment of long-range interactions. Nevertheless, such binding pattern may increase the probability of association because the interacting factors can dimerize, thus acting as a bridge between the LCR and the promoter (Song et al. 2007). How various erythroid transcription factors and their partners coordinately work together during differentiation to reorganize the globin locus and establish dynamic enhancer-promoter interactions is still unknown. Interestingly, EKLF not only mediates cis interactions within the globin locus but is also able to establish a more global network with genes located in the same or different chromosomes (Schoenfelder et al. 2010). De Laat and collaborators have recently explored the mechanisms by with the LCR can interact with sequences located in the same or different chromosomes by using transgenic mice in which the human β-globin LCR is inserted into a gene-dense region of mouse chromosome 8. Using 4C, the human LCR was shown to contact genes controlled by EKLF and GATA-1 both in cis and trans (Noordermeer et al. 2011).
It appears that the use of long-range enhancer-promoter interactions is a general phenomenon in the activation of gene expression. Activation of target genes by regulation of long-range interactions has also been reported in many other genes, in different model organisms, and involving a variety of proteins (Kagey et al. 2010; Lin et al. 2007; Melnikova et al. 2008; Ong and Corces 2011; Ren et al. 2011). It is not clear whether different enhancer-bound transcription factors use the same mechanistic principles to interact with components of the transcriptional machinery at the promoter. This will likely determine the frequency and stability of enhancer-promoter contacts and the contribution of these interactions to a heritable pattern of this aspect of nuclear organization.
Insulator-mediated interactions
The ability of enhancers to interact with and activate promoters over long linear distances raises the question of how the genome prevents inappropriate interaction of an enhancer with non-target genes. Insulators appear to regulate this facet of genome function by ensuring that enhancers target the appropriate promoter, by preventing inappropriate interactions between enhancers and promoters or by preventing the spreading of repressive or active chromatin (Wallace and Felsenfeld 2007; Yang and Corces 2011; Zhao and Dean 2005). Barrier insulators have been shown to interfere with the spreading of repressive chromatin by recruiting chromatin remodeling enzymes, whereas enhancer-blocking insulators appear to function by mediating intra- or inter-chromosomal interactions. We will limit our remarks to insulators that function by the latter mechanism.
Observations at multiple gene loci in vertebrate cells suggest that enhancer-blocking insulators can come into physical contact through interactions mediated by the protein CTCF, which can dimerize and form loops of the intervening DNA. In experiments in which CTCF separates the enhancer and promoter, these two sequences become allocated to different loops and the enhancer is unable to contact the promoter (Figure 1B) (Hou et al. 2008; Yusufzai et al. 2004). Drosophila contains several different insulators named after their DNA binding proteins, including Su(Hw), dCTCF and BEAF; each of these insulators also contains the CP190 protein, which is necessary to mediate interactions among individual insulator sites (Bushey et al. 2008; Bushey et al. 2009). It has been shown that a Drosophila insulator containing the dCTCF and CP190 proteins is induced at the Eip75B gene after cells are treated with the hormone ecdysone. This insulator prevents an ecdysone enhancer from activating transcription of genes that are not regulated by this hormone (Wood et al. 2011). In other situations, two CTCF sites located between a distal enhancer and a promoter can interact to bring the enhancer close to the promoter and activate transcription (Figure 1B) (Handoko et al. 2011; Xu et al. 2011). The effect of insulators is not limited to the regulation of enhancer function. In Drosophila, two separate Su(Hw) insulators can form a loop to bring an upstream PRE element together with a downstream target gene to mediate repression (Comet et al. 2011). These results suggest that the outcome of insulator-mediated interactions is context-dependent, and it differs depending on the location of interacting insulator sites with respect to other regulatory sequences.
Insights into the molecular mechanisms by which insulators establish and/or maintain these loops have come from the finding that CTCF co-localizes with cohesin at many sites in the genome of vertebrate cells (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008). This observation has led to the suggestion that cohesin, whose best known function is to maintain chromatids together between S phase and anaphase, may play a similar role in maintaining together CTCF-based loops (Figure 1B). Other recent evidence also suggests that CTCF and cohesin can work independently or coordinately to mediate long-range interactions at different gene loci during development and differentiation (Degner et al. 2011; Dorsett 2011; Hou et al. 2010; Kagey et al. 2010; Seitan et al. 2011). CTCF and cohesin colocalize and mediate long-range interactions to activate transcription of the developmentally regulated cytokine IFNG gene, which is disrupted when cohesin is knocked down without affecting the binding of CTCF (Hadjur et al. 2009). CTCF and cohesin were also shown to be inter-dependent in the establishment of long-range interactions in a 2 Mb region of human chromosome 11 encompassing the β-globin locus and flanking olfactory receptor genes (Hou et al. 2010). Interestingly, cohesin can also mediate functional interactions independent of CTCF. In ES cells, cohesin was found to colocalize with mediator at many enhancers and promoters, suggesting that these proteins can more directly facilitate communication between enhancers and promoters in a CTCF-independent manner Figure 1A and 1B) (Kagey et al. 2010). In Drosophila, interactions between multiple insulators appear to come together at specific nuclear locations to form insulator bodies (Figure 1C) (Bushey et al. 2009). Given the extent of the involvement of insulators in mediating interactions among different sequences in the genome, these elements are likely to be one of the main contributors to the establishment of the three-dimensional organization of the chromatin fiber.
The effect of insulators on nuclear function is not limited to transcription, in agreement with the idea that the role of insulators is to mediate interactions and that the outcome depends on the nature of the sequences involoved in these contacts. For example, insulators regulate V(D)J recombination at the Igh locus by bringing together distant sequences to undergo specific patterns of recombination. DNA rearrangements in the Igh locus of pro-B cells are under temporal and spatial regulation during B cell development in a process that initiates with DH to JH rearrangement followed by rearrangement of a VH gene segment to DHJH. CTCF insulators are critical for the implementation of this complex pattern of DNA rearrangements. Around 60 CTCF sites are present throughout the VH region as well as 2 additional clusters within other parts of the Igh locus in pro-B cells(Degner et al. 2011; Guo et al. 2011; Guo et al. 2011). These two clusters are present next to DH and the 3' regulatory region of JH, and they strongly interact in pre-pro-B and pro-B cells to stimulate the selection of DH over VH promoters before initiation of DH-JH. These CTCF sites also interact with an intronic enhancer (Eμ), which is required for the antisense transcription of DH. Antisense transcription through the DH locus precedes DH-JH rearrangement and is probably important to make the DH region accessible for subsequent recombination. Thus, CTCF-mediated interactions select DH over VH promoters for antisense transcription and bring together DH and JH instead of VH. Later, in pro-B cells, the locus compacts to bring VH genes close to the DH-JH region through interactions that also depend on CTCF(Degner et al. 2011; Guo et al. 2011; Guo et al. 2011). As a consequence, insulator-mediated chromosome interactions regulate V(D)J recombination both spatially and temporally.
Polycomb-mediated long-range repressive interactions
Long-range interactions have also been shown to underlie the process by which Polycomb (Pc) represses transcription (Figure 2). The mechanisms and significance of these interactions have been best characterized in Drosophila (Bantignies et al. 2011; Lanzuolo et al. 2007; Tiwari et al. 2008). The Pc complex is involved in the repression of Drosophila Hox genes during development; these genes are located in two different clusters separated by more than 10 Mb on the same chromosome arm (Figure 2A). Polycomb-Group (PcG) proteins co-localize in the nucleus in structures named Pc bodies, where the Hox genes are present and are co-regulated to maintain the identity of body segments in the anterior-posterior axis (Figure 2B). A recent study found that Hox genes only colocalize in Pc bodies in tissues where these genes are repressed. For example, the Antennapedia (Antp in the ANT-C domain) and abdominal-B (abdB in the BX-C domain) genes colocalize in the nucleus and this colocalization depends on PcG proteins (Figure 2C). Experiments using 4C have revealed that the long-range interactions between Antp and abdB are mediated by two Pc Response Elements (PREs), Fab7 and Mcp, present in the abdB gene. Deletion of Fab-7 results in a reduction of the interactions between Antp and abd-B while at the same time decreasing the expression of genes in the ANT-C domain (Bantignies et al. 2011). Interactions mediated by PREs and PcG proteins that result in repression of Hox gene expression is a conserved feature of Hox gene regulation in both Drosophila and mammalian cells (Bantignies and Cavalli 2011).
Figure 2.
Organization of the Drosophila bithorax Complex (BX-C). A. linear arrangement of the three genes in the BX-C, which are indicated by green arrows; orange ovals represent the transcription complex at the promoter of each gene whereas red spheres represent PREs and associated proteins. B. Interactions among the PREs and promoters of the genes result in a specific three-dimensional arrangement of the locus that results in repression of transcription. C. Multiple Hox gene loci can be co-repressed and associate at nuclear locations termed Pc bodies.
The results discussed above suggest that Pc proteins bound to PREs can mediate interactions among Pc target genes within a region of several hundred kilobases and also over tens of megabases. Is it then possible that Pc proteins can contribute to the formation of a genome-wide interaction network among PREs? To answer this question, 4C studies on several PcG target genes were conducted and results of these studies demonstrated that an extensive interacting network specific for PcG target genes does exist in the nucleus. Interestingly, interactions in this network are mostly limited to genes within the same chromosome arm. Using a Drosophila strain carrying an inversion between the left and right arms of the third chromosome, it was found that interactions were only formed within each new arm. These results suggest that each Drosophila chromosome arm is a spatially distinct domain that potentially limits interactions between chromosome arms on either side of the centromere (Tolhuis et al. 2011).
Long-range interactions and the regulation of imprinted genes
As is the case of co-repressed Hox genes, groups of co-regulated imprinted genes can form an imprinting interactome. Long range intra- and inter-chromosomal interactions between various regulatory sequences have been linked to parent of origin specific regulation of gene expression. This phenomenon has been studied in detail in the mouse Igf2/H19 locus. The imprinting control region (ICR) in this locus is responsible for the regulation of allele-specific expression of Igf2 from the paternal allele and H19 from the maternal allele (Edwards and Ferguson-Smith 2007). The DNA of the Igf2/H19 ICR is methylated on the paternal but not on the maternal alleles. CTCF can bind to the DNA when its recognition sequence is not methylated. As a consequence, the differential methylation status of the maternal and the paternal chromosomes results in distinct patterns of three-dimensional arrangements of the DNA at this locus. One arrangement favors expression of H19 from the maternal allele whereas an alternative organization has the opposite effect and favors Igf2 expression from the paternal allele. Results from 3C experiments show that the ICR on the mouse maternal allele interacts with CTCF sites in the upstream differentially methylated region 1 (DMR1) and downstream MAR3 sites that flank the Igf2 gene. The structure thus formed prevents the accessibility of the enhancer to Igf2, which is enclosed in a separated domain (Kurukuti et al. 2006; Murrell et al. 2004). In human cells, the maternal ICR interacts with a CTCF site located downstream of the shared enhancer, creating a loop that encloses the enhancer and prevents its interaction with Igf2 (Nativio et al. 2009). This interaction between the ICR and CTCF does not take place in the paternal allele. In mouse cells, without CTCF bound to the ICR on the paternal allele, the ICR contacts the DMR2 site located downstream of Igf2, which allows the interaction between the enhancer and the Igf2 promoter (Kurukuti et al. 2006; Murrell et al. 2004). These results show that the imprinted expression of Igf2/H19 is dependent on a specific three-dimensional organization unique for each allele, which in turn is due to different CTCF binding patterns effected by distinct DNA methylation profiles.
CTCF-mediated interactions are not limited to sequences located within an imprinted locus. Using 4C and modified 3C experiments, several groups have found the existence of trans interactions between Igf2/H19 and imprinted genes on other chromosomes. Examples of sequences that interact with Igf2/H19 are the intergenic region between the Wsb1 and Nf1 genes on paternal chromosome 11, the Abcg2 gene on chromosome 6 and Osbpl1a on chromosome 18 (Ling et al. 2006; Zhao et al. 2006). Knocking down CTCF or mutation of the maternal Igf2/H19 ICR abolishes the interactions among the imprinted regions. Moreover, loss of CTCF binding to the maternal Igf2/H19 ICR also leads to miss-regulation of imprinted genes normally associated in trans (Zhao et al. 2006). Interestingly, imprinted loci were found overrepresented among the regions involved in inter-chromosome interactions with the H19 ICR. The clustering of imprinted genes by inter-chromosome interactions, termed the imprinting interactome, may facilitate the regulation of these genes in trans. Therefore, CTCF appears to function as a central mediator that brings these various imprinted genes together. Interestingly, CTCF binding sites are not notably enriched at these imprinted loci. A plausible explanation for this phenomenon is that CTCF might mediate the imprinted trans network of genes through interactions with unidentified protein factors associated with these imprinted genes.
Inter-chromosomal interactions during X-chromosome inactivation in mammals
Interphase pairing of homologous chromosomes is rare in metazoans but it does take place. Phenomena such as transvection and paramutation have been know for many years. One of the best studied cases at the molecular level, and of special interest in the context of CTCF function, is X chromosome inactivation in mammalian female embryonic cells during the early stages of development (Augui et al. 2011; Wutz 2011). Before deciding which X chromosome to inactivate, the two X chromosomes need to pair briefly through an unknown mechanism (Augui et al. 2007; Bacher et al. 2006; Xu et al. 2006). Random collision facilitated by related protein factors could be the initial force driving the two X chromosomes to contact each other (Scialdone and Nicodemi 2008). However, specific pairing at the X-pairing region (Xpr), though controversial (Sun et al. 2010), followed by pairing at the X inactivation center (XIC) suggests that a sophisticated mechanism exists (Augui et al. 2007). Autosomes carrying insertions of either Xpr or XIC can undergo pairing with the X chromosome at the same developmental stage while reducing the X: X pairing rate (Augui et al. 2007; Xu et al. 2006). Although the mechanism by which the transient X:X pairing is initiated is still elusive, it has been found that knocking down either CTCF or Oct4 prevents homologous association. Moreover, depletion of CTCF results in a loss of X inactivation, whereas lack of Oct4 leads to silencing of both X chromosomes (Donohoe et al. 2009; Xu et al. 2007). Although further work is necessary to ensure that the role of CTCF is direct, these results support a function for this protein in the establishment of inter-chromosomal interactions.
Towards a global nuclear interactome
Observations described in the previous sections point to the existence of a multitude of interactions among specific sequences in the genome. Some of these interactions are intra-chromosomal and serve to bring together regulatory sequences of individual genes in order to activate or repress their transcription. Other interactions are inter-chromosomal and serve to bring together genes that are co-activated or co-repressed. These interactions either precede or are a consequence of transcription. There may be other physical contacts among DNA sequences that serve an organizational role, with the goal of facilitating the transition between various phases of the cell cycle or to maintain broad patterns of epigenetic memory required for the establishment of specific cell fates. Finding these interactions require genome-wide maps of physical contacts at kb resolution.
Towards reaching this goal, a genome-wide interactome of estrogen receptor alpha (ER-alpha) binding sites has been recently completed using chromatin interaction analysis by paired-end tag (ChIA-PET) Results from this analysis suggest that ER-alpha functions by extensive chromatin looping to bring genes together for coordinated transcriptional regulation. These studies further support the idea that interactions among multiple sites in the genome constitute a primary mechanism for regulating transcription (Fullwood et al. 2009).
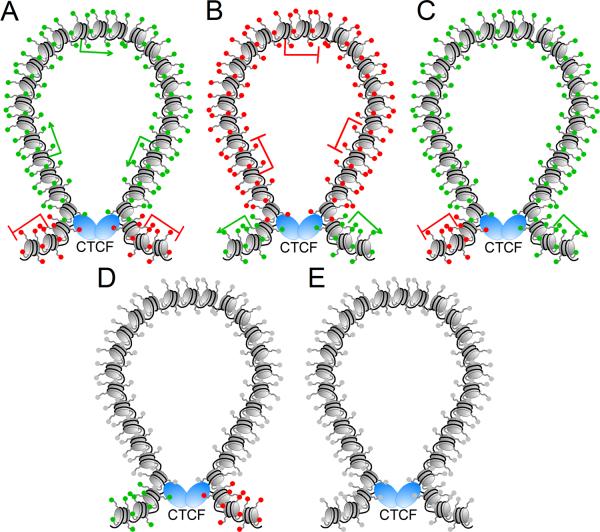
More recently, experiments to obtain a genome-wide CTCF-chromatin interactome in mouse ES cells have been carried out using ChIA-PET (Handoko et al. 2011). A total of 1,480 cis- and 336 trans-interacting loci were identified in this study. These contacts may represent just a small fraction of all the loops mediated by CTCF and its partner proteins in the nucleus. These interactions establish five distinct chromatin domains by delineating the boundaries of various linearly arranged active and repressive chromatin regions (Figure 3). Out of these five chromatin domains, no significant percentage of loops is established within the same active or repressive chromatin region, which further confirms the boundary function of CTCF. Another interesting finding is that CTCF-mediated loops shorten the distance between enhancers or PREs and promoters, thus facilitating activation or repression of transcription as earlier found at specific genes (Comet et al. 2011; Xu et al. 2011). These results confirm the hypothesis that interactions mediated by CTCF are not limited to insulator function but represent a general mechanism to bring together various regulatory sequences (Phillips and Corces 2009; Yang and Corces 2011). These findings present an interesting though static picture of the three-dimensional organization of the genome in one cell type. Genome-wide mapping of CTCF in different cell types revealed that, although many sites are constant across different cells, a fraction of them change from one cell type to another (Barski et al. 2007). This may suggest that part of the CTCF interactome could be conserved while some of the interactions are cell type-specific and change during cell differentiation to mediate specific functions.
Figure 3.
Domains created by interactions between CTCF insulators in mouse embryonic stem cells. Actively transcribed genes are represented by a green arrow and silenced genes by a red line; nucleosomes and the histone tails are represented in grey, with active histone modifications indicated as green spheres and repressive modifications as red spheres. DNA is represented in black and CTCF as blue ovals. A. CTCF forms a loop to separate a domain containing active histone modifications and transcribed genes from repressive marks and silenced genes. B. CTCF forms a loop to separate a domain containing repressive histone modifications and silenced genes from active marks and transcribed genes. C. CTCF forms a loop containing nucleosomes enriched in mono and dimethylated H3K4, and trimethylated H3K4 at the boundaries of the loops, whereas the active transcription modification H3K36me3 and repressive H3K27me3 mark are observed outside the loops on opposite sides. D. A fourth class of loops formed by CTCF lack specific histone modifications, while active H3K4 methylation marks are observed in one side and repressive H3K9, H3K20 and H3K27 methylation modifications are present in the other side. E. The rest of the loops formed by CTCF do not show specific chromatin modifications.
Concluding remarks
The use of 3C-based techniques has allowed great progress in mapping interactions between different sites in the genome. Until now, this effort has been limited, with a few exceptions, to the analysis of relatively close intra-chromosomal interactions within specific loci. Although 4C and 5C techniques could expand the size of the genomic regions under study, the complexity of the computational and statistical analysis of the results will probably preclude the general use of these approaches until these computational methods become standardized. In the meantime, advances in next generation sequencing combined with increased affordability will allow the use of Hi-C to map the three-dimensional organization of a specific cell type with a resolution of a few kilobases. Given the complexity of the interactions involved and biases in the HiC procedure, including variability in the distances between restriction sites, GC content differences of ligation junctions and sequence uniqueness, analysis of HiC sequence data remains a challenge. An important step in solving these issues has come from the use of an integrated probabilistic background model that allows the normalization of HiC sequence data(Yaffe and Tanay 2011). This approach has allowed the mapping of long-range interactions between active promoters and interactions between CTCF sites. Nevertheless, unraveling the significance of these interactions may first require mapping those contacts mediated by specific proteins using antibodies to select subsets of these interactions.
It is likely that this new information will revolutionize the way in which we think about nuclear metabolism by allowing the identification of new sequences and proteins involved in the regulation of various nuclear functions. Nevertheless, many challenges still lie ahead. Being able to separate meaningful from accidental interactions and causal from correlative are just a few of these challenges.
ACKNOWLEGMENTS
Work in the authors' laboratory is supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
References
- Ansari A, Hampsey M. A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011 doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci U S A. 2011;108:2294–2299. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci U S A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Smith S, Zhou J. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc Natl Acad Sci U S A. 2007;104:3237–3242. doi: 10.1073/pnas.0605730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF Mediates Interchromosomal Colocalization Between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3' end processing and transcription initiation. Mol Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Melnikova L, Kostuchenko M, Silicheva M, Georgiev P. Drosophila gypsy insulator and yellow enhancers regulate activity of yellow promoter through the same regulatory element. Chromosoma. 2008;117:137–145. doi: 10.1007/s00412-007-0132-6. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D, Greaves DR. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007;90:407–415. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ren X, Siegel R, Kim U, Roeder RG. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol Cell. 2011;42:342–355. doi: 10.1016/j.molcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Nicodemi M. Mechanics and dynamics of X-chromosome pairing at X inactivation. PLoS Comput Biol. 2008;4:e1000244. doi: 10.1371/journal.pcbi.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukov YG, Sural TH, Kuroda MI, Sedat JW. Evidence of activity-specific, radial organization of mitotic chromosomes in Drosophila. PLoS Biol. 2011;9:e1000574. doi: 10.1371/journal.pbio.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Fukue Y, Nolen L, Sadreyev RI, Lee JT. Characterization of Xpr (Xpct) reveals instability but no effects on X-chromosome pairing or Xist expression. Transcr. 2010;1:46–56. doi: 10.4161/trns.1.1.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces V. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Molecular Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol. 2011;18:372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011 doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Yang J, Corces VG. Chromatin insulators: a role in nuclear organization and gene expression. Adv Cancer Res. 2011;110:43–76. doi: 10.1016/B978-0-12-386469-7.00003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao H, Dean A. Organizing the genome: enhancers and insulators. Biochem Cell Biol. 2005;83:516–524. doi: 10.1139/o05-054. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Singh Sandhu K, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]