Abstract
KRAS and BRAF mutations lead to the constitutive activation of EGFR signaling through the oncogenic Ras/Raf/Mek/Erk pathway. Currently, KRAS is the only potential biomarker for predicting the efficacy of anti-EGFR monoclonal antibodies (mAb) in colorectal cancer (CRC). However, a recent report suggested that the use of cetuximab was associated with survival benefit among patients with p.G13D-mutated tumors. Furthermore, although the presence of mutated BRAF is one of the most powerful prognostic factors for advanced and recurrent CRC, it remains unknown whether patients with BRAF-mutated tumors experience a survival benefit from treatment with anti-EGFR mAb. Thus, the prognostic or predictive relevance of the KRAS and BRAF genotype in CRC remains controversial despite several investigations. Routine KRAS/BRAF screening of pathological specimens is required to promote the appropriate clinical use of anti-EGFR mAb and to determine malignant phenotypes in CRC. The significance of KRAS/BRAF mutations as predictive or prognostic biomarkers should be taken into consideration when selecting a KRAS/BRAF screening assay. This article will review the spectrum of KRAS/BRAF genotype and the impact of KRAS/BRAF mutations on the clinicopathological features and prognosis of patients with CRC, particularly when differentiating between the mutations at KRAS codons 12 and 13. Furthermore, the predictive role of KRAS/BRAF mutations in treatments with anti-EGFR mAb will be verified, focusing on KRAS p.G13D and BRAF mutations.
Keywords: Anti-EGFR monoclonal antibody, BRAF, Cetuximab, Chemotherapy, Clinicopathological features, Colorectal cancer, Driver mutation, EGFR, KRAS codon 12, KRAS codon 13, KRAS p.G13D, Panitumumab, Predictive marker, Prognostic biomarker, Screening.
INTRODUCTION
The development of colorectal cancer (CRC) is a multistep process that occurs because of the accumulation of several genetic alterations, including chromosomal abnormalities, gene mutations, and epigenetic modifications involving several genes that regulate proliferation, differentiation, apoptosis, and angiogenesis [1, 2].
Of the various genetic alterations, an important molecular target for metastatic CRC treatment is the epidermal growth factor receptor (EGFR). EGFR, also known as HER1 or ErbB, is a 170-kD receptor tyrosine kinase and belongs to the ErbB receptor family. There are four members in the ErbB receptor family: ErbB1 (EGFR, HER1), ErbB2 (HER2/neu), ErbB3 (HER3), and ErbB4 (HER4). The binding of several specific ligands, such as EGF, TGF-α, or amphiregulin, results in the dimerization of EGFR and subsequent phosphorylation of several tyrosine residues [3, 4]. These phosphorylated tyrosines serve as binding sites for several signal transducers that initiate multiple signaling pathways, including the Ras/Raf/MAP/MEK/ERK and/or PTEN/PI3K/Akt pathways. Although EGFR plays important roles in cell differentiation and proliferation in normal cells, the activation of EGFR signaling is frequently observed in CRC cells, where it results in cell proliferation, migration and metastasis, evasion of apoptosis, or angiogenesis [5]. Approximately 35% CRC tissues carry a mutation at codon 12 or 13 of KRAS that leads to the constitutive activation of EGFR downstream pathways [6-10].
Information on the KRAS/BRAF genotype is also extremely useful when selecting systemic chemotherapy for advanced and recurrent patients with CRC, where it can help identify patients with poor prognoses. KRAS and BRAF are currently under focus as potential prognostic and predictive biomarkers in patients with metastatic diseases treated with anti-EGFR monoclonal antibodies (mAb), such as cetuximab and panitumumab [11-14]. Several retrospective analyses revealed that cetuximab treatment is ineffective in patients with KRAS mutations, thereby suggesting that the KRASgenotype is a useful predictive biomarker for cetuximab or panitumumab therapy in CRC [11-13, 15]. It has also been suggested that wild-type BRAF is required for a successful response to panitumumab or cetuximab therapies in patients with metastatic CRC [9, 10, 16, 17]. However, the prognostic relevance of the KRAS genotype in CRC remains controversial despite several multi-institutional investigations since the 1990s [18-22].
In this article, I will review the spectrum of the KRAS/BRAF genotype and the clinical outcomes of KRAS/BRAF mutations in patients with CRC. The prognostic and/or predictive impact of KRAS/BRAF mutations will then be discussed, focusing on the difference between mutations at KRAS codons 12 and 13.
POTENTIAL PREDICTIVE BIOMARKERS FOR ANTI-EGFR THERAPY
The molecular mechanisms underlying response or resistance to anti-EGFR mAb still remain largely unknown. However, the clinical predictive factors that indicate the response or resistance to anti-EGFR therapy should be identified before beginning such a treatment in patients with CRC to prevent drug-induced toxicity and avoid unnecessary expenses. The main research areas in this setting have been focusing on the role of (i) EGFR protein expression, (ii) EGFR gene copy number, (iii) EGFR gene mutations, (iv) overexpression of EGFR ligands (such as epiregulin and amphiregulin), (v) methylation of the EGFR promoter, and (vi) markers of EGFR downstream signaling [8, 9, 23-28].
For initial clinical trials, patients with metastatic CRC were selected if they had tumors positive for the expression of EGFR as detected by immunohistochemistry (IHC). However, cetuximab is also effective in patients with CRC having tumors that do not express EGFR when examined by IHC [29]. Indeed, EGFR is overexpressed in 30%–85% patients with CRC. Therefore, the level of EGFR protein expression has proved to be poorly associated with sensitivity to anti-EGFR mAb. Inconsistent methodology and interpretation of EGFR IHC expression in tumor samples may be an explanation for this. Inter-observer variability in the definitions of the expression EGFR may depend on the tissue fixation technique used, possibly leading to false negative samples by IHC using paraffin-embedded tumor tissues. Significant differences in EGFR IHC expression between a patient’s primary tumor and their metastatic tissue specimen may be another explanation. The primary tumor is frequently used to establish the patient’s EGFR status, but metastases are treated with cetuximab. A third explanation is that high-affinity EGFRs are the predominant biologically active receptors that lead to the activation of protein tyrosine kinase, thereby contributing significantly to signal transduction [30]. However, the anti-EGFR antibodies that are most commonly used do not distinguish between high-affinity and low-affinity EGFRs [31]. Another potential explanation may be the potential of cetuximab to induce antibody-dependent cell-mediated cytotoxicity (ADCC) despite an equivalent pharmacological EGFR blockade.
In a small fraction of CRCs, the overexpression of EGFR is frequently associated with amplification of the gene. The EGFR gene copy number evaluated by quantitative PCR does not appear to correlate with the clinical outcome of patients, whereas the result of the analysis by fluorescence in situ hybridization (FISH) appears to be associated with an increase in treatment response [32]. However, the predictive value is uncertain, and further studies are required to assess the increase of EGFR gene copy number as a predictive biomarker of response to anti-EGFR therapy. Activating mutations in the EGFR catalytic domain plays an important role in determining the responsiveness to anti-EGFR therapy in lung cancer. However, mutations in the EGFR tyrosine kinase domain are considered to be extremely rare in patients with CRC [33] and they are not significantly associated with the clinical response of metastatic CRC to anti-EGFR mAb [24].
The overexpression of alternative EGFR ligands, such epiregulin and amphireguline, may promote tumor growth and survival by an autocrine loop [34]. Several studies have correlated the expression of these ligands with sensitivity to cetuximab monotherapy. The results showed a statistically longer progression-free survival (PFS) period among patients with high expression of epiregulin. The exclusive use of an amphiregulin or epiregulin gene expression profile does not, however, result in the selection of patient populations benefiting from cetuximab treatment [35].
Scartozzi et al., investigated the correlation between the efficacy of irinotecan plus cetuximab therapy and methylation status in the EGFR promoter [36]. Patients with tumors harboring the hypermethylating EGFR promoter experienced a worse clinical outcome in terms of progression-free survival (PFS) and overall survival (OS), suggesting that the methylation of EGFR promoter plays a role in determining the efficacy of anti-EGFR mAb. Thus, the hypermethylation of EGFR promoter may be a valuable and important indicator that should be considered in further investigations of the role of EGFR as a therapeutic target in patients with CRC.
Collectively, the predictive value of alterations in EGFR expression level remains unconvincing in the use of anti-EGFR therapy. Therefore, the focus has shifted to alterations of the key signaling pathway downstream of EGFR.
BIOMARKERS DOWNSTREAM OF EGFR
The constitutive activation of signaling pathways downstream of EGFR drive the growth and progression of CRC and provide an escape mechanism that allows tumors to overcome the pharmacological blockade induced by anti-EGFR mAb [37]. KRAS, BRAF, PTEN, and PI3KCA mutations have been highlighted as the mechanisms that activate EGFR signaling pathway.
KRAS is a proto-oncogene encoding a small 21-kD guanosine triphosphate (GTP)/guanosine diphosphate (GDP) binding protein involved in the regulation of the cellular response to many extracellular stimuli [38]. After binding and activation by GTP, KRAS recruits the oncogene BRAF, which phosphorylates MAP2K (mitogen-activated protein kinase kinase), thereby initiating MAPK signaling leading to the expression of the protein involved in cell proliferation, differentiation, and survival [39]. KRAS is the mostly commonly mutated gene in this pathway, and 35%–45% patients with CRCs carry this mutation, which is an early event in colon tumorigenesis [40]. KRAS mutations frequently induce glycine-to-valine substitutions at the catalytic sites of amino acids, which leads to the loss of GTPase activity and subsequent continuous binding of GTP to RAS. This constitutive activation of RAS results in the dysregulation of the downstream RAS-ERK signaling pathway independently of EGFR. Similarly, the kinase activity of the BRAF mutant protein is greatly elevated, which also constitutively stimulates downstream ERK activity independently of RAS and EGFR. Thus, the constitutive activation of KRAS or BRAF mutation leads to EGFR-independent tumorigenicity in patients with CRC. Therefore, the oncogenic activation of the RAS signaling pathway impairs the response of colorectal cancer cells to cetuximab [6-10, 41].
The PTEN/PI3K/Akt pathway also affects several cellular processes such as cell proliferation, apoptosis, and invasion [42]. Signal transduction through this pathway is mediated by conversion of phosphatidylinositol bisphosphate (PIP2) to phosphatidylinositol triphosphate (PIP3) by phosphatidylinositol 3 kinases (PI3K) following their activation, and this reaction is antagonized by phosphatase and a tensin homolog deleted on chromosome ten (PTEN). The PTEN mutation is known to correlate with microsatellite instability (MSI-H) in patients with CRC [43, 44]. Of the genes that encode the enzymatic subunit of PI3K heterodimers, the PIK3CA gene that encodes the p110 subunit of PI3K has been found to be most frequently activated by its mutations in some human cancers [42]; this promotes AKT1 phosphorylation to activate a parallel intracellular axis [45].
Several reports have suggested that there is cross-talk between Ras/Raf/MAP/MEK/ERK and/or PTEN/PI3K/Akt pathways. Specifically, PIK3CA can be activated via interaction with the RAS protein [46].
SPECTRUM OF THE KRAS AND BRAF GENOTYPES IN PATIENTS WITH CRC
Estimates of the KRAS mutation frequency in metastatic CRCs are based on selective clinical studies or drug admission trials with variable inclusion criteria. According to previous investigations on the spectrum of the KRAS genotype in our database of CRC cases, the most frequent mutations at the KRAS codon 12 were G12D, G12V, G12R, G12C, G12S, and G12A, which accounted for more than 95% of the codon 12 mutations. The G13D and G13C mutations at codon 13 and the G61H, G61L, G61E, and G61K mutations at codon 61 were the most common mutations that occurred at these codons [47]. All these KRAS mutations have been previously described as oncogenically active and they are present in the COSMIC (catalog of somatic mutations in cancer) database [48]. Data from a large Japanese population of patients with advanced and recurrent CRC revealed that KRAS mutations were present in approximately 35% patients with CRC of which 25% patients had mutations at codon 12 and 10% patients had mutations at codon 13. This observation was consistent with that of previous studies on selected cohorts that reported frequencies in the range of 30%–42% [47].
Although more than 40 somatic mutations have been described in the BRAF kinase domain, the most common mutation across various cancers is the classic GTG→GAG substitution at position 1799 of exon 15, which results in the V600E amino acid change and subsequent constitutive activation of the EGFR signaling pathway. Functionally, this is the most important mutation involved in the receptor-independent aberrant activation of the EGFR signaling pathway and CRC carcinogenesis. Recent studies in western countries suggested that BRAF mutations occur in 10%–20% of patients with sporadic diseases [8, 9, 10, 49, 50]. BRAF V600E mutation in the Japanese population was observed in 4.7% patients with CRC; this appeared to be lower than that found in western populations. None of the patients with CRC carried both KRAS and BRAF mutations, supporting the hypothesis that KRAS and BRAF mutations are mutually exclusive [51-53]. One possible explanation for the comparatively low frequency of BRAF mutations might be the difference in ethnicities. Indeed, several studies reported that the mutation rates of DNA mismatch repair (MMR) genes, such as hMSH2 and hMLH1, in hereditary non-polyposis colorectal cancer (HNPCC) varies across countries. Therefore, geographical variation may account for the differences in the mutation spectrum of BRAF, as observed for MMR genes [54-56].
KRAS TESTING IS A SCREEN FOR DRIVER MUTATION, BUT NOT FOR SUPER-RESPONDER
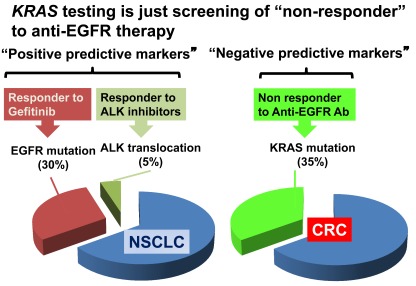
Routine KRAS/BRAF screening should be performed before initiating anti-EGFR therapy in patients with CRC to predict non-responsiveness to anti-EGFR therapy and to prevent drug-induced toxicity. In addition, limiting the use of anti-EGFR mAb to patients with wild-type, i.e., non-mutated, KRAS testing may result in avoiding heavy expenses [57]. Therefore, optimal KRAS/BRAF geno-typing procedures with pathological specimens are necessary. The significance of KRAS/BRAF mutations as predictive markers in patients with CRC should be considered while selecting a method for KRAS testing. Patients with non-small cell lung cancer (NSCLC) show 30% EGFR mutations and 5% ALK translocations, which are driver mutation targeted by molecular target agents such as Gefitinib or ALK inhibitors. Because the populations with these gene alterations are super responders to specific molecular target therapies, screening for driver mutations is essential and requires a technique with high sensitivity. In patients with CRC, KRAS testing is also a screen for a driver mutation. However, unlike NSCLC, the main purpose of KRAS genotyping in patients with CRC is screening for absolute non-responders to anti-EGFR mAb (Fig. 1).
Fig. (1).
KRAS testing is a screening of non-responders to anti-EGFR therapy. Abbreviations: NSCLC, non-small cell lung cancer; CRC, colorectal cancer.
Allele-specific PCR methods such as TaqMan MGB Probes or ScorpionsARMS are available as commercial test kits, and some clinical trials have applied this method for KRAS genotyping. ScorpionsARMS is believed to be more sensitive than both direct sequencing and cycleave PCR and can detect mutations in samples containing 1% mutant allele sequences. However, optimal KRAS genotyping methods may not necessarily be highly sensitive. The most critical mistake that should be avoided is an overestimation of the population with KRAS mutations that would lead to depriving true responders of the benefits of anti-EGFR mAb. Furthermore, allele-specific PCR methods such as TaqMan MGB Probes or ScorpionsARMS are too expensive to be used as routine diagnostic methods for KRAS genotyping [47].
Taken together, appropriate methods should be selected for KRAS genotyping, taking into consideration KRAS mutations as negative predictive biomarkers.
ASSOCIATION OF BRAF/KRAS MUTATIONS WITH CLINICOPATHOLOGICAL FEATURES
Several reports suggested that tumors harboring BRAF mutations have different clinical and histopathological features compared with tumors harboring KRAS mutations. BRAF mutations occur more frequently in right-sided tumors [58-61]. A study evaluating the correlation between KRAS/BRAF mutational status and clinicopathological features in advanced and recurrent CRC also found that in 60% patients with CRCs having BRAF mutations, the tumor metastasized to the peritoneum compared with approximately 15% patients with CRCs with other subtypes. Furthermore, 60% BRAF mutation-positive specimens belonged to poorly differentiated adenocarcinoma or mucinous carcinoma subtypes [61]. It was recently reported that mucinous histology indicates a poor response to oxaliplatin- and/or irinotecan-based chemotherapies and is correlated with poor OS [62]. Because BRAF mutations are more frequent in mucinous carcinomas than in non-mucinous carcinomas as demonstrated by the present study and a previous study [63], the poor prognosis associated with mucinous histology may be at least partially explained by the poor prognosis of patients with CRC having BRAF mutations. These specific clinicopathological features support the hypothesis that BRAF mutation-mediated carcinogenesis in patients with CRC is initiated by altered BRAF function as an early step in the serrated pathway [64] that leads to the activation of EGFR signaling. In contrast to BRAF mutations, no significant differences have been observed in clinicopathological parameters based on the KRAS genotype in many studies, probably due to the lack of differentiation between KRAS12 mutations and KRAS13 mutations. However, an analysis in which a population of patients with CRC was categorized into four subtypes—KRAS and BRAF (wild/wild), KRAS12 mutations, KRAS13 mutations, and BRAF mutations (V600E), suggested that KRAS13 mutations were also associated with right-sided tumors [61]. This suggests the possibility that KRAS13 may have a phenotype distinct from that of other KRAS genotypes.
PROGNOSTIC ROLE OF KRAS MUTATIONS
The prognostic value of KRAS mutations in patients with CRC remains controversial. Although the prognostic role of KRAS mutations has been previously investigated, no definitive conclusions have been drawn [65]. This may be because of differences in terms of study size, patient selection, tumor sampling, use of archival versus fresh/frozen material, laboratory methods, and data analyses. Furthermore, such prognostic analyses are performed mostly in homogeneous groups of metastatic patients with CRC treated with a specific chemotherapy regimen with or without cetuximab [14, 66] (Table 1).
Table 1.
KRAS Mutation and Prognosis
| Trial | Population | Therapy | Overall Survival (month) | HR | P-value | Prognostic ? | |
|---|---|---|---|---|---|---|---|
| KRAS wt | KRAS mut | ||||||
| CO.17 | 3rd line | BSC | 4.8 | 4.6 | 1.01 | 0.97 | No |
| CAIRO-2 | 1st line | CapeOX+BV | 22.4 | 24.9 | 0.82 | No | |
| N0147 | Stage III | FOLFOX+Cmab | *72.3 % | *64.2 % | 0.7 | 0.004 | Yes |
| COIN | 1st line | FU+OX+/-Cmab | 17.5 | 14.4 | <0.0001 | Yes | |
| Fariña-Sarasqueta, A. et al. | Stage III | - | ? | ? | 0.03 | Yes | |
| PETACC-3 | Stage II/III | FU/LV+/-CPT-11 | 1.09 | 0.48 | No | ||
| FOCUS | 1st line sequential | FU+/-CPT-11/OX | ? | ? | 1.24 | 0.008 | Yes |
| Van Cutsem, E. et al. | 3rd line | BSC | 7.6 | 4.4 | **N.S. | No | |
| EPIC | 2nd line | CPT-11 | 11.56 | 10.68 | **N.S. | No | |
3 year Disease free survival
statistically not significant
Abbreviations: BSC, best supportive care; Cape, capecitabine; OX, oxaliplatin; Cmab, cetuximab; mut, mutated.
A recent translational study by Roth et al., suggested that the prognostic value for KRAS mutation status for PFS and OS was lacking in PETACC-3, EORTC 40993, and SAKK 60-00 trials of patients with stage II and III resected colon cancer [22]. However, it has been reported that stage III patients having KRAS mutations displayed significantly worse disease-free survival compared with those having wild-type KRAS [50]. Furthermore, an N0147 trial assessing the potential benefit from cetuximab treatment combined with FOLFOX in patients with resected stage III CRC showed that the three-year disease-free survival in patients with wild-type KRAS was significantly better than that in patients with KRAS mutants (72.3% versus 64.2%, HR = 0.7, p = 0.004) (Table 1). These analyses suggest that KRAS mutations are independent prognostic factors [67]. A Medical Research Council (MRC) COIN trial assessed the effects of cetuximab combined with oxaliplatin and fluoropyrimidine chemotherapy as a first-line treatment of patients with advanced CRC. This trial found that the median OS was significantly shorter in patients with KRAS, NRAS, or BRAF mutations (n = 706, 13.6 months) compared with those with wild types for KRAS, NRAS, and BRAF (n = 581, 20.1 months), irrespective of the treatment [68].
More importantly, few studies have differentiated KRAS mutations at codon 12 from those at codon 13 with respect to clinicopathological features and survival. Recent findings have suggested that CRC with a KRAS mutation is not clinically homogeneous but heterogeneous population [69]. This hypothesis may be supported by the fact that NSCLCs harboring alterations in the EGFR gene are biologically and pharmacologically heterogeneous. Indeed, there are differences in the transforming potential and EGFR tyrosine kinase inhibitor (TKI) sensitivity associated with EGFR somatic mutations L858R and the deletion mutant Del (746–750) in NSCLCs [70].
Collaborative RASCAL II studies were conducted to investigate the prognostic role of KRAS mutations in CRC progression. To explore the effect of KRAS mutations at different stages of CRC, 3493 patients were recruited in this multivariate analysis. RASCAL studies showed that tumors carrying a substitution of glycine to valine at codon 12, which was found in 8.6% patients, had a statistically significant impact on worse PFS (p = 0.0004, HR = 1.3) and OS (p = 0.008, HR = 1.29) [40]. This clinical data was supported by the finding that KRAS12 mutations confer a more aggressive transforming phenotype than KRAS13 mutations through a significant increase in the activation of AKT and expression of bcl-2, and a significant decrease in the expression of RhoA [71]. However, multivariate analysis by Bazan et al., revealed that KRAS13 mutations, but not other mutations, were independently related to the risk of relapse or death in a consecutive series of 160 untreated patients (median of follow up period = 71 months) who underwent resective surgery for primary CRC [72]. Consistent with this study, Yokota et al., examined 229 patients with advanced and recurrent CRC who were treated with systemic chemotherapy, and demonstrated that the OS for patients with KRAS13 mutations was significantly worse than for those who had wild-type KRAS and wild-type BRAF, whereas KRAS12 mutation did not affect patient OS [61]. Furthermore, KRAS/BRAF genotype was analyzed in a large subgroup of 845 patients with metastatic CRCs who received FOLFIRI and FOLFOX chemotherapy with or without cetuximab as the first-line treatment in the CRYSTAL and OPUS studies, respectively [66]. The results revealed that KRAS13D mutations are associated with poor prognosis. Therefore, the finding that stage III patients with KRAS mutations displayed significantly worse disease-free survival than those with wild-type KRAS [50, 64, 67], might be partially explained by the impact of either KRAS12 or KRAS13 mutations on prognosis.
Taken together, differences in KRAS mutations at codons 12 and 13 may result in different biological, biochemical, and functional consequences that could influence the prognosis of CRC [72]. Larger studies are required to confirm whether a specific KRAS mutation might lead to a clinically relevant prognostic effect in patients with CRC.
ARE KRAS MUTATIONS NEGATIVE PREDICTIVE BIOMARKERS FOR ANTI-EGFR mAB?
Several retrospective analyses have revealed that patients with KRAS mutations receiving first and subsequent lines of treatment do not respond to cetuximab or panitumumab, and that they show no survival benefit from such treatments [11-13, 15, 73]. KRAS mutations have emerged as a major predictor of resistance to anti-EGFR mAb in the clinical setting. Therefore, patients with metastatic CRC with KRAS codon 12- or KRAS codon 13-mutated tumors are presently excluded from treatment with anti-EGFR mAb.
However, one patient with a mutated KRAS tumor (1.2%) had a response in the CO.17 trial comparing cetuximab monotherapy with best supportive care (BSC) in patients with chemotherapy-refractory metastatic CRC [11]. Furthermore, a recent retrospective analysis by De Roock et al., examined 579 patients with chemotherapy-refractory CRC who received cetuximab treatment, and revealed that patients with p.G13D-mutated tumors showed a trend toward a higher response rate than other KRAS-mutated tumors (6.3% versus 1.6%, p = 0.15). Strikingly, patients with KRAS codon p.G13D mutations who received cetuximab experienced longer progression-free and overall survival compared with BSC alone. In contrast, patients with other KRAS mutations did not appear to benefit from cetuximab. The authors suggested that p.G13D-mutated tumors may have a worse prognosis, based on the finding that patients with KRAS p.G13D mutations who received BSC alone showed significantly shorter survival compared with those with other KRAS mutations in the CO.17 study [74].
Furthermore, the association of KRAS p.G13D mutation with clinical outcome was investigated in a pooled analysis of patients from the CRYSTAL and OPUS studies. The population consisted of 689 patients in each treatment arm, including 447 versus 398 with wild-type KRAS, 41 versus 42 with KRAS p.G13D, and 201 versus 249 with other KRAS mutations, for chemotherapy alone and cetuximab plus chemotherapy arms, respectively. A heterogeneous treatment effect was observed with significant treatment interaction with the KRAS mutation status for response (p < 0.0001), PFS (p < 0.0001), and OS (p = 0.0219). In particular, the response rate in patients with KRAS p.G13D treated with cetuximab plus chemotherapy was better than that in those treated with chemotherapy alone (40.5% versus 22.0%; 95% CI, 0.90 to 6.45, p = 0.0748). The hazard ratio for PFS among patients with KRAS p.G13D was 0.60 (95% CI, 0.32 to 1.12, p = 0.1037), while that for OS among patients with KRAS p.G13D was 0.80 (95% CI, 0.49 to 1.30) in favor of the cetuximab plus chemotherapy arm. Although treatment effects were not statistically significant, patients with KRAS p.G13D had a similar relative treatment effect compared with patients with wild-type KRAS.
Taken together, these data may suggest that KRAS p.G13D mutations are poor prognostic biomarker, and the use of cetuximab may affect prolonged survival in patients receiving first-line chemotherapy and those with chemotherapy-refractory metastatic colon cancer. The clinical benefit of anti-EGFR therapy in patients with KRAS mutations, which are rare, may be partially explained by the benefit in p.G13D-mutated group. Further prospectively generated clinical investigations are necessary to confirm these data, because whether the KRAS p.G13D mutation is an effective negative predictive biomarker remains controversial.
PROGNOSTIC ROLE OF BRAF MUTATIONS
A BRAF mutation (V600E) has been studied in recent years for a better understanding of its possible role in prognosis and predicting the response to anti-EGFR mAb.
While few studies investigated the impact of KRAS12 and KRAS13 mutations on CRC prognosis, a series of recent studies confirmed the potential adverse prognostic impact of BRAF mutations (Table 2). Yokota et al., identified BRAF V600E mutation as an independent prognostic factor for survival in a representative cohort of 229 patients with advanced and recurrent CRC. The presence of this BRAF mutation was associated with a significantly higher risk of dying from cancer-related causes, independently of other factors such as age, gender, PS, KRAS status, pathological finding, number of metastases, and metastatic sites [61]. This finding is consistent with those of other recent studies using patients with both stage II and III disease and patients across all disease stages [21, 22, 50]. For example, an analysis of stage II and stage III patients with CRC [22, 50] was consistent with the finding that 44% population had recurrent disease [61]. Furthermore, BRAF mutations were correlated with survival in a heterogeneous group of patients with CRC that included all disease stages [20] (Table 2). Furthermore, BRAF mutations are prognostic biomarkers for OS, particularly in patients with microsatellite instability (MSI), both low (MSI-L) and stable (MSI-S) tumors. In the high (MSI-H) subpopulation, a prognostic value of KRAS and BRAF mutation status was not found for RFS and OS [22] (Table 4). Whereas BRAF mutations had no prognostic value in the relapse-free survival of stage II-III CRC, BRAF mutation is a strong determinant of OS after relapse [22].
Table 2.
BRAF Mutation and Prognosis
| Population | HR (95% CI) | Reference | Prognostic ? | |
|---|---|---|---|---|
| Ann Oncol. 2010; 21(12):2396-402 | stage II / III | 0.45 (0.25–0.8) | Mutant | Yes |
| Gut 2009; 58: 90-96 | All stage | 1.20 (0.79–1.80) | Wild | Yes |
| PETACC-3 | stage II / III | 1.19 (0.84-1.69) | Wild | Yes |
| Br J Cancer 2011;104:856-62 | Recurrent and advanced | 4.25 (2.08–8.67) | Wild | Yes |
Abbreviations: CT, chemotherapy; CB, CapeOX/bevacizumab; CBC, CapeOX/bevacizumab plus cetuximab.
Table 4.
Are KRAS/BRAF Mutations Predictive and/or Prognostic?
| KRAS mut | BRAF mut | ||||
|---|---|---|---|---|---|
| Codon 12 mutant | Codon 13 mutant | Codon 61 mutant | MSI | MSS | |
| Predictive marker | Negative predictive | Negative predictive? | Negative predictive? | Positive predictive? | |
| Prognostic marker | No | Yes? | ? | No | Yes |
Abbreviations: MSI, microsatellite unstable; MSS, microsatellite stable.
Furthermore, the prognostic value of BRAF was analyzed in patients with CRC treated with specific chemotherapy regimens in clinical trials that evaluated a combination of cetuximab with chemotherapy (Table 3). The CAIRO-2 study investigated a large series of metastatic patients with CRC treated with chemotherapy and bevacizumab with or without cetuximab in a subgroup of 520 patients. This study revealed that patients with CRC having BRAF mutations show a worse outcome, both in terms of PFS and OS, irrespective of the addition of cetuximab to the treatment [14]. The pooled analysis of the abovementioned CRYSTAL and OPUS studies revealed that the outcome of patients with CRC having BRAF mutations is worse than that of patients with CRC having wild-type BRAF, independently of treatment with cetuximab [66]. These findings further support the hypothesis that BRAF mutations are negative prognostic biomarkers. The BRAF genotype might be an additional stratification factor for future clinical trials of advanced and recurrent CRC.
Table 3.
BRAF Mutation and Prognosis in the Clinical Trials Evaluating Combination of Cetuximab with Chemotherapy
| Population | Therapy | BRAF wt | BRAF mut | P-value | Prognostic ? | |
|---|---|---|---|---|---|---|
| ASCO2010, Abstract No. 3506 | 1st line CRYSTAL/OPUS | CT | 21.1 | 9.9 | - | Yes |
| CT+Cmab | 24.8 | 14.1 | - | |||
| N Engl J Med. 2009; 361: 98-99 | 1st line CAIRO-2 | CB | 24.6 | 15.0 | 0.002 | Yes |
| CBC | 21.5 | 15.2 | 0.001 |
Abbreviations: CT, chemotherapy; CB, CapeOX/bevacizumab; CBC, CapeOX/bevacizumab plus cetuximab.
PREDICTIVE ROLE OF BRAF MUTATIONS
Di Nicolantonio et al., retrospectively analyzed objective tumor responses and survival, and the mutational status of KRAS and BRAF in 113 patients with metastatic CRC treated with cetuximab or panitumumab [9]. None of the BRAF-mutated patients responded to the treatment, while none of the responders carried BRAF mutations. BRAF-mutated patients had significantly shorter progression-free survival and overall survival than wild-type patients. The effect of BRAF V600E mutation on cetuximab or panitumumab response was also assessed using cellular models of CRC. The introduction of BRAF V600E allele impaired the therapeutic effect of cetuximab or panitumumab. Similarly, Souglakos et al., addressed the predictive value of BRAF in 100 patients treated with cetuximab, including 8 in the first line, 37 in the second, and 55 in the third or higher, always in combination with chemotherapy [10]. No patients with a BRAF-mutant tumor responded to cetuximab, whereas objective responses were observed in 17% patients with wild-type BRAF. Patients with BRAF mutation also had a shorter PFS, regardless of whether cetuximab was administered in the second or third or higher lines. The effects of BRAF status on the efficacy of cetuximab plus chemotherapy were retrospectively analyzed in patients with metastatic CRC having wild-type KRAS [16, 17], indicating that the presence of BRAF mutation was significantly correlated with lower response rate than wild-type BRAF, with a response rate of 8.3% (2/24) in carriers of BRAF mutations versus 38.0% in BRAF wild types [17]. These results suggest that wild-type BRAF is required for the response to anti-EGFR mAb in metastatic CRC. However, these studies lacked data on BRAF-mutated patients treated with chemotherapy alone, so they failed to directly compare the efficacy of adding cetuximab or panitumumab to chemotherapy with that of chemotherapy alone in a cohort of BRAF-mutated patients. Therefore, whether BRAF and KRAS mutations are negative predictive biomarkers for anti-EGFR mAb cannot be ascertained.
In the pooled analysis of CRYSTAL and OPUS, patients with BRAF mutations seemed to benefit from the addition of cetuximab, with an increase of OS and a doubling of PFS rates, although this was not statistically significant [66]. The addition of cetuximab to FOLFIRI or FOLFOX regimens showed a trend towards better survival compared with FOLFIRI or FOLFOX alone. This result raises the possibility that the use of cetuximab might be effective for disease control at least as the first-line chemotherapy for patients with wild-type KRAS and mutant BRAF.
Taken together, the association of BRAF mutations with the efficacy of anti-EGFR therapy remains controversial, but its significant negative prognostic value has been established. Such discrepant results among studies might be partially explained by the differential significance of BRAF mutations as predictive biomarkers for anti-EGFR mAb in the first-line and second-line or higher line chemotherapy. The relatively low frequency of BRAF mutations in patients with CRC makes it relatively difficult to draw absolute conclusions, but the present observations should be confirmed by examining an increased number of patients with BRAF mutations.
CONCLUSIONS
The activation of EGFR signaling, such as Ras/Raf/MAP/MEK/ERK and/or PTEN/PI3K/Akt pathways, plays an important role in tumorigenesis and the tumor progression of CRC. Two predominant EGFR inhibitors have been developed including monoclonal antibodies that target the extracellular domain of EGFR and small molecule TKIs that target the receptor catalytic domain of EGFR. Although both classes of agents show clear antitumor activity, only the anti-EGFR mAb has been approved for clinical use in the treatment of patients with metastatic CRC. Because the predictive value of alterations in EGFR expression level is unclear in the use of anti-EGFR mAb, the focus has shifted to alterations of key signaling pathways downstream of EGFR. In particular, KRAS and BRAF mutations have been highlighted as the activating mechanisms of the EGFR signaling pathway. Routine screening for KRAS/BRAF genotype is extremely important for identifying patients with shorter survival in response to systemic chemotherapy, regardless of the use of anti-EGFR mAb, and for predicting patients who would benefit from anti-EGFR mAb treatment, which is costly and potentially toxic. However, the significance of KRAS/BRAF mutations as prognostic and/or predictive biomarkers in patients with CRC should be considered while selecting a method for KRAS genotyping.
KRAS mutations were observed in approximately 35% patients with CRC, of which 25% patients had mutations at codon 12 and 10% patients had mutations at codon 13. The KRAS genotype is a useful predictive biomarker for patients with metastatic CRC that is treated with anti-EGFR mAb. Recent reports have raised the possibility that KRAS13 may have a specific phenotype that is different from other KRAS genotypes. Therefore, differences in KRAS mutations at codons 12 and 13 may result in different biological, biochemical, and functional consequences and clinical features, which may also influence the prognosis of CRC. Indeed, several retrospective analyses have suggested that KRAS mutations at codon 13, particularly KRAS p.G13D, as well as BRAF mutations are prognostic factors. Furthermore, a recent major research finding is that patients with KRAS p.G13D, but not other mutations, may experience a survival benefit from treatment with cetuximab plus chemotherapy. These findings also support the hypothesis that patients with CRC having KRAS mutations constitute a heterogeneous population. Since the prognostic and/or predictive role of KRAS13 mutations continues to remain controversial, further prospective clinical investigations are warranted.
Several reports have suggested that tumors harboring BRAF mutations have distinct clinicopathological features. Importantly, BRAF mutations are significant negative prognostic biomarkers in patients with recurrent CRC across all disease stages. Moreover, the prognostic value of BRAF mutations has been confirmed in patients with CRC treated with specific chemotherapy regimens in clinical trials evaluating a combination of cetuximab with chemotherapy. However, whether BRAF mutations are negative predictive biomarkers for anti-EGFR mAb has not been ascertained, because the controlled study, which directly compared the efficacy of adding anti-EGFR mAb to chemotherapy with that of chemotherapy alone, is lacking in a small population with BRAF mutations. The application of novel strategies targeting BRAF kinase is warranted for the treatment of patients with CRC with BRAF mutations to improve their poor survival.
The mechanism of how anti-EGFR mAb functions is now being revealed. However, clinical data suggest that the Ras/Raf/ERK pathway is insufficient for completely predicting the response to anti-EGFR mAbs. Therefore, other factors, such as PIK3CA/PTEN deregulation and/or the expression status of epiregulin or amphiregulin, should also be focused on as possible predictive biomarkers for anti-EGFR mAb.
ACKNOWLEDGEMENTS
None declared.
CONFLICT OF INTEREST
None declared.
REFERENCES
- 1.Russo A, Rizzo S, Bronte G, Silvestris N, Colucci G, Gebbia N, Bazan V, Fulfaro F. The long and winding road to useful predictive factors for anti-EGFR therapy in metastatic colorectal carcinoma: the KRAS/BRAF pathway. Oncology. 2009;77(Suppl 1):57–68. doi: 10.1159/000258497. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectaltumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 3.Arteaga CL. Targeting HER1/EGFR: a molecular approach to cancer therapy. Semin. Oncol. 2003;30:3–14. [PubMed] [Google Scholar]
- 4.Arteaga CL. Overview of epidermal growth factor receptor biology and its role as a therapeutic target in human neoplasia. Semin. Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.35642. [DOI] [PubMed] [Google Scholar]
- 5.Fang JY, Richardson BC. The MAPK signaling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler KW, Vogelstein B. Colorectal tumors. In: Kinzler KW, Vogelstein B, editors. The genetic basis of human cancer. London (U.K.): McGraw-Hill; 1999. pp. 565–587. [Google Scholar]
- 7.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of BRAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 8.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 9.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 10.Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br. J. Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 13.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J. Clin. Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 14.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N. Engl. J. Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 15.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 16.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J. Clin. Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 17.De Roock W, Claes B, Bernasconi D, De Schutter J, Bies-mans B, Fountzilas G, Kalogeras KT, Kotoula V, Papa-michael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 18.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter ‘‘RASCAL’’ study. J. Natl. Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 19.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin. Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakar S, Deng G, Sahai V, Matsuzaki K, Tanaka H, Miura S, Kim YS. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Arch. Pathol. Lab. Med. 2008;132:958–964. doi: 10.5858/2008-132-958-CCCIMP. [DOI] [PubMed] [Google Scholar]
- 21.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 23.Spindler KL, Lindebjerg J, Nielsen JN, Olsen DA, Bisgård C, Brandslund I, Jakobsen A. Epidermal growth factor receptor analyses in colorectal cancer: a comparison of methods. Int. J. Oncol. 2006;29:1159–1165. [PubMed] [Google Scholar]
- 24.Moroni M, Sartore-Bianchi A, Benvenuti S, Artale S, Bardelli A, Siena S. Somatic mutation of EGFR catalytic domain and treatment with gefitinib in colorectal cancer. Ann. Oncol. 2005;16:1848–1849. doi: 10.1093/annonc/mdi356. [DOI] [PubMed] [Google Scholar]
- 25.Scartozzi M, Bearzi I, Pierantoni C, Mandolesi A, Loupakis F, Zaniboni A, Catalano V, Quadri A, Zorzi F, Berardi R, Biscotti T, Labianca R, Falcone A, Cascinu S. Nuclear factor-kB tumor expression predicts response and survival in irinotecan-refractory metastatic colorectal cancer treated with cetuximab-irinotecan therapy. J. Clin. Oncol. 2007;25:3930–3935. doi: 10.1200/JCO.2007.11.5022. [DOI] [PubMed] [Google Scholar]
- 26.Ng K, Zhu AX. Targeting the epidermal growth factor receptor in metastatic colorectal cancer. Crit. Rev. Oncol/Hematol. 2008;65:8–20. doi: 10.1016/j.critrevonc.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E, Canestrari E, Funel N, Schiavon G, Petrini I, Magnani M, Tonini G, Campani D, Floriani I, Cascinu S, Falcone A. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J. Clin. Oncol. 2009;27:2622–2629. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 28.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann. Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 29.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J. Clin. Oncol. 2005;23:1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 30.Mattoon D, Klein P, Lemmon MA, Lax I, Schlessinger J. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc. Natl. Acad. Sci. USA. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Defize LH, Arndt-Jovin DJ, Jovin TM, Boonstra J, Meisenhelder J, Hunter T, de Hey HT, de Laat SW. A431 cell variants lacking the blood group A antigen display increased high affinity epidermal growth factor-receptor number, protein-tyrosine kinase activity, and receptor turnover. J Cell Biol. 1988;107:939–949. doi: 10.1083/jcb.107.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooi A, Takehana T, Li X, Suzuki S, Kunitomo K, Iino H, Fujii H, Takeda Y, Dobashi Y. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod. Pathol. 2004;17:895–904. doi: 10.1038/modpathol.3800137. [DOI] [PubMed] [Google Scholar]
- 33.Barber T D, Vogelstein B, Kinzler K W, Velculescu V E. Somatic Mutations of EGFR in Colorectal Cancers and Glioblastomas. N. Engl. J. Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, Fieuws S, Vandesompele J, Peeters M, Van Laethem JL, Humblet Y, Pénault-Llorca F, De Hertogh G, Laurent-Puig P, Van Cutsem E, Tejpar S. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J. Clin. Oncol. 2009;27:5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 35.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbi-son CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA, 3rd, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J. Clin. Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 36.Scartozzi M, Bearzi I, Mandolesi A, Giampieri R, Faloppi L, Galizia E, Loupakis F, Zaniboni A, Zorzi F, Biscotti T, Labianca R, Falcone A, Cascinu S. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br. J. Cancer. 2011;104(11):1786–1790. doi: 10.1038/bjc.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J. Natl. Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disordersand cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 39.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 40.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SNL, Lövig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Sena-gore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br. J. Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boughdady IS, Kinsella AR, Haboubi NY, Schofield PF. K-ras gene mutation in colorectal adenomas and carcinomas from familial adenomatous polyposis patients. Surg. Oncol. 1992;1:269–274. doi: 10.1016/0960-7404(92)90087-2. [DOI] [PubMed] [Google Scholar]
- 42.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3(10):1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 43.Danielsen SA, Lind GE, Bjørnslett M, Meling GI, Rognum TO, Heim S, Lothe RA. Novel mutations of the suppressor gene PTEN in colorectal carcinomas stratified by microsatellite instability- and TP53 mutation- status. Hum. Mutat. 2008;29(11):E252–262. doi: 10.1002/humu.20860. [DOI] [PubMed] [Google Scholar]
- 44.Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomäki P, de la Chapelle A, Aaltonen LA, Eng C. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002;161(2):439–447. doi: 10.1016/S0002-9440(10)64200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 46.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 47.Yokota T, Shibata N, Ura T, Takahari D, Shitara K, Muro K, Yatabe Y. Cycleave polymerase chain reaction method is practically applicable for V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS)/V-raf murine sarcoma viral oncogene homolog B1 (BRAF) genotyping in colorectal cancer. Transl. Res. 2010;156:98–105. doi: 10.1016/j.trsl.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 48.COSMIC (Catalogue Of Somatic Mutations In Cancer) Database, S. Institute, Welcome Trust Genome Campus, Hinxton, Cambridge, http://www.sanger.ac.uk/genetics/CGP/cosmic/
- 49.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 50.Fariña-Sarasqueta A, an Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann. Oncol. 2010;21(12):2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 51.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 52.Frattini M, Balestra D, Suardi S, Oggionni M, Alberici P, Radice P, Costa A, Daidone MG, Leo E, Pilotti S, Bertario L, Pierotti MA. Different genetic features associated with colon and rectal carcinogenesis. Clin. Cancer Res. 2004;10:4015–4021. doi: 10.1158/1078-0432.CCR-04-0031. [DOI] [PubMed] [Google Scholar]
- 53.Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA. RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia. 2008;10:680–686. doi: 10.1593/neo.08312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei SC, Yu CY, Tsai-Wu JJ, Su YN, Sheu JC, Wu CH, Wang CY, Wong JM. Low mutation rate of hMSH2 and hMLH1 in Taiwanese hereditary non-polyposis colorectal cancer. Clin. Genet. 2003;64:243–251. doi: 10.1034/j.1399-0004.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- 55.Lee SC, Guo JY, Lim R, Soo R, Koay E, Salto-Tellez M, Leong A, Goh BC. Clinical and molecular characteristics of hereditary non-polyposis colorectal cancer families in Southeast Asia. Clin. Genet. 2005;68:137–145. doi: 10.1111/j.1399-0004.2005.00469.x. [DOI] [PubMed] [Google Scholar]
- 56.Goldberg Y, Porat RM, Kedar I, Shochat C, Sagi M, Eilat A, Mendelson S, Hamburger T, Nissan A, Hubert A, Kadouri L, Pikarski E, Lerer I, Abeliovich D, Bercovich D, Peretz T. Mutation spectrum in HNPCC in the Israeli population. Fam. Cancer. 2008;7:309–317. doi: 10.1007/s10689-008-9191-y. [DOI] [PubMed] [Google Scholar]
- 57.Mancl EE, Kolesar JM, Vermeulen LC. Clinical and economic value of screening for Kras mutations as predictors of response to epidermal growth factor receptor inhibitors. Am. J. Health Syst. Pharm. 2009;66:2105–2112. doi: 10.2146/ajhp090036. [DOI] [PubMed] [Google Scholar]
- 58.Kim IJ, Kang HC, Jang SG, Kim K, Ahn SA, Yoon HJ, Yoon SN, Park JG. Oligonucleotide microarray analysis of distinct gene expression patterns in colorectal cancer tissues harboring BRAF and K-ras mutations. Carcinogenesis. 2006;27:392–404. doi: 10.1093/carcin/bgi237. [DOI] [PubMed] [Google Scholar]
- 59.Deng G, Kakar S, Tanaka H, Matsuzaki K, Miura S, Sleisenger MH, Kim YS. Proximal and distal colorectal cancers show distinct gene-specific methylation profiles and clinical and molecular characteristics. Eur. J. Cancer. 2008;44:1290–1301. doi: 10.1016/j.ejca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 60.Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and KRAS-mutated colorectal cancer and implications for prognosis. Int. J. Cancer. 2010;127:367–380. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 61.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, Yatabe Y. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Catalano V, Loupakis F, Graziano F, Torresi U, Bisonni R, Mari D, Fornaro L, Baldelli AM, Giordani P, Rossi D, Alessandroni P, Giustini L, Silva RR, Falcone A, D'Emidio S, Fedeli SL. Mucinous histology predicts for poor response rate and overall survival of patients with colorectal cancer and treated with first-line oxaliplatin- and/or irinotecan-based chemotherapy. Br. J. Cancer. 2009;100:881–887. doi: 10.1038/sj.bjc.6604955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogino S, Brahmandam M, Cantor M, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod. Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 64.Bennecke M, Kriegl L, Bajbouj M, Retzlaff K, Robine S, Jung A, Arkan MC, Kirchner T, Greten FR. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell. 2010;18:135–146. doi: 10.1016/j.ccr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Castagnola P, Giaretti W. Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim. Biophys. Acta. 2005;1756:115–125. doi: 10.1016/j.bbcan.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Bokemeyer C, Köhne CH, Rougier P, Stroh C, Schlichting M, Van Cutsem E. (2010) Cetuximab with chemotherapy (CT) as first-line treatment for metastatic colorectal cancer (mCRC): Analysis of the CRYSTAL and OPUS studies according to KRAS and BRAF mutation status. ASCO Annual Meeting, Abstract No. 3506. J. Clin. Oncol. 2010;28:15s. [Google Scholar]
- 67.Alberts S R, Sargent D J, Smyrk T C, Shields A F, Chan E, Goldberg R M, Gill S, Kahlenberg M S, Thibodeau S N, Nair S. Adjuvant mFOLFOX6 with or without cetuxiumab (Cmab) in KRAS wild-type (WT) patients (pts) with resected stage III colon cancer (CC): Results from NCCTG Intergroup Phase III Trial N0147. 2010 ASCO Annual Meeting, Abstract No. 3507. J. Clin. Oncol. 2010;28:18s. [Google Scholar]
- 68.Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL, Kay E, Mitchell JK, Madi A, Jasani B, James MD, Bridgewater J, Kennedy MJ, Claes B, Lambrechts D, Kaplan R, Cheadle JP MRC COIN Trial Investigators. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377(9783):2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Connell M J, Lavery I C, Gray R G, Quirke P, Kerr D J, Lopatin M, Yothers G A, Lee M, Clark-Langone K, Wolmark N. Comparison of molecular and pathologic features of stage II and stage III colon cancer in four large studies conducted for development of the 12-gene colon cancer recurrence score. 2010 ASCO Annual Meeting, Abstract No. 3503. J. Clin. Oncol. 2010;28:15s. [Google Scholar]
- 70.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 71.Guerrero S, Casanova I, Farré L, Mazo A, Capellà G, Mangues R. K-ras Codon 12 Mutation Induces Higher Level of Resistance to Apoptosis and Predisposition to Anchorage-independent Growth Than Codon 13 Mutation or Proto-Oncogene Overexpression. Cancer Res. 2000;60(23):6750–6756. [PubMed] [Google Scholar]
- 72.Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G, Salerno S, Tomasino RM, Labianca R, Gebbia N, Russo A. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann. Oncol. 2002;13:1438–1446. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 73.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 74.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Pi-essevaux H, Van Cutsem E, O'Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S. Association of RAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]