Abstract
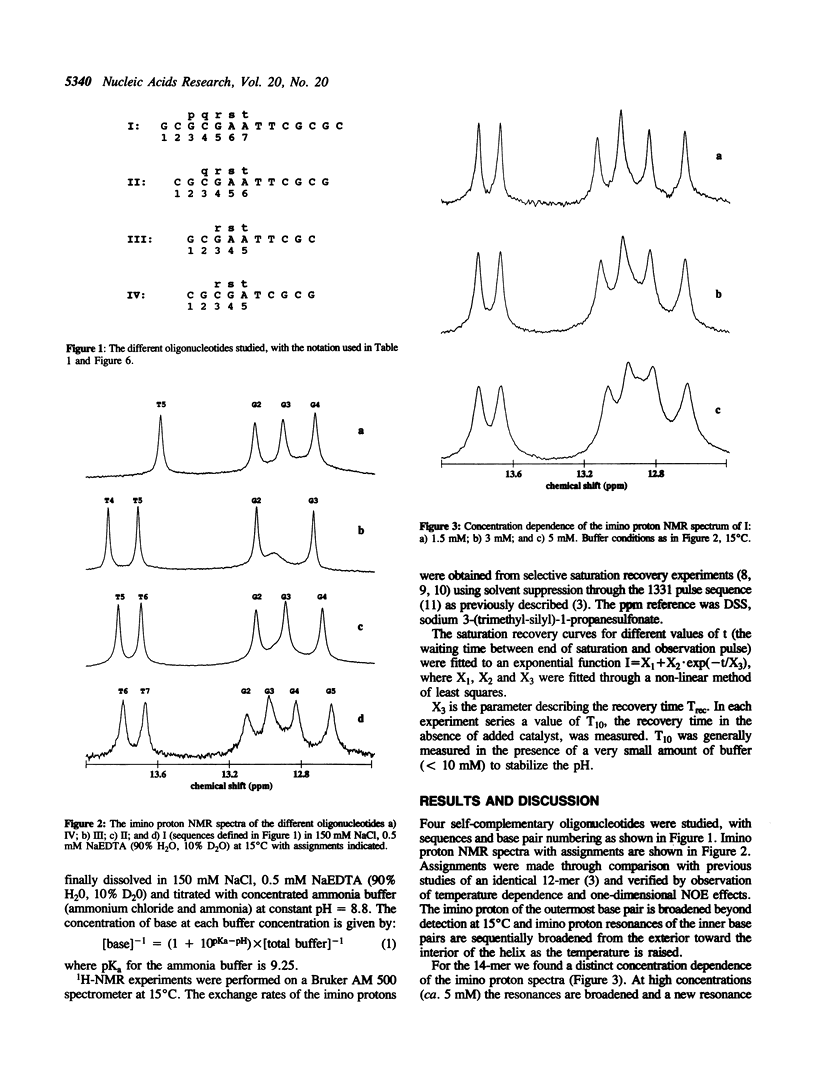
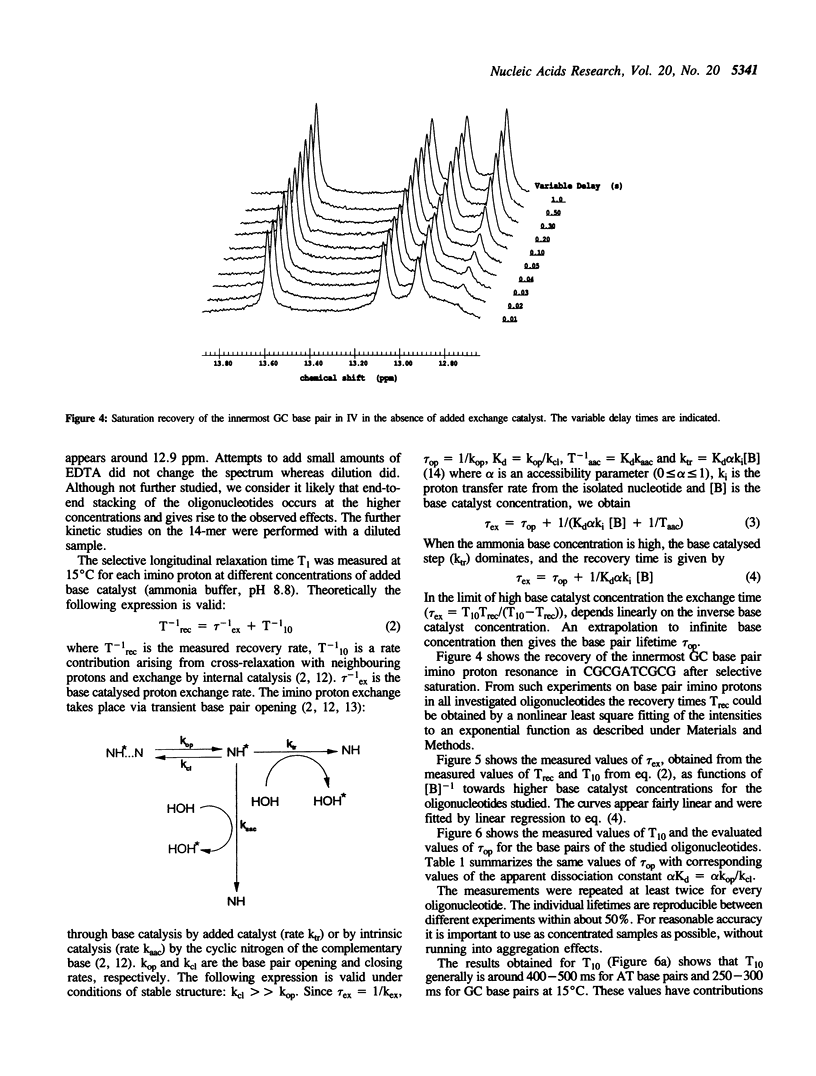
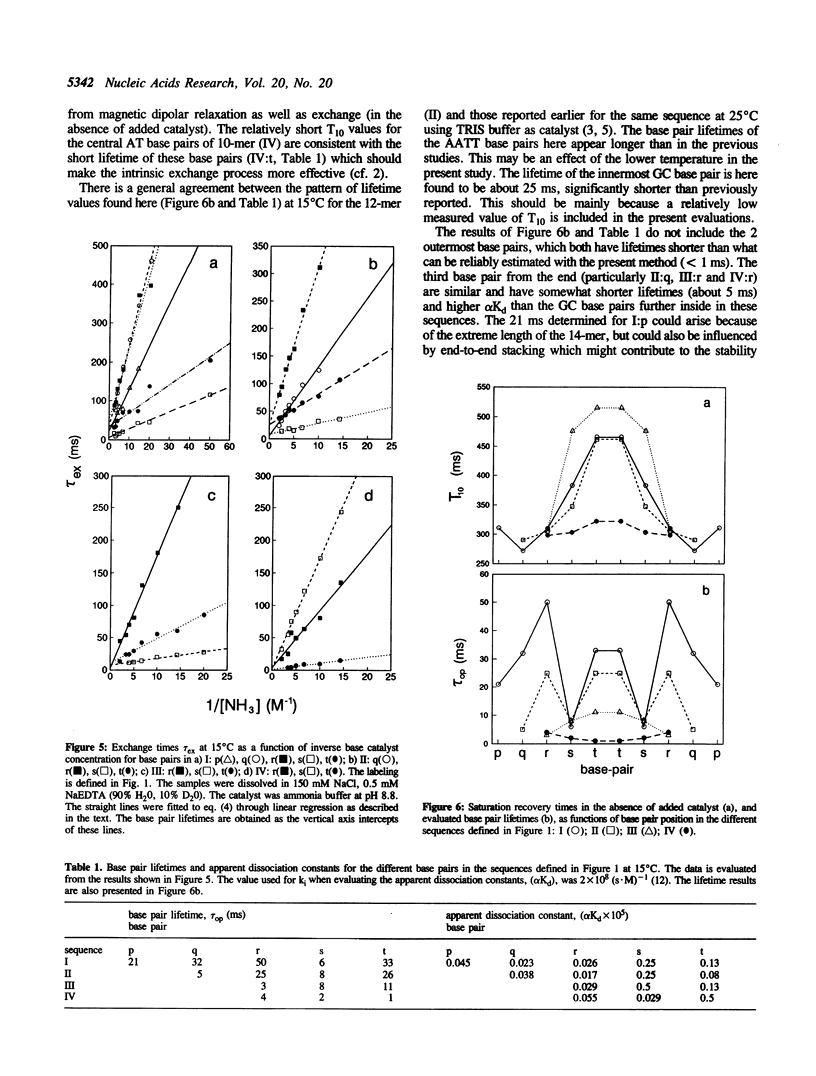
The base catalysed imino proton exchange in DNA oligonucleotides of different sequences and lengths was studied by 1H-NMR saturation recovery experiments. The self-complementary sequences studied were GCGCGAATTCGCGC (I), CGCGAATTCGCG (II), GCGAATTCGC (III), and CGCGATCGCG (IV). The evaluation of base pair lifetimes was made after correction for the measured 'absence of added catalyst' effect which was found to be characterized by recovery times of 400-500 ms for the AT base pairs and 250-300 ms for the GC base pairs at 15 degrees C. End effects with rapid exchange is noticeable up to 3 base pairs from either end of the duplexes. The inner hexamer cores GAATTC of sequences I-II show similar base pair lifetime patterns, around 30 ms for the innermost AT, 5-10 ms for the outer AT and 20-50 ms for the GC base pairs at 15 degrees C. The shorter sequences III and particularly IV show much shorter lifetimes in their central AT base pairs (11 ms and 1 ms, respectively).
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braunlin W. H., Bloomfield V. A. 1H NMR study of the base-pairing reactions of d(GGAATTCC): salt and polyamine effects on the imino proton exchange. Biochemistry. 1988 Feb 23;27(4):1184–1191. doi: 10.1021/bi00404a018. [DOI] [PubMed] [Google Scholar]
- Guéron M., Kochoyan M., Leroy J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987 Jul 2;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. An NMR study of the exchange rates for protons involved in the secondary and tertiary structure of yeast tRNA Phe. Nucleic Acids Res. 1977 Oct;4(10):3599–3615. doi: 10.1093/nar/4.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Study of transfer ribonucleic acid unfolding by dynamic nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3996–4006. doi: 10.1021/bi00517a008. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Broseta D., Guéron M. Proton exchange and base-pair kinetics of poly(rA).poly(rU) and poly(rI).poly(rC). J Mol Biol. 1985 Jul 5;184(1):165–178. doi: 10.1016/0022-2836(85)90050-6. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Charretier E., Kochoyan M., Guéron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988 Dec 13;27(25):8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Gao X. L., Guéron M., Patel D. J. Proton exchange and internal motions in two chromomycin dimer-DNA oligomer complexes. Biochemistry. 1991 Jun 11;30(23):5653–5661. doi: 10.1021/bi00237a003. [DOI] [PubMed] [Google Scholar]
- Lycksell P. O., Gräslund A., Claesens F., McLaughlin L. W., Larsson U., Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987 Nov 11;15(21):9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe J. G., Russu I. M. Proton exchange and base-pair opening kinetics in 5'-d(CGCGAATTCGCG)-3' and related dodecamers. Nucleic Acids Res. 1990 Feb 25;18(4):821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Pardi A., Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982 May 7;216(4546):581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- Teitelbaum H., Englander S. W. Open states in native polynucleotides. I. Hydrogen-exchange study of adenine-containing double helices. J Mol Biol. 1975 Feb 15;92(1):55–78. doi: 10.1016/0022-2836(75)90091-1. [DOI] [PubMed] [Google Scholar]



