Abstract
Caspase-3 is synthesized as a dormant proenzyme and is maintained in an inactive conformation by an Asp-Asp-Asp “safety-catch” regulatory tripeptide contained within a flexible loop near the large-subunit/small-subunit junction. Removal of this “safety catch” results in substantially enhanced autocatalytic maturation as well as increased vulnerability to proteolytic activation by upstream proteases in the apoptotic pathway such as caspase-9 and granzyme B. The safety catch functions through multiple ionic interactions that are disrupted by acidification, which occurs in the cytosol of cells during the early stages of apoptosis. We propose that the caspase-3 safety catch is a key regulatory checkpoint in the apoptotic cascade that regulates terminal events in the caspase cascade by modulating the triggering of caspase-3 activation.
Apoptosis is mediated by a proteolytic cascade in which upstream “activator” caspases (group III enzymes) initiate and amplify the maturation of “effector” caspases (group II enzymes) that, in turn, cleave a discrete subset of cellular polypeptides to manifest the apoptotic phenotype (1–3). The activation of proximal caspases in the apoptotic cascade (e.g., caspase-8 and -9) is thought to occur by an autocatalytic mechanism. For example, ligation of the CD95 (Fas, APO-1) receptor recruits procaspase-8 to an oligomeric complex via the adapter protein FADD/MORT1 (4, 5). The proximity of multiple caspase-8 proenzymes results in maturation and activation, presumably by autolytic proteolysis. Similarly, oligomerization of procaspase-9 is mediated by APAF-1 after a cytochrome c-stimulated conformational change (6). As with procaspase-8, procaspase-9 maturation probably occurs as a consequence of autolytic proteolysis. An autolytic mechanism of activation presupposes that the proenzymes themselves have sufficient catalytic activity to mediate self-maturation under the appropriate conditions. Whereas autolytic activation has been demonstrated for caspase-1, -8, and -9 after oligomerization (7–10), procaspase-3 appears to be particularly resistant to activation by this mechanism (ref. 7 and unpublished data). Because activator (group III) caspases require effector (group II) enzymes such as caspase-3 to mediate apoptosis, the resistance of procaspase-3 to autolytic maturation may safeguard cells from accidental apoptosis and serve as a terminal checkpoint in the apoptotic cascade. In this report, we demonstrate that the caspase-3 proenzyme is catalytically competent but under strict regulatory self-control by an Asp-Asp-Asp tripeptide contained within the proenzyme itself. This “safety-catch” regulatory tripeptide accounts for procaspase-3 resistance to autolytic maturation as well as resistance to proteolytic activation by upstream proteases such as caspase-9 and granzyme B. These results suggest that the safety-catch regulatory tripeptide plays an important role in modulating the sensitivity of cells to apoptosis and that release of the caspase-3 safety catch may be an important determinant of apoptotic competency.
Materials and Methods
Patient Samples.
Protein extracts from colon tissue specimens from 20 patients with adenocarcinoma were prepared as described (11).
Immunoblotting.
Lysates were analyzed by SDS/PAGE followed by transfer to nitrocellulose and immunoblotting, using an antibody (MF#R280) raised against the large subunit of recombinant human caspase-3, essentially as described (12).
Synthesis of Irreversible Active-Site Probe (biotin-DEVD-aomk).
Acyloxymethylketones react with and covalently modify the catalytic cysteine of caspases (13). To design a DEVDase-directed, active-site probe, FMOC-Glu(OTBU)-OH was coupled (HOBT,DCI) with Val-OALL in CHCL3 at 0oC for 2.5 h and purified on silica gel with hexane/ethyl acetate (60:40). FMOC-Glu(OTBU) Val-OALL was deprotected with pyrrolidine/(Ph3P)4Pd, coupled (HOBT,DCI), and purified as described above. FMOC-Asp(OTBU) Glu(OTBU) Val-OALL was deprotected with pyrrolidine/(Ph3P)4Pd and coupled with biotin (HOBT, DCI) in dimethylformamide at 0°C for 2 days. After purification on silica gel with dichloromethane/methanol (95:5), biotin-Asp(OTBU) Glu(OTBU) Val-OALL was deprotected with pyrrolidine/(Ph3P)4Pd in tetrahydrofuran, coupled as described (14, 15), and purified on silica gel with dichloromethane/methanol (94:6). The final deprotection was done with trifluoroacetic acid in dichloromethane to obtain biotin-DEVD-aomk (L772,094).
Biotinylation of Caspase-3.
Biotinylation of catalytically active caspase-3 was carried out by incubating lysates with the active-site probe biotin-DEVD-aomk (L772,094) in ICE buffer III [50 mM Hepes-KOH, pH 7.0/2 mM EDTA/0.1% (wt/vol) CHAPS/10% (wt/vol) sucrose/5 mM DTT] at 37°C for 30 min. Biotinylated proteins were resolved by SDS/PAGE, transferred to nitrocellulose, and visualized by using streptavidin coupled to horseradish peroxidase (Amersham Pharmacia) followed by enhanced chemiluminescence detection (ECL system; Amersham Pharmacia). Immunoprecipitation of biotinylated proteins was carried out by using the anti-caspase-3 antibody MF397 in TENT buffer (50 mM Tris⋅HCl, pH 8.0/2 mM EDTA/150 mM NaCl/1% Triton X-100) and protein A-Sepharose beads. Immune precipitates were visualized by SDS/PAGE and blotted by using streptavidin coupled to horseradish peroxidase.
Mutagenesis of Procaspase-3.
The cDNA for human procaspase-3, flanked by either BspHI sites or XbaI sites (introduced by PCR), was subcloned in the NcoI site of the prokaryotic expression vector pET11d (Novagen) or the XbaI site of the eukaryotic expression vector pTracer-CMV (Invitrogen), respectively. Mutagenesis of procaspase-3 was carried out by using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene). The mutations and entire sequence integrity were confirmed by dideoxy sequencing. (Note that the pET11d cloning strategy substitutes a Ser for the Glu present immediately after the initiating Met.)
Preparation of Bacterial Lysates Expressing Procaspase-3.
Wild-type and mutant procaspase-3 cDNAs, subcloned in the prokaryotic expression vector pET11d, were transformed into the protease-deficient strain of Escherichia coli, BL21 (DE3) pLysS (Novagen). A single colony was used to inoculate 10 ml of M9 medium containing 100 μg/ml carbenicillin and 34 μg/ml chloramphenicol. When the culture reached an OD600 of 0.6, protein expression was induced with 1 mM isopropyl β-d-thiogalactoside. Aliquots of 2.5 ml were removed at the indicated times and centrifuged, and bacterial pellets were lysed by sonication in ICE buffer III and then cleared by centrifugation.
Ac-DEVD-AMC Cleavage Assay.
The catalytic activity of caspase-3 was determined by measuring the proteolytic cleavage of the fluorogenic substrate Ac-DEVD-AMC in a reaction mixture containing 10 μM substrate in ICE buffer III. Production of AMC was measured continuously in a Cytofluor 96-well plate reader by using an excitation wavelength of 380 nm and an emission wavelength of 460 nm. In some experiments, the Ac-DEVD-AMC cleavage activity was determined after treatment for 1 h at 37°C with molar excess amounts of granzyme B, purified as described (3), to assess the total amount of enzyme that could be activated. The amount of active procaspase-3 used in the cleavage assays performed in Fig. 4d was determined by measuring the Ac-DEVD-AMC cleavage activity in an E. coli extract expressing an uncleavable form of procaspase-3 (D9E, D28E, D175E). An uncleavable form of procaspase-3 was chosen to ensure that no mature caspase-3 was contaminating the extract. The specific activity of procaspase-3 was shown (S.R., E. Peterson, N.A.T., and D.W.N., unpublished results) to be identical to the specific activity of mature caspase-3 (1 pmol AMC cleaved per min per μl = 1 nM).
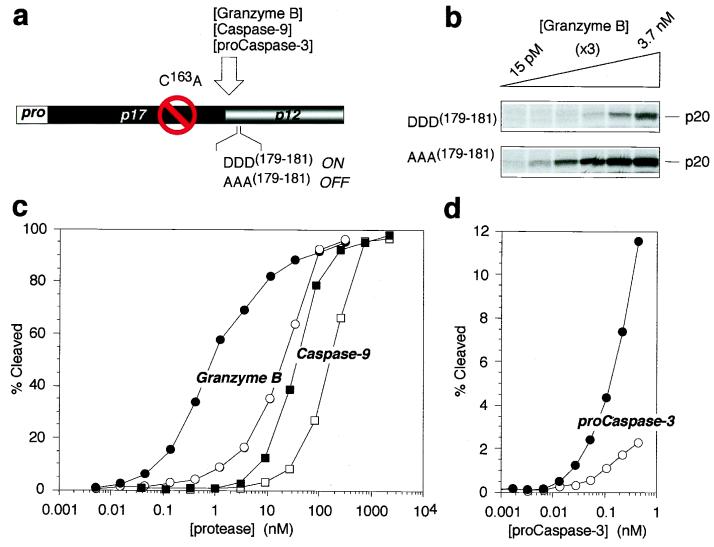
Figure 4.
The safety catch modulates vulnerability of procaspase-3 to activation by upstream proteases. (a) Diagram of procaspase-3 mutants used to test vulnerability to upstream proteases. (b) Representative fluorogram of granzyme B-mediated cleavage of 35S-labeled procaspase-3 with intact (DDD(179–181)) or disabled (AAA(179–181)) safety catch. (c) Quantitation of procaspase-3 DDD(179–181) (open objects) and AAA(179–181) (solid objects) cleavage by granzyme B (circles) and caspase-9 (squares). (d) Quantitation of procaspase-3 DDD(179–181) (●) and AAA(179–181) (○) cleavage by a catalytically active form of procaspase-3 expressed in E. coli (D9E, D28E, D175E).
Transfection of Procaspase-3.
HeLa cells seeded on 60-mm dishes (0.75 × 106 cells per dish) were transfected with the pTracer-CMV (Invitrogen) constructs by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. After 48 h, cells were scraped from the dish, pooled with the culture medium, and collected by centrifugation. Cell pellets were lysed in 200 μl of a buffer containing 62.5 mM Tris⋅HCl (pH 6.8), 4 M urea, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.003% bromophenol blue followed by sonication for 1 min, and lysates (5 μl) were analyzed by SDS/PAGE and immunoblotting by using the anti-caspase-3 antibody MF#R280 as described above.
In Vitro Cleavage of Procaspase-3 Mutants.
[35S]Methionine-labeled procaspase-3 C163A containing an intact “safety catch” (DDD179–181) or a mutated safety catch (AAA179–181) was generated by coupled in vitro transcription/translation using the Promega TNT reticulocyte lysate system and tested in an in vitro cleavage assay (12) by using either recombinant human caspase-9 (16), purified granzyme B (3), or an E. coli extract expressing a catalytically active procaspase-3 (D9E, D28E, D175E), prepared as described above.
Maturation of Procaspase-3 by Acidification.
Lysates were prepared from the E. coli strain BL21(DE3) pLysS as described above or from the human neuronal precursor cell line NT2 (Stratagene) according to the procedure described (6). Five microliters of the bacterial lysate or 35 μg of the NT2 lysate was incubated in buffer A (25 mM Mes/25 mM Pipes/25 mM Tricine/10 mM KCl/1.5 mM MgCl2/1 mM EDTA/1 mM EGTA/5 mM DTT), adjusted to the indicated pH with KOH. (Mes, Pipes, and Tricine were combined as buffers because they have distinct pKa values that cover a broad pH range.) After a 2-h incubation at 37°C, the maturation of caspase-3 was assessed by (i) anti-caspase-3 immunoblotting and/or (ii) measuring proteolytic cleavage of the substrate Ac-DEVD-AMC, as described above.
Results
Procaspase-3 Is Catalytically Competent but Resists in Vivo Autoactivation.
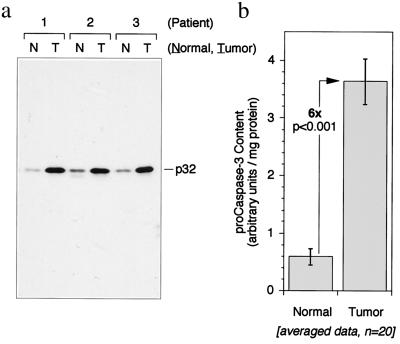
Most procaspases undergo rapid autolytic activation when accumulated by mammalian cell transfection, overexpression in bacteria, or induced oligomerization (refs. 7–10 and unpublished observations). One notable exception is procaspase-3 (the major effector caspase in most cell types), which is remarkably resistant to autoactivation under most experimental conditions. This resistance to activation also appears to extend to the enzyme in vivo because we have observed that high levels of procaspase-3 are present in colonic adenocarcinomas from human cancer patients without evidence of proteolytic maturation or apoptosis (Fig. 1). Because the resistance of procaspase-3 to proteolytic activation may play a critical role in determining the sensitivity of cells to apoptosis and thus may contribute to the attenuated apoptosis observed in many cancers, we sought to determine the molecular basis for this resistance.
Figure 1.
The caspase-3 proenzyme is overexpressed in human colon adenocarcinomas. (a) α-Caspase-3 immunoblot of normal human colonic mucosa (N) and autologous tumor tissue (T). Three representative patients are shown. (b) Quantitative analysis of caspase-3 expression in tumor and adjacent normal mucosa from 20 patients. Mean and SE are shown.
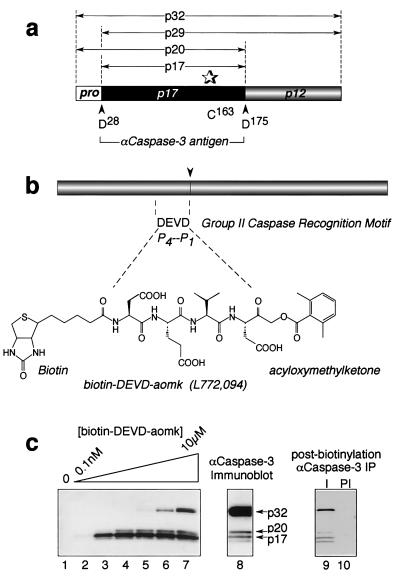
One obvious possibility that could account for the inability of procaspase-3 to autoactivate under physiological conditions is a lack of catalytic activity within the proenzyme. This would be in contrast to other caspases whose proforms clearly contain a catalytic activity that is sufficient for full proteolytic maturation when the proenzymes are recruited into oligomeric complexes (e.g., caspase-8, -9, and others). To determine whether caspase-3 proenzyme activity could be detected in its native mammalian environment, a sensitive caspase-3-directed, active-site probe was synthesized (biotin-DEVD-aomk; Fig. 2b) that requires catalytic turnover to covalently biotinylate the thiol of the active-site cysteine. In Jurkat cells that were stimulated to undergo apoptosis, where part of the procaspase-3 pool was converted to its mature form (Fig. 2c, lane 8), both the mature enzyme plus a fraction of the p32 precursor were labeled with the active-site probe (lane 7). The identity of the biotinylated 32-kDa polypeptide as procaspase-3 was confirmed by immunoprecipitation with an antibody to caspase-3 (lane 9). Labeling was not observed in nonapoptotic cells (data not shown), suggesting that the proenzyme acquires catalytic competency as a consequence of signals or conditions that accompany apoptotic stimulation.
Figure 2.
The caspase-3 proenzyme exhibits catalytic activity in apoptotic Jurkat cell extracts. (a) Caspase-3 proenzyme organization. (b) Structure of the active-site probe, biotin-DEVD-aomk (L772,094). (c) Biotinylation of procaspase-3 by L772,094 in apoptotic extracts from Jurkat cells. I, α-caspase-3 immune; PI, preimmune.
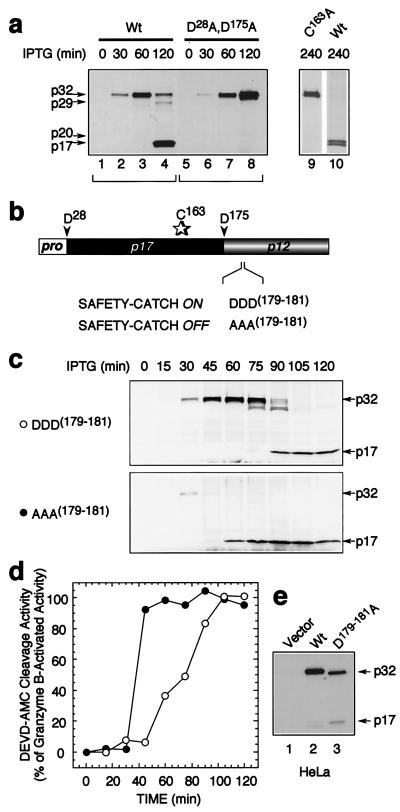
All mammalian cells contain endogenous caspases and, thus, are unsuited for studying procaspase autocatalysis. We therefore expressed procaspase-3 in a protease-deficient strain of E. coli from a vector containing an isopropyl β-d-thiogalactoside-inducible promoter. Full expression of the p32 caspase-3 proenzyme occurred after 1 h of induction, with latent conversion to the mature enzyme within 2 h (Fig. 3a, lanes 1–4). Maturation was autocatalytic and not a result of residual bacterial proteases because a point mutant of the active-site cysteine (C163A) remained entirely unprocessed even after 4 h (Fig. 3a, lane 9 vs. 10). This indicates that the caspase-3 proenzyme harbors sufficient catalytic activity to mediate its own proteolytic maturation and is not processed by any other activity in these bacteria. Procaspase-3 is processed at two sites in vivo to yield the fully mature protease; cleavage after D175 separates the large and small subunits whereas cleavage after D28 removes the short prodomain from the amino terminus of the large subunit (see Fig. 1a). To verify that the autoprocessing of the enzyme in bacteria faithfully reproduced the cleavage events that the enzyme undergoes in mammalian cells in vivo, we mutated these sites (D28A,D175A) and observed complete ablation of autoproteolytic maturation (Fig. 1a, lanes 5–8). This system therefore could be used to examine the resistance of procaspase-3 to autoactivation in the absence of other components of the apoptotic machinery that otherwise would be present in mammalian cells.
Figure 3.
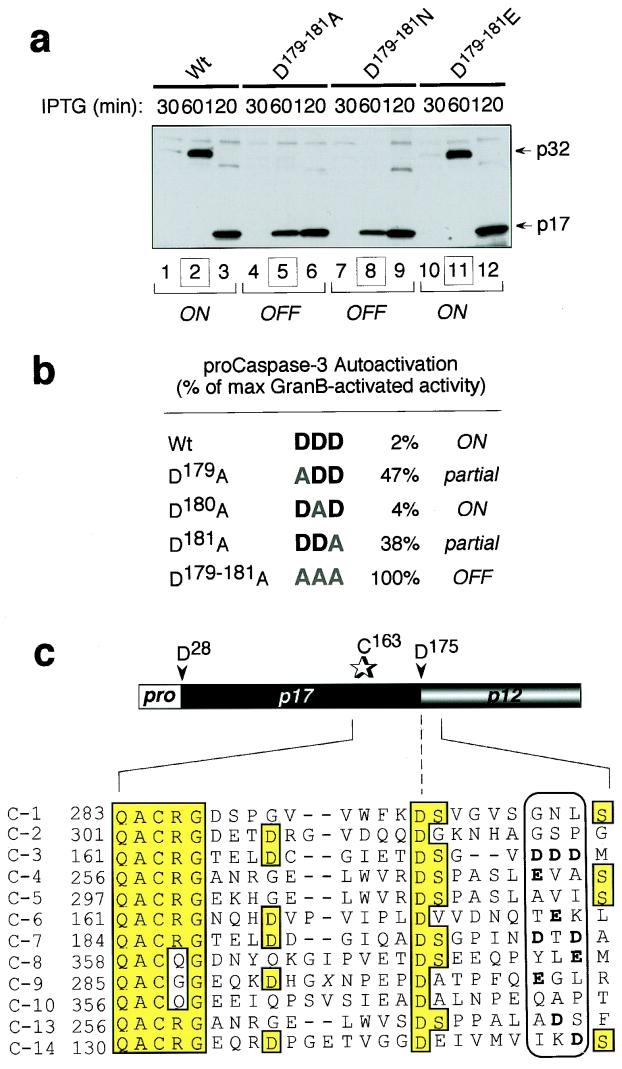
Identification of a safety-catch regulatory tripeptide responsible for the latent autocatalytic activation of procaspase-3. (a) Latent autocatalytic maturation of the caspase-3 proenzyme when expressed in E. coli, as visualized by α-caspase-3 immunoblotting. (b) Diagram of procaspase-3 showing the safety-catch regulatory tripeptide D179–181. (c and d) α-Caspase-3 immunoblot (c) and corresponding Ac-DEVD-AMC cleavage activity (d) of time course of autolytic maturation of wild-type and mutant procaspase-3 D179–181A expressed in E. coli. (e) Accelerated autoactivation of the procaspase-3 mutant D179–181A after transfection in HeLa cells, as visualized by α-caspase-3 immunoblotting.
Identification of a Regulatory Safety-Catch Tripeptide That Mediates Procaspase-3 Dormancy.
The latency of maturation in bacteria, even when procaspase-3 was expressed at very high levels, suggests that the major determinant for resistance to autoactivation is harbored within the proenzyme itself and possibly could be a regulatory peptide segment as observed in other protease superfamilies. We therefore set out to determine the nature of procaspase-3 dormancy and establish the molecular basis for resistance to autocatalytic maturation. Most proteases are synthesized as inactive precursors (zymogens) and require removal of an “activation segment,” usually contained within the prodomain, to achieve full catalytic competency (reviewed in ref. 17). The inhibitory peptide segment in procaspase-3 was not attributable to the prodomain because its removal did not affect the rate of autocatalytic maturation after isopropyl β-d-thiogalactoside-induced expression of pro-caspase-3 in bacteria or activation after transfection into mammalian cells (data not shown). To identify the putative inhibitory peptide segment, we performed extensive mutagenesis along the entire procaspase-3 polypeptide to search for residues that might contribute to proenzyme dormancy. This was aided by observations that the latent autocatalytic maturation of procaspase-3 in bacterial cultures after prolonged incubation (>60 min; see Fig. 3a) was coincident with medium acidification and preliminary studies that indicated that acidification facilitated autoactivation through the disruption of ionic bridges. We therefore used the x-ray crystal structure of caspase-3 (18) to home in on candidate residues that (i) would bear ionic charge and be capable of forming salt bridges and (ii) occurred in solvent-exposed flexible loops. After extensive mutagenesis, a sequence of three adjacent aspartic acids (residues 179–181), immediately downstream of the cleavage site between the large and small subunits, was identified as the regulatory peptide (Fig. 3b). Mutagenic elimination of the tripeptide by conversion to Ala residues (D179–181A) resulted in accelerated autocatalytic maturation (Fig. 3c) with 100% of the expressed protein becoming catalytically competent (Fig. 3d) (e.g., granzyme B treatment did not unmask latent activity as it did for the wild-type protease). The relevance of the tripeptide and its putative regulatory role was substantiated further in mammalian cells by demonstrating that transfection of HeLa cells with the procaspase-3 mutant (D179–181A), in which the regulatory tripeptide was eliminated, resulted in substantial maturation of the proenzyme whereas transfected wild-type procaspase-3 remained entirely unprocessed (Fig. 3e), as is commonly observed in mammalian cells that overexpress wild-type procaspase-3. Because the regulatory tripeptide modulates the propensity for autolytic triggering of caspase-3 activity, we named it the caspase “safety catch” to reflect its role in obviating caspase-3 activation.
The Safety Catch Modulates Procaspase-3 Vulnerability to Activation by Upstream Proteases.
Procaspase-3 shows resistance to autocatalytic activation when expressed in bacteria as well as in mammalian cells, and this latency can be overcome by removal of the safety-catch regulatory tripeptide by mutagenesis (described above). We next wanted to determine whether this was also true for trans-activation of procaspase-3 by other proteases that lie upstream of caspase-3 in the apoptotic cascade. An autocatalytically silent mutant of procaspase-3 was synthesized in reticulocyte lysate and used as a substrate for determining the ability of other proteases to cleave the proenzyme at the activation junction between the large and small subunits (Fig. 4a). Removal of the caspase-3 regulatory safety catch increased the vulnerability of the proenzyme to maturation by cytotoxic T lymphocyte-derived granzyme B (ca. 20-fold; Fig. 4 b and c), as well as by the upstream initiator of the “intrinsic” cell death pathway, caspase-9 (ca. 10-fold; Fig. 4c) and by catalytically competent procaspase-3 (ca. 10-fold; Fig. 4d). Surprisingly, the cleavage of procaspase-3 by the upstream initiator of the receptor-mediated “extrinsic” cell death pathway, caspase-8, was unaffected by the presence or absence of the safety-catch tripeptide within procaspase-3 (data not shown). Collectively, these findings suggest that the status of the safety catch governs the sensitivity of procaspase-3 to most apoptotic initiator mechanisms, with the notable exception of the caspase-8 pathway.
Components of the Regulatory Safety Catch.
Individual components of the safety-catch tripeptide were examined next to delineate the key elements of its inhibitory activity. Mutation of the tripeptide in procaspase-3 to isosteric Asn (D179–181N) recapitulated the rapid autoactivation seen with the Ala-substituted (D179–181A) enzyme whereas substitution of the Asps with another anionic tripeptide, Glu (D179–181E), restored the wild-type phenotype (Fig. 5a). The ability of the isoelectronic Glu, but not the isosteric Asn, to substitute for Asp as the regulatory tripeptide prompted us to examine the possibility that oppositely charged amino acids such as Arg or Lys were forming an ionic interaction with the safety-catch regulatory tripeptide, resulting in inhibition of caspase-3 autoactivation. Arg and Lys residues play a critical role in forming the caspase-3 active site (18), but these could not be probed by mutagenesis because of loss of catalytic activity. The x-ray crystal structure of caspase-3 was used to identify potential, surface-exposed cationic-binding partners proximal to the Asp-Asp-Asp tripeptide. These, however, were excluded by mutagenesis because none of the mutations (including K38Q, R144,147Q, R238,241Q, and K271Q) affected the rate of procaspase-3 autoactivation (data not shown). Within the safety catch itself, the single amino acid mutations D179A and D181A, but not D180A, were intermediate in their rate of autoactivation, suggesting the involvement of multiple ionic contacts (Fig. 5b). The function of the regulatory safety catch therefore is endowed by the peripheral Asp residues of the DDD tripeptide. Interestingly, the corresponding tripeptide in procaspase-7 (another effector caspase and the caspase most related to caspase-3) is DTD, with the two key aspartic acids that contribute to the safety catch in procaspase-3 being conserved (Fig. 5c). This peripheral anionic pair is not present in the corresponding region from any other human caspases.
Figure 5.
Two peripheral anionic residues within the safety-catch tripeptide are sufficient to mediate the resistance of procaspase-3 to activation. (a) α-Caspase-3 immunoblot of time course of autolytic maturation of wild-type (Wt) and mutant (D179–181A, D179–181N, D179–181E) procaspase-3 in E. coli. (b) Ac-DEVD-AMC cleavage activity of Wt and mutant procaspase-3 (D179A, D180A, D181A, and D179–181A) expressed as a percentage of total granzyme B-activated activity. (c) Conservation of the safety-catch regulatory tripeptide in procaspase-3 and -7.
Modulation of the Safety Catch by Intracellular pH.
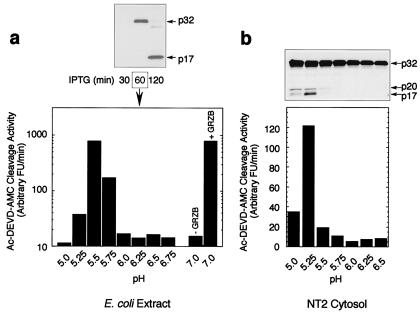
We next sought to determine how the caspase-3 safety catch might be modulated in vivo. An initial clue came from the observation that the latent autocatalytic maturation of procaspase-3 in bacterial cultures after prolonged incubation (>60 min; see Fig. 3a) was coincident with medium acidification. Acidification-induced autoactivation could be reproduced in vitro by incubating recombinant procaspase-3, derived from bacteria, in buffers at low pH (Fig. 6a). The same was true for the endogenous procaspase-3 present in neuronal-precursor NT2 cell cytosols (Fig. 6b) as well as purified homogeneous procaspase-3(His)6 (data not shown). The observation that acidification induces the autoactivation of endogenous procaspase-3 in NT2 cell cytosols suggests that the safety-catch Asp-Asp-Asp tripeptide plays a critical role in maintaining the dormancy of procaspase-3 in eukaryotic cells. The triggering of procaspase-3 autoactivation by acidification is consistent with disruption of intramolecular salt bridges, an activation mechanism that has been observed previously in other protease families (17). Alternatively, acidification would increase the degree of protonation of the peripheral Asp residues, rendering them neutral and, hence, similar to the D179–181N mutant, which lost safety-catch behavior. These mechanisms are likely to be significant in vivo as well because intracellular acidification is known to occur during apoptosis (19–22). Significant levels of apoptosis also have been observed by using a proton ionophore under conditions in which intracellular pH is clamped to 6.0 or below, but not to 6.75 (23), consistent with the pH ranges necessary for procaspase-3 autoactivation. Hyperacidification of the mitochondrial intermembrane space, where a substantial portion of the procaspase-3 pool is contained in some cell types (24), also has been demonstrated during apoptosis (25). Finally, mitochondria-induced acidification of the cytosol occurs as an early event during apoptosis and strongly favors caspase activation by the “intrinsic” cell death pathway (26). The caspase-3 proenzyme therefore may use the safety catch to sense the physiological environment of the cell and arm the enzyme for rapid activation when the pH of the cell declines during the early stages of apoptosis (Fig. 7a). We propose that this is an important regulatory checkpoint that both precludes the accidental activation of procaspase-3 in healthy cells having stable intracellular pH while facilitating proteolytic activation of caspase-3 in damaged or stressed cells in which homeostatic maintenance of normal cellular pH is perturbed. In this context, the ability of caspase-8 to ignore the status of the safety catch may have evolved as a mechanism to mediate the killing of perfectly healthy cells, as is required during “death receptor”-mediated culling of cell populations.
Figure 6.
Acidification disables the safety catch and promotes procaspase-3 activation. (a) Ac-DEVD-AMC cleavage activity in procaspase-3 expressing bacterial extracts after incubation in buffers at varying pH levels. (b) α-Caspase-3 immunoblot and corresponding Ac-DEVD-AMC cleavage activity in cytosolic extracts from the neuronal precursor cells NT2 treated as described in a.
Figure 7.
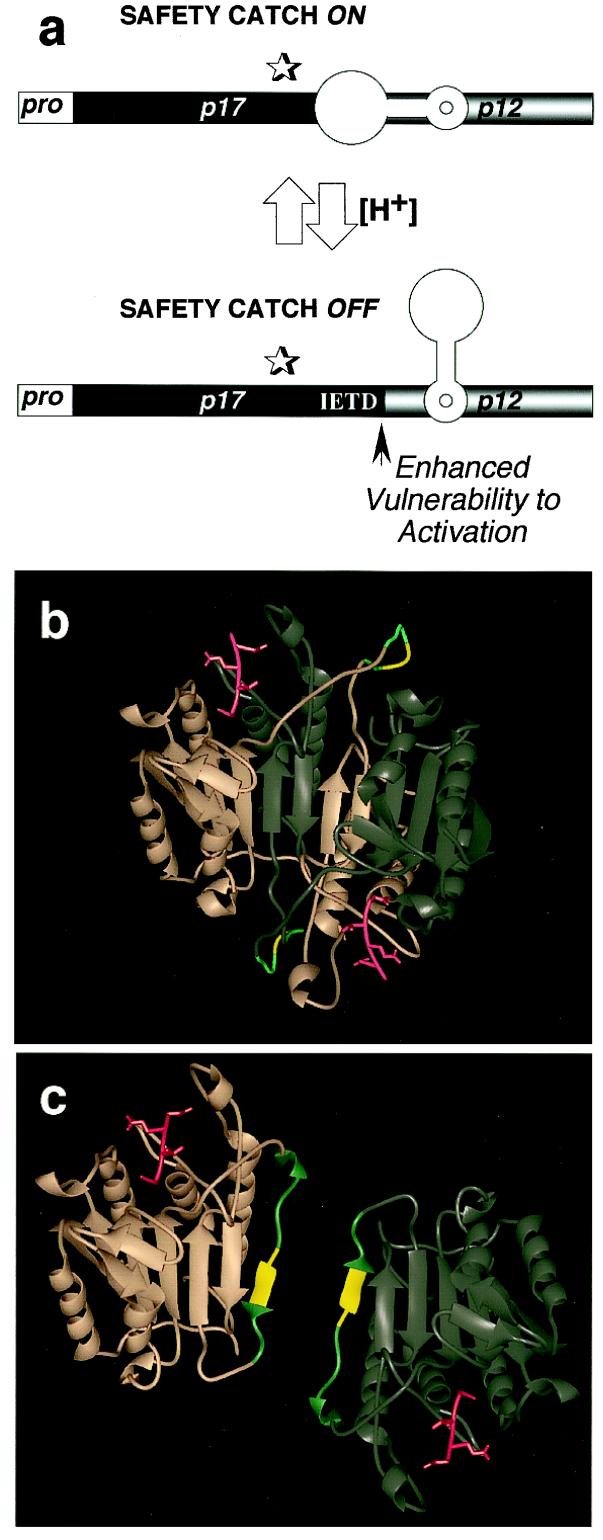
Modeling the safety catch. (a) Diagram of procaspase-3 showing the safety catch in the on and off positions. (b and c) Two proenzyme structures are proposed based on the x-ray crystal structure of mature caspase-3 complexed with the tetrapeptide–aldehyde inhibitor Ac-DEVD-CHO (18), in red. Modeled (in teal) are the 12 aa (residues 174–185) omitted from the x-ray crystal structure of mature caspase-3, with the safety catch Asp-Asp-Asp in yellow. (b) The first model shows the interdigitation of two caspase-3 proenzymes (in copper and pewter), with the 12-aa flexible loop assuming a pH-sensitive configuration that renders inaccessible the IETD175 cleavage site. (c) The second model shows each procaspase-3 as independent, with the 12-aa flexible loop capping the two edges of the hydrophobic β-sheets that normally interact to form the caspase-3 dimer, structuring the IETD175 cleavage site to make it unavailable.
Discussion
In summary, we have identified an unexpected mechanism for caspase regulation that involves a self-contained regulatory tripeptide, composed of three sequential Asp residues, that maintains the dormancy of the caspase-3 proenzyme. This tripeptide, which we have termed the safety catch, must be dissociated by disruption of ionic interactions by acidification or other unknown factors to permit full commitment to apoptosis.
The subunit organization of mature caspase-3, based on the x-ray crystal structure, suggests two potential mechanisms by which the safety-catch regulatory tripeptide might occlude the IETD-processing site at the subunit junction within the proenzyme. One possibility is that the safety-catch tripeptide is contained within two, interdigitating caspase-3 proenzymes in a highly flexible loop between the large and small subunits and assumes a pH-sensitive conformation that renders the IETD cleavage site inaccessible under normal conditions and vulnerable after cellular acidification (Fig. 7b). An alternative possibility is that the safety-catch regulatory tripeptide causes the IETD-processing site to cap the hydrophobic β-sheets that normally come together in the mature caspase-3 heterotetramer, rendering it inaccessible to cleavage (Fig. 7c). The mutual repulsion between the two DDD tripeptides in such a configuration also would be expected to interfere with dimerization and proximity-induced trans-activation. In both of these cases, the availability of the processing site between the large and small subunits would be encumbered and thus would interfere with the vulnerability of procaspase-3 to upstream proteases and to autoactivation.
The strict control of procaspase-3 activation bestowed by the safety catch raises the question of how this might be regulated in vivo. Disabling of the safety catch, for example, appears to be assisted by the intracellular acidification that is known to accompany apoptosis. In contrast, resistance to apoptosis may be conferred by as-yet-unknown molecules that stabilize the safety catch. We have observed, for example, that XIAP abolishes acidification-triggered procaspase-3 autoactivation in vitro (IC50 = 95 nM; data not shown), although we were unable to distinguish between safety-catch stabilization vs. direct inhibition of proenzyme catalytic activity.
Finally, some therapeutic opportunities may exist for accelerating apoptosis (i.e., in cancer treatment) if the safety catch tripeptide can be bound with cationic-binding partners and thereby be precluded from its regulatory role in maintaining procaspase-3 dormancy. It has been demonstrated, for example, that the proapoptotic properties of RGD-containing peptides are mediated in part by their ability to induce caspase-3 autolytic maturation (27). This was suggested to be a result of conformational changes mediated by the binding of RGD peptides to a RGD-binding motif (DDM) in procaspase-3. The two aspartic acids in the proposed RGD-binding motif are part of the safety-catch tripeptide described herein. Although there is no evidence that RGD-containing proteins modulate caspases in vivo, the likelihood that intracellular acidification assists in the activation of the apoptotic cascade via the caspase-3 safety catch is compelling. This mechanism therefore adds an important level of regulatory control over a critical component of the cell death machinery and suggests potential opportunities for apoptosis sensitization.
Acknowledgments
We thank Dr. M. Garcia-Calvo and J. Tam for providing recombinant human caspase-9 and purified granzyme B, respectively, as well as J. Vaillancourt for preparing the caspase amino acid alignment.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nicholson D W. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw W C, Martins L M, Kaufmann S H. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 3.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 4.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Chinnaiyan A M, Kischkel F C, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 6.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Chang H Y, Baltimore D. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 9.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 10.Martin D A, Siegel R M, Zheng L, Lenardo M J. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 11.Kargman S L, O'Neill G P, Vickers P J, Evans J F, Mancini J A, Jothy S. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]
- 12.Gervais F G, Thornberry N A, Ruffolo S C, Nicholson D W, Roy S. J Biol Chem. 1998;273:17102–17108. doi: 10.1074/jbc.273.27.17102. [DOI] [PubMed] [Google Scholar]
- 13.Thornberry N A, Peterson E P, Zhao J J, Howard A D, Griffin P R, Chapman K T. Biochemistry. 1994;33:3934–3940. doi: 10.1021/bi00179a020. [DOI] [PubMed] [Google Scholar]
- 14.Chapman K T. Bioorg Med Chem Lett. 1992;2:613–618. [Google Scholar]
- 15.Krantz A, Copp L J, Coles P J, Smith R A, Heard S B. Biochemistry. 1991;30:4678–4687. doi: 10.1021/bi00233a007. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Calvo M, Peterson E P, Rasper D M, Vaillancourt J P, Zamboni R, Nicholson D W, Thornberry N A. Cell Death Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 17.Khan A R, James M N. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotonda J, Nicholson D W, Fazil K M, Gallant M, Gareau Y, Labelle M, Peterson E P, Rasper D M, Ruel R, Vaillancourt J P, et al. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 19.Furlong I J, Ascaso R, Lopez Rivas A, Collins M K. J Cell Sci. 1997;110:653–661. doi: 10.1242/jcs.110.5.653. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds J E, Li J, Eastman A. Cytometry. 1996;25:349–357. doi: 10.1002/(SICI)1097-0320(19961201)25:4<349::AID-CYTO6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Wolf C M, Reynolds J E, Morana S J, Eastman A. Exp Cell Res. 1997;230:22–27. doi: 10.1006/excr.1996.3401. [DOI] [PubMed] [Google Scholar]
- 22.Gottlieb R A, Nordberg J, Skowronski E, Babior B M. Proc Natl Acad Sci USA. 1996;93:654–658. doi: 10.1073/pnas.93.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle K M, Irwin J P, Humes B R, Runge S W. J Cell Biochem. 1997;67:231–240. [PubMed] [Google Scholar]
- 24.Mancini M, Nicholson D W, Roy S, Thornberry N A, Peterson E P, Casciola-Rosen L A, Rosen A. J Cell Biol. 1998;140:1485–1495. doi: 10.1083/jcb.140.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Heiden M G, Chandel N S, Williamson E K, Schumacker P T, Thompson C B. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 26.Matsuyama S, Llopis J, Deveraux Q L, Tsien R Y, Reed J C. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 27.Buckley C D, Pilling D, Henriquez N V, Parsonage G, Threlfall K, Scheel-Toellner D, Simmons D L, Akbar A N, Lord J M, Salmon M. Nature (London) 1999;397:534–539. doi: 10.1038/17409. [DOI] [PubMed] [Google Scholar]