Abstract
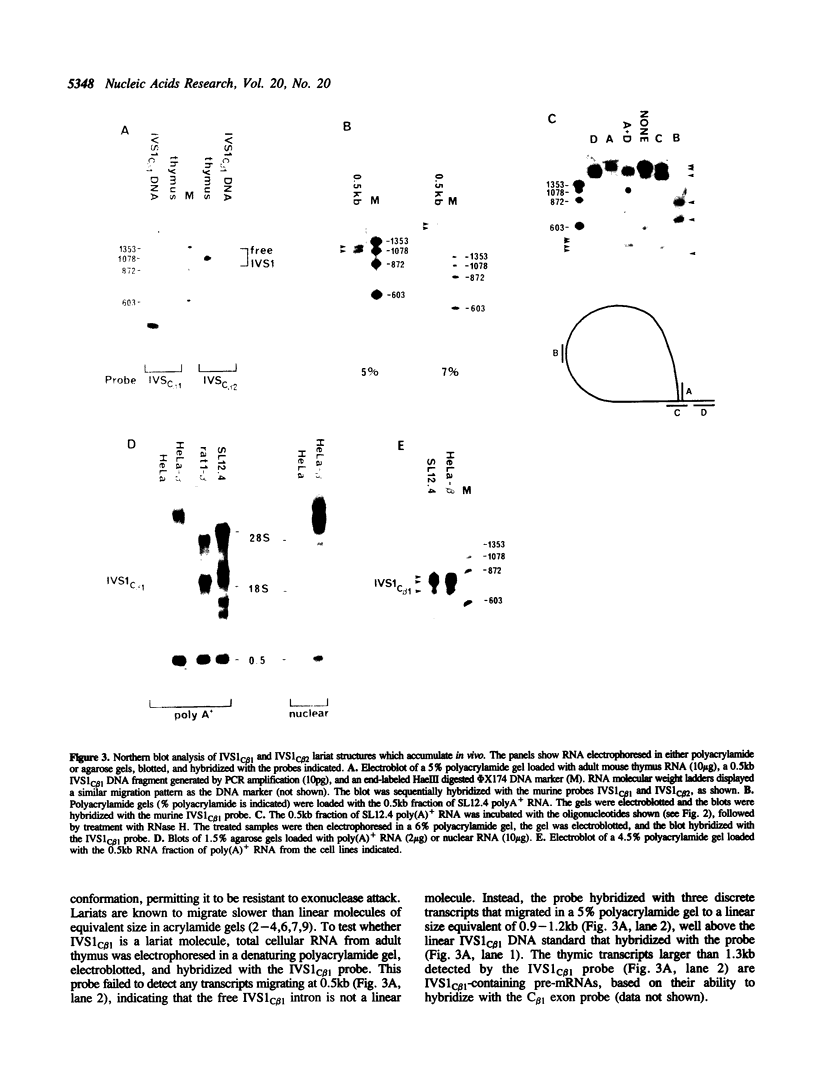
The vast majority of mammalian genes are interrupted by non-coding segments of DNA termed introns. Introns are spliced out of RNA transcripts as lariat structures, and then are typically debranched and rapidly degraded. Here, we described an unusual spliced intron from the constant region of the T cell receptor-beta (TCR-beta) locus that is relatively stable in mammalian cells. This intron, IVS1C beta 1, accumulates as a set of lariat RNA structures with different length tails in the nucleus of T cells. The accumulation of this spliced intron is developmentally regulated during murine thymocyte ontogeny. The property of stability appears to be evolutionarily conserved since the human version of this intron also accumulates in T cells. The stability is selective since other spliced TCR-beta introns do not detectably accumulate in T cells. The unusual stability of this intron does not depend on T cell specific factors since non-T cells transfected with TCR-beta gene constructs also accumulate spliced IVS1C beta 1. The discovery of a mammalian intron that accumulates as a lariat in vivo provides an opportunity to elucidate mechanisms that regulate intron debranching, stability, and nuclear localization.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman S. A., Bursztajn S., Bowen B., Gilbert W. Localization of an acetylcholine receptor intron to the nuclear membrane. Science. 1990 Jan 12;247(4939):212–214. doi: 10.1126/science.1688472. [DOI] [PubMed] [Google Scholar]
- Chapman K. B., Boeke J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991 May 3;65(3):483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- Coleclough C., Wood D. Introns excised from immunoglobulin pre-mRNAs exist as discrete species. Mol Cell Biol. 1984 Oct;4(10):2017–2022. doi: 10.1128/mcb.4.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. M. Molecular genetics of the T cell-receptor beta chain. Annu Rev Immunol. 1985;3:537–560. doi: 10.1146/annurev.iy.03.040185.002541. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Farrell M. J., Dobson A. T., Feldman L. T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink P. J., Matis L. A., McElligott D. L., Bookman M., Hedrick S. M. Correlations between T-cell specificity and the structure of the antigen receptor. Nature. 1986 May 15;321(6067):219–226. doi: 10.1038/321219a0. [DOI] [PubMed] [Google Scholar]
- Gascoigne N. R., Chien Y., Becker D. M., Kavaler J., Davis M. M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature. 1984 Aug 2;310(5976):387–391. doi: 10.1038/310387a0. [DOI] [PubMed] [Google Scholar]
- Harpold M. M., Evans R. M., Salditt-Georgieff M., Darnell J. E. Production of mRNA in Chinese hamster cells: relationship of the rate of synthesis to the cytoplasmic concentration of nine specific mRNA sequences. Cell. 1979 Aug;17(4):1025–1035. doi: 10.1016/0092-8674(79)90341-6. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Maxwell E. S. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990 Nov 25;18(22):6565–6571. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Minard K., Mjolsness S., Kronenberg M., Goverman J., Hunkapiller T., Prystowsky M. B., Yoshikai Y., Fitch F., Mak T. W. Mouse T cell antigen receptor: structure and organization of constant and joining gene segments encoding the beta polypeptide. Cell. 1984 Jul;37(3):1101–1110. doi: 10.1016/0092-8674(84)90444-6. [DOI] [PubMed] [Google Scholar]
- Michaeli T., Pan Z. Q., Prives C. An excised SV40 intron accumulates and is stable in Xenopus laevis oocytes. Genes Dev. 1988 Aug;2(8):1012–1020. doi: 10.1101/gad.2.8.1012. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Qian L., Wilkinson M. DNA fragment purification: removal of agarose 10 minutes after electrophoresis. Biotechniques. 1991 Jun;10(6):736–738. [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Konarksa M. M., Grabowski P. J., Lamond A. I., Marciniak R., Seiler S. R. Splicing of messenger RNA precursors. Cold Spring Harb Symp Quant Biol. 1987;52:277–285. doi: 10.1101/sqb.1987.052.01.033. [DOI] [PubMed] [Google Scholar]
- Toyonaga B., Yoshikai Y., Vadasz V., Chin B., Mak T. W. Organization and sequences of the diversity, joining, and constant region genes of the human T-cell receptor beta chain. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8624–8628. doi: 10.1073/pnas.82.24.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A., Kefford R., Milstein C., Forster A., Rabbitts T. H. Sequence and evolution of the human T-cell antigen receptor beta-chain genes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5068–5072. doi: 10.1073/pnas.82.15.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. F., MacLeod C. L. Induction of T-cell receptor-alpha and -beta mRNA in SL12 cells can occur by transcriptional and post-transcriptional mechanisms. EMBO J. 1988 Jan;7(1):101–109. doi: 10.1002/j.1460-2075.1988.tb02788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M., Doskow J., Lindsey S. RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res. 1991 Feb 11;19(3):679–679. doi: 10.1093/nar/19.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]