Abstract
Cohesin is a protein complex originally identified for its role in sister chromatid cohesion, although increasing evidence portrays it also as a major organizer of interphase chromatin. Vertebrate cohesin consists of Smc1, Smc3, Rad21/Scc1 and either stromal antigen 1 (SA1) or SA2. To explore the functional specificity of these two versions of cohesin and their relevance for embryonic development and cancer, we generated a mouse model deficient for SA1. Complete ablation of SA1 results in embryonic lethality, while heterozygous animals have shorter lifespan and earlier onset of tumourigenesis. SA1-null mouse embryonic fibroblasts show decreased proliferation and increased aneuploidy as a result of chromosome segregation defects. These defects are not caused by impaired centromeric cohesion, which depends on cohesin-SA2. Instead, they arise from defective telomere replication, which requires cohesion mediated specifically by cohesin-SA1. We propose a novel mechanism for aneuploidy generation that involves impaired telomere replication upon loss of cohesin-SA1, with clear implications in tumourigenesis.
Keywords: cancer, chromosome segregation, cohesion, embryonic development, mouse model
Introduction
Cohesin was originally identified as a mediator of sister chromatid cohesion (Guacci et al, 1997; Michaelis et al, 1997; Losada et al, 1998). Its cohesive function is essential for accurate chromosome segregation and for homologous recombination (HR)-mediated DNA repair in G2 (Nasmyth and Haering, 2009). More recent evidence suggests that cohesin acts as a chromatin loop organizer in the interphase nucleus and thereby contributes to regulate gene expression, at least of certain loci (Hadjur et al, 2009; Mishiro et al, 2009; Nativio et al, 2009; Kagey et al, 2010; Seitan et al, 2011). It also participates in the spatial organization of DNA replication factories, which in turn determines origin activation and thus the efficiency of the duplication process (Guillou et al, 2010). Cohesin is composed of a heterodimer of Smc1 and Smc3, the kleisin subunit Rad21 (also known as Scc1/Mcd1) and a protein called Scc3 in yeast and stromal antigen (SA) in vertebrates, that form a ring-like structure that embraces the chromatin fibre(s) (Anderson et al, 2002; Haering et al, 2008). Two additional proteins named Pds5 and Wapl interact closely with cohesin to modulate its binding to chromatin and are sometimes considered cohesin subunits (Losada et al, 2005; Gandhi et al, 2006; Kueng et al, 2006; Gause et al, 2010). Another cohesion factor named Sororin associates with and stabilizes the cohesin rings embracing two sister chromatids after DNA replication (Lafont et al, 2010; Nishiyama et al, 2010).
In somatic vertebrate cells, there are two versions of the Scc3/SA subunit, SA1 and SA2 (Carramolino et al, 1997; Losada et al, 2000; Sumara et al, 2000). They are ∼1250 amino acid long proteins with >75% sequence identity along their central core, which contains two long regions of α-rod repeats (Palidwor et al, 2009). The N-terminal and C-terminal domains are more divergent. Cohesin-SA2 (i.e., cohesin containing the SA2 subunit) is 3–10 times more abundant than cohesin-SA1 in human (HeLa) and Xenopus somatic cells, whereas Xenopus eggs contain 10 times more cohesin-SA1 than cohesin-SA2 (Losada et al, 2000; Holzmann et al, 2010). The functional differences between the two complexes are yet to be clarified. Immunofluorescent staining indicates that SA1 and SA2 distribute throughout interphase chromatin in an indistinguishable manner (Losada et al, 2000; Sumara et al, 2000). Genome-wide analyses using chromatin immunoprecipitation (ChIP) techniques also suggest that there are no significant differences between the distribution of a common cohesin subunit like Rad21 and either SA2 (Wendt et al, 2008) or SA1 (Rubio et al, 2008). Both cohesins are released in prophase and only a small fraction is left by metaphase to ensure chromosome alignment (Vagnarelli et al, 2004; Toyoda and Yanagida, 2006). SA1 and SA2 can be detected at the centromeres of mitotic chromosomes from Xenopus egg extracts and human cells, respectively (Losada et al, 2000; Hauf et al, 2005) and Sgo1-PP2A, the protector of centromeric cohesin, can reverse the phosphorylation of both SA subunits by Polo, at least in vitro (Rivera and Losada, 2009). However, downregulation of SA1 or SA2 in HeLa cells by siRNA has led to suggest that cohesin-SA2 is specifically required for centromeric cohesion whereas cohesin-SA1 is responsible for arm and telomere cohesion (Canudas and Smith, 2009).
To gain insight into the functional specificity of SA1 and SA2-containing cohesin complexes and their importance for cell viability, we have generated mouse embryos deficient for SA1. This approach ensures a complete absence of the protein and gives us the opportunity to transcend the cellular level in our studies and address the relevance of SA1 functions in embryonic development. This is particularly important in view of the existence of a human developmental disorder affecting 1 in 10 000 newborns, known as Cornelia de Lange syndrome (CdLS), caused by heterozygous mutations in the gene encoding the cohesin loader Nipbl/Scc2 (Liu and Krantz, 2009). Moreover, inactivation of SA2 has been very recently proposed to drive aneuploidy in human cancer (Solomon et al, 2011). Here, we show that SA1 deficiency results in delayed embryo development and lethality, which demonstrates that the functions of cohesin-SA1 and cohesin-SA2 are not redundant. We report for the first time that defects in cohesin-SA1 function generate aneuploidy and result in increased incidence and earlier onset of tumourigenesis. Unlike cohesin-SA2, lack of cohesin-SA1 leads to aneuploidy in the absence of precocious separation of sister chromatids (PSSC) in metaphase. Instead, chromosome segregation defects are caused by defective telomere replication in the absence of telomere cohesion, which is mediated exclusively by cohesin-SA1.
Results
Ablation of SA1 results in late embryonic lethality
We obtained from the German Gene Trap Consortium an embryonic stem (ES) cell line in which a rFlpROSA-geo cassette containing a splicing acceptor site and a polyadenylation sequence was inserted between exons 3 and 4 of the murine Stag1 gene encoding the SA1-cohesin subunit (Figure 1A). The insertion creates a null allele and mouse embryos carrying this allele turn blue upon incubation in X-gal (Figure 1B). Heterozygous mice are viable and fertile but they do not produce homozygous progeny, indicating that SA1 function is essential for viability (Supplementary Table S1). Homozygous mutant embryos can be found at Mendelian ratios by day 11.5 of gestation (E11.5), allowing us to obtain mouse embryonic fibroblasts (MEFs) lacking SA1 mRNA (Figure 1C) or protein (Figure 1D). SA1 transcripts are 1–3 times more abundant than those of SA2 in MEFs, ES cells and in cells from different mouse tissues (Supplementary Figure S1A and B) whereas SA2 protein levels are three times higher than SA1 in MEFs (Supplementary Figure S1C). The levels of SA2 mRNA and SA2 protein remain virtually unchanged in the SA1-null MEFs (Figure 1C and D, respectively), suggesting that no compensatory upregulation of the other SA subunit occurs. SA1-null MEFs show clear proliferation defects (Figure 1E) that cannot be attributed to an obvious cell-cycle arrest or to senescence (Figure 1F and data not shown), and are highly aneuploid even in very early passages (Figure 1G). Importantly, in cells obtained from fetal livers at E14.5, without any passage in culture, aneuploidy is also higher in the SA1-null liver cells (Figure 1H). Viability of SA1-null embryos strongly decreases by E12.5 but some embryos that survive to E18.5 are much smaller than their wild-type littermates (Figure 1I) and show a substantial reduction of BrdU incorporation in most embryonic tissues (Figure 1J). They present a clear developmental delay and additional features that are reminiscent of CdLS, such as impaired lipid metabolism and delayed ossification (Remeseiro et al, 2012). Thus, some functions of cohesin-SA1 that cannot be performed by cohesin-SA2 are essential for cell proliferation and, thereby, to fulfil embryonic development.
Figure 1.
A knockout mouse model for cohesin-SA1 subunit. (A) Schematic representation of the Stag1-knockout (KO) allele used in this study. The murine Stag1 locus encoding SA1 contains 34 exons. The precise location of the gene trap cassette is indicated as well as the position of the primers used for genotyping. LTR, long terminal repeat; SA, splice acceptor; βgeo, β-galactosidase/neomycin phosphotransferase fusion gene; pA, polyadenylation sequence; triangles represent target sites for FLPe and Cre recombinases (Schnutgen et al, 2005). (B) X-gal staining of whole embryos carrying the KO allele in heterozygosis (left) and homozygosis (right), and PCR analysis of DNA purified from cells of the indicated genotypes. (C) Quantitative RT–PCR analysis to evaluate the mRNA levels of SA1 and SA2 in the indicated MEFs. Values are given as picograms per 2 μg of total RNA. (D) Western blot analysis of whole-cell extracts prepared from MEFs. Tubulin is used as loading control. (E) Growth curves of primary MEFs of the indicated genotypes (left) and representative images of the cultures by day 6 after plating the same number of cells. (F) DNA content profile of asynchronous wild-type and SA1-null primary MEFs. Percentage of cells in each phase of the cell cycle is shown. (G) Graph showing the distribution in the number of chromosomes of at least 100 metaphases from two clones of wild-type, two clones of heterozygous and six clones of SA1-null primary MEFs. (H) Same analysis carried out in cells from fetal livers. At least 150 metaphases from three wild-type and three SA1-null embryos were examined. (I) E18.5 embryos from the same litter were photographed and genotyped. Notice the reduced size of the SA1-null embryos. (J) BrdU staining of skin and kidney sections from E17.5 embryos of the indicated genotypes. Arrowheads point to proliferative areas, such as hair follicles in the skin (top panels) and the outer layer of the kidney (bottom panels). Automated quantification of the relative BrdU-positive area in whole embryo sections with Definiens Software shows a clear reduction of 23.4±0.7% in SA1-null embryos with respect to wild-type (n=4 wild-type embryos and n=5 SA1-null E17.5 embryos were analysed). Scale bars, 200 μm (top) and 100 μm (bottom).
SA1 heterozygous animals have increased risk of cancer but are protected against acute tumourigenesis
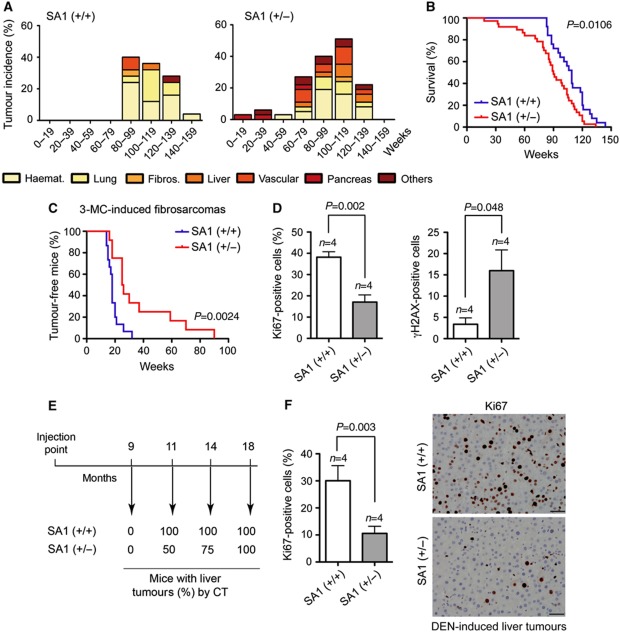
SA1 heterozygous mice have increased incidence of spontaneous tumours, shorter lifespan than their wild-type littermates and some features of premature ageing such as acute kyphosis (Figure 2A and B; Supplementary Figure S2A, respectively). Monoallelic inactivation of SA1 accelerates the onset of tumourigenesis and changes the spectrum of tumours from that observed in wild-type mice. Increased incidence of hepatocellular carcinomas and some other carcinomas, as well as vascular tumours, is observed in SA1 heterozygous mice. Most remarkable is the early appearance of cystic papillary neoplasm (labelled as ‘pancreas’ in Figure 2A), which resembles human pancreatic intraductal papillary mucinous neoplasm and is extremely infrequent in mice (Supplementary Figure S2B, top panel). Tumours of mesenchymal origin, such as haematopoietic tumours (lymphomas and histiocytic sarcomas), are common in ageing wild-type mice of this genetic background, but appear earlier in the SA1 heterozygous animals (see examples in Supplementary Figure S2B). To further address the contribution of SA1 haploinsufficiency to tumourigenesis, we induced tumour formation both genetically and chemically. In terms of survival and tumour incidence, we did not observe significant differences between wild-type and SA1 heterozygous mice in p53-null and PTEN heterozygous backgrounds, probably due to the dominant effect that the loss of function of these tumour suppressors has in tumourigenesis (Supplementary Figure S2C). Strikingly, young SA1 heterozygous mice showed clear resistance to carcinogenesis upon treatment with 3-methyl-colanthrene (3-MC) and diethyl-nitrosamine (DEN) to induce fibrosarcomas and liver tumours, respectively (Figure 2C and E). Increased DNA damage signalling (γH2AX staining) and lower proliferation rates (Ki67 staining) could be observed in fibrosarcoma samples from SA1 heterozygous mice (Figure 2D; Supplementary Figure S2D). DEN-induced liver tumours from these animals also showed reduced number of Ki67-positive cells (Figure 2F). Thus, we suspect that the reduced proliferative capacity of SA1-deficient cells delays the growth of acute induced tumours in the young animals. However, SA1 haploinsufficiency increases susceptibility to spontaneous tumours and accelerates death in ageing mice by promoting aneuploidy.
Figure 2.
Reduced lifespan and increased incidence of spontaneous tumours in SA1 heterozygous mice, but higher resistance against chemically induced tumours. (A) Tumour incidence in wild-type (n=25) and SA1 heterozygous mice (n=37) relative to animal age in weeks. Note that SA1 heterozygous mice present higher tumour incidence and earlier onset of tumourigenesis. Haematopoietic tumours, fibrosarcomas, lung, liver, vascular and pancreas tumours are represented; the category ‘others’ includes osteomas, papillomas and mammary gland tumours. (B) Kaplan–Meier survival curves for wild-type (blue) and SA1 heterozygous mice (red) (n=25 and 37, respectively). (C) Kaplan–Meier curves showing tumour-free survival for wild-type and SA1 heterozygous mice injected with 3-MC to induce fibrosarcomas (n=15 animals per genotype). (D) Quantification of cells showing positive staining for Ki67 and γH2AX on tissue sections from fibrosarcomas like those shown in Supplementary Figure S2D. Five fields were counted per mouse of a total of four mice of each genotype. (E) Wild-type and SA1 heterozygous 15-day-old male mice (n=4 each) were injected with DEN and appearance of liver tumours was assessed by computed tomography (CT) at the indicated times after injection. (F) Quantification of Ki67-positive cells (left) in tissue sections of the liver tumours induced by DEN (right). Five fields were counted per mouse of a total of four mice of each genotype. Scale bars, 50 μm.
Telomere-specific cohesion defects in SA1-null cells
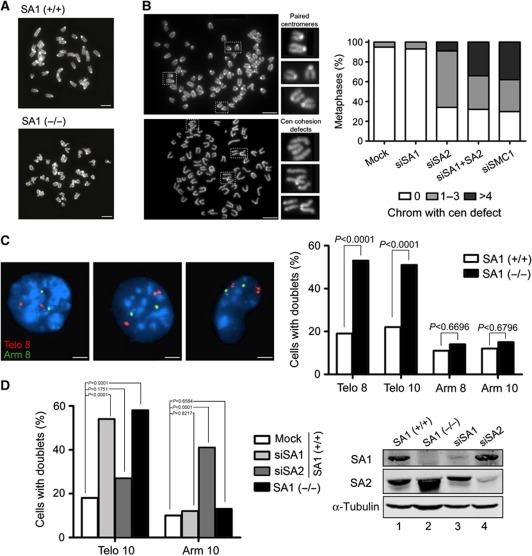
Metaphase spreads from SA1-null cells present chromosomes with normally paired centromeres (Figure 3A), suggesting that cohesin-SA1 is not essential to maintain centromere cohesion in mouse chromosomes. Treatment of the mouse C2C12 cell line with siRNAs against SA1, SA2, both SA1 and SA2 or SMC1 further confirmed this result (Figure 3B; western blot analysis in Supplementary Figure S3A). The lack of only SA1 does not affect centromere cohesion, whereas the absence of SA2 does. Double depletion of SA1 and SA2, or depletion of SMC1 increases by almost four-fold the percentage of metaphases with more than four chromosomes showing clear centromere cohesion defects (Figure 3B). This result suggests that both cohesin-SA1 and cohesin-SA2 contribute to arm cohesion so that in the absence of both separation of the sister centromeres becomes more evident. Since arm and telomere cohesion are difficult to assess in metaphase spreads, we performed instead fluorescence in situ hybridization (FISH) of interphase cells with probes from subtelomere and arm regions. Cohesion defects, revealed by the appearance of doublets, are significantly increased in the SA1-null MEFs for the subtelomeric probes, but not for the arm probes, suggesting that SA1 has a specific role in telomere cohesion in mouse cells (Figure 3C). Downregulation of SA1 or SA2 in MEFs by siRNA also showed that telomere cohesion specifically depends on SA1 whereas arm cohesion in the loci examined was only affected by the absence of SA2 (Figure 3D). Taken all together, our results indicate that centromeric cohesion relies on cohesin-SA2 whereas cohesin-SA1 plays a unique role in telomere cohesion in mouse cells.
Figure 3.
A specific role of SA1 in telomere cohesion. (A) Metaphase spreads from wild-type and SA1-null MEFs showing proper centromere cohesion. Scale bars, 10 μm. (B) Metaphase spreads from mouse C2C12 cells treated with control (top panel) and siRNA against SA2 (bottom panel) are shown as examples of proper and defective centromere cohesion, respectively. Scale bars, 10 μm. On the right, bar graph with the quantification of metaphase cells showing none, 1–3 or ⩾4 chromosomes with split centromeres after treatment with the indicated siRNAs (n⩾200 metaphases per condition from two independent experiments). (C) FISH analysis of wild-type or SA1-null primary MEFs in interphase with probes from the subtelomeric regions of chromosome 8 and 10 (Telo 8 and Telo 10) and arm regions of the same chromosomes (Arm 8 and Arm 10). n⩾100 cells per clone from two independent clones per genotype. Scale bars, 5 μm. (D) FISH analysis performed as in (C) in wild-type primary MEFs untransfected or transfected with siRNAs against SA1 (siSA1) or SA2 (siSA2). n⩾100 cells per condition.
Aberrant telomeres in cells lacking SA1
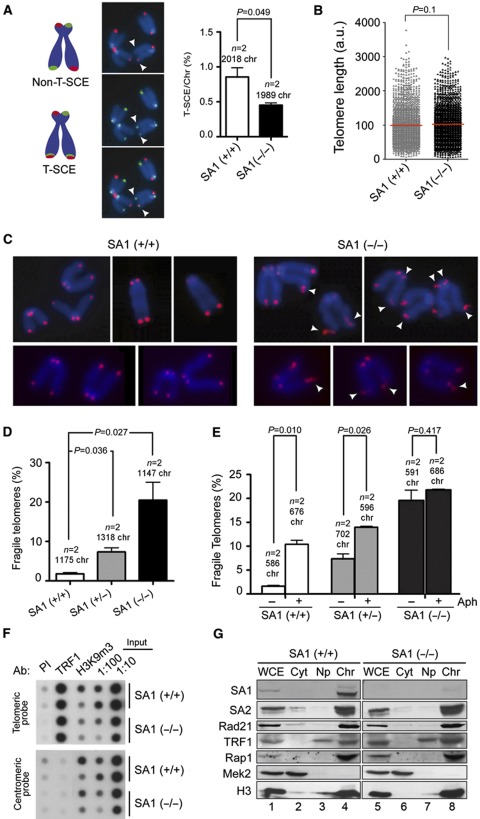
We next looked at sister chromatid exchange at telomeres (T-SCE), an event that needs cohesion, by means of a FISH technique involving the use of telomeric probes specific for the lagging and the leading strands, chromosome orientation FISH or CO-FISH (Bailey et al, 2004). Consistent with the loss of cohesion, we found a two-fold decrease in T-SCE in cells lacking SA1 (Figure 4A). Telomere length was similar in wild-type and SA1-null MEFs (Figure 4B), whereas the number of telomere fusions was higher in the absence of SA1 (1.2 fusion events per 100 chromosomes compared with 0.17 in wild-type cells, n=2 clones per genotype, at least 1100 chromosomes counted). More striking was the high incidence of aberrant telomere structures in SA1-null mitotic chromosomes, as judged by the appearance of an irregularly shaped, not round but elongated or split telomeric FISH signal (Figure 4C and D). SA1 heterozygous MEFs showed an intermediate phenotype (Figure 4D, grey bar). These aberrant structures, known as fragile telomeres, have been described in mouse cells deficient for components of the shelterin complex that binds the TTAGGG repeats and safeguards the ends of mammalian chromosomes (Munoz et al, 2005; Palm and de Lange, 2008; Martinez et al, 2009; Sfeir et al, 2009). They receive this name for their resemblance to common fragile sites, chromosomal regions that challenge the replication machinery and are often visualized as breaks or gaps in metaphase chromosomes upon partial inhibition of DNA synthesis (Durkin and Glover, 2007). Indeed, treatment of wild-type MEFs with low doses of the DNA replication inhibitor aphidicolin increases significantly the frequency of fragile telomeres in control cells and even in SA1 heterozygous cells (Figure 4E, white and grey bars, respectively). However, this is not the case for SA1-null cells, maybe because a further increase in the already high incidence of fragile telomeres prevents progression to mitosis (Figure 4E, black bars). Telomere fragility in SA1-null cells does not result from impaired recruitment of shelterin to telomeres, as shown by both chromatin fractionation and ChIP-dot blot assays (Figure 4F and G). Importantly, expression of full-length SA1 in SA1-null MEFs rescues telomere fragility almost completely (Supplementary Figure S4), confirming that the telomere defect is due to impaired cohesin-SA1 function.
Figure 4.
Telomere fragility in the absence of SA1. (A) T-SCE measured by CO-FISH (chromosome orientation FISH) in the telomeres of primary MEFs. The drawing on the left explains how T-SCE are visualized. The images on the middle show an example in which the telomeres indicated by arrowheads have undergone exchange since they are labelled by both the leading and the lagging strand-specific telomeric probes (green and red, respectively). Quantification of exchange events is shown in the bar graph on the right. (B) Telomere length measured by Q-FISH (quantitative FISH) for telomeres from wild-type and SA1-null MEFs (12 metaphase cells from two clones for each genotype). (C) Metaphase chromosomes from wild-type and SA1-null MEFs stained with a telomeric repeat probe (red) and DAPI (blue). Arrowheads point to fragile telomeres. (D) Quantification of fragile telomeres in chromosomes from two clones each of wild-type, SA1 (+/−) and SA1-null MEFs. (E) Telomere fragility measured in wild-type, SA1 (+/−) and SA1-null MEFs either untreated (−) or treated (+) with 0.5 μM aphidicolin for 24 h. In all cases two clones of each genotype were used. (F) ChIP-dot blot analysis of two clones each of wild-type and SA1-null MEFs with preimmune serum as negative control (PI), anti-TRF1 and anti-H3K9m3 as positive control. The chromatin obtained was transferred to a membrane and hybridized with a telomeric probe and a centromeric probe (major satellite) as control. TRF1 is present only at telomeres and its abundance is not affected by the lack of SA1. (G) Chromatin fractionation followed by immunoblotting with antibodies against cohesin subunits and shelterin proteins in wild-type and SA1-null cells. Mek2 cytoplasmatic kinase and histone H3 are used as control for the fractionation procedure. WCE, whole-cell extract; Cyt, cytoplasm; Np, nucleoplasm; Chr, chromatin fraction. The amount of TRF1 and Rap1 in chromatin does not depend on SA1 (lanes 4 and 8).
Deficient telomere replication in SA1-null cells
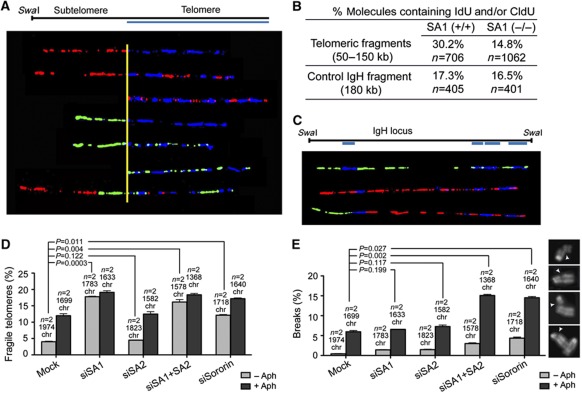
In order to assess telomere replication, we performed single-molecule analysis of replicated DNA (SMARD) on telomeric regions (Norio and Schildkraut, 2001; Sfeir et al, 2009) in primary wild-type and SA1-null MEFs. By counting the number of telomeric molecules containing a labelled nucleotide (IdU and/or CldU), we observed a highly significant reduction in the fraction of replicating telomeres in the SA1-null MEFs (P<0.0001; Figure 5A and B). Among the labelled telomeres, twice as many molecules were doubly labelled in the sample from SA1-null cells (13.9%) compared with wild-type cells (6.6%), which also reflects slower replication. Cell-cycle profiles of the two populations and BrdU incorporation at the time point studied were identical (Supplementary Figure S5), suggesting that there is not a general defect in S-phase progression. The specificity of this defect at telomeres is further supported by the SMARD analysis of the IgH locus as a control, which showed no evidence for altered replication dynamics in the absence of SA1 (P=0.78; Figure 5B and C). Thus, SA1 is specifically required for proper replication of telomeres.
Figure 5.
SMARD analysis of telomeric DNA reveals defective replication in SA1-null MEFs upon loss of telomere cohesion. (A) Examples of stretched telomeric DNA molecules of variable lengths (50–150 kb), identified by FISH with a telomeric probe (blue), that incorporated IdU (red) and/or CldU (green) during the time of the pulses. (B) Results of the SMARD analysis for telomeres (top) and molecules containing the IgH locus as control (bottom) from wild-type and SA1-null MEFs. (C) Examples of IdU/CldU (red/green) incorporation patterns in SwaI-digested fragments (180 kb) corresponding to the IgH locus, as identified by FISH with the indicated probes (blue). (D) Quantification of fragile telomeres in mouse C2C12 cells treated with no siRNA (mock) or siRNAs against SA1 (siSA1), SA2 (siSA2) or both (siSA1+SA2) or Sororin (siSororin), either in the absence (grey bars) or in the presence of aphidicolin (black bars) as in Figure 4E. (E) Quantification of breaks along the chromosome arms in the indicated cells either untreated (grey bars) or treated with aphidicolin (black bars). The images on the right show examples of the broken chromosomes.
Cohesion mediated by cohesin-SA1 contributes to efficient replication of telomeres
We next asked what is the mechanistic contribution of cohesin-SA1 to telomere replication. On one hand, cohesin affects origin activation most likely by promoting the proper organization of replication factories by intra-chromatid looping (Guillou et al, 2010). On the other, cohesion mediated by cohesin could contribute to HR required to restart stalled forks frequently found at telomeres (Sfeir et al, 2009; Badie et al, 2010). To distinguish between these two possibilities, we checked the effect of Sororin on telomere fragility. Sororin is a cohesin-interacting factor required for sister chromatid cohesion but not for origin activation (Guillou et al, 2010; Nishiyama et al, 2010). As in MEFs, telomere fragility occurred specifically upon treatment of C2C12 cells with siRNAs against SA1, but not SA2 (Figure 5D, grey bars), or upon addition of aphidicolin (Figure 5D, black bars). Importantly, cells with reduced Sororin showed a significant increase in telomere fragility (see Supplementary Figure S3B for western blot analysis). Thus, the cohesive function of cohesin-SA1 is required for efficient fork progression through subtelomere/telomere regions. In addition, we observed that depletion of SA1 or SA2 alone did not increase the percentage of chromosomes with breaks upon aphidicolin treatment compared with control cells, whereas simultaneous depletion of both or depletion of Sororin did (Figure 5E, black bars). This result indicates that both cohesin-SA1 and cohesin-SA2 contribute to cohesion and prevent fragile site breakage along chromosome arms, while only cohesin-SA1 can perform this function at telomeres.
Defective chromosome segregation in SA1-null cells leads to aneuploidy
To explore the mechanism that generates aneuploidy, we followed progression through mitosis of wild-type and SA1-null MEFs by live-cell imaging. Previous results in human cells had shown that depletion of cohesin causes a Mad2-dependent metaphase arrest triggered by the lack of tension in the absence of proper centromeric cohesion (Toyoda and Yanagida, 2006). In contrast, the time that cells spend in mitosis before starting anaphase is similar in wild-type and SA1-null MEFs (median is 40 and 45 min, respectively; Figure 6A, blue lines), consistent with the finding of robust centromeric cohesion in these cells. However, almost 30% of mitotic cells lacking SA1 fail to complete mitosis appropriately and either are unable to complete cytokinesis and end up as binucleated cells (Figure 6A, green lines; Figure 6B, middle), or start anaphase but then collapse into one single tetraploid nucleus (Figure 6A, yellow lines), or die in mitosis before decondensing their chromosomes (mitotic catastrophe; Figure 6A, black lines; Figure 6B, bottom). Thus, cells in the two former categories become tetraploid after traversing mitosis. Staining of fixed cells also showed a five-fold increase in the number of binucleated cells among SA1-null cells compared with wild-type cells (Supplementary Figure S6). We have observed increased frequency of both lagging chromosomes and chromatin bridges among the SA1-null cells (Figure 6C, black bars). The presence of these structures could account for the observed cleavage furrow regression that results in either mitotic catastrophe or tetraploidization. While the former phenotype may contribute to the decreased proliferation rates of SA1-null cells, the latter is a reported initiator of aneuploidy (Ganem et al, 2007). We also found increased frequency of defective anaphases in SA1 heterozygous cells, both ex vivo (in MEFs; Figure 6C, grey bars) and in vivo (in tissue sections; Supplementary Figure S7). This finding further supports the idea that aneuploidy resulting from faulty chromosome segregation promotes tumourigenesis in SA1 heterozygous animals.
Figure 6.
Chromosome segregation defects in SA1-null cells. (A) Graphical summary of the fates of mitotic cells from wild-type (n=88) and SA1-null MEFs (n=86) observed by live-cell imaging. Each line represents the progression through mitosis of a single cell and it is coloured according to the legend shown below the graph. (B) Examples of wild-type and SA1-null MEFs progressing through mitosis after transfection with H2B-mCherry (red) to label chromatin. The time after NEBD for each frame is indicated. Examples of a normal mitosis (top), a mitosis that leads to the formation of a binucleated cell (middle) and a mitosis that ends up in cell death (bottom) are shown. (C) Aberrant anaphases in SA1 (+/−) and SA1-null MEFs. Examples of a proper anaphase (top), an anaphase with a lagging chromosome (middle) and an anaphase with a chromatin bridge (bottom) are shown (n⩾50 cells per clone from two independent clones per genotype).
Telomere replication defects cause chromosome missegregation in SA1-null cells
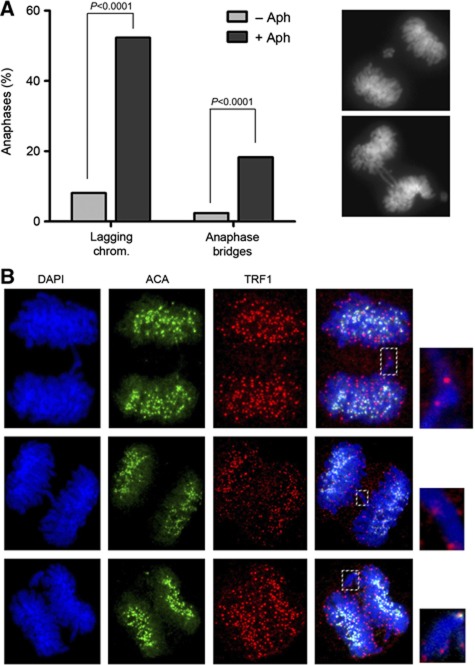
We next tested whether replicative stress could lead to chromosome segregation defects in MEFs. Indeed, wild-type MEFs treated with low doses of aphidicolin show high incidence of both lagging chromosomes and anaphases bridges (Figure 7A). Since SA1-deficient cells present replication defects specifically at telomeres, it is reasonable to propose that the chromosome segregation defects described in the previous paragraph result from impaired telomere replication. Immunostaining of SA1-null MEFs going through anaphase reveals that out of 21 anaphases displaying chromatin bridges none showed centromere (ACA) staining at the bridge, whereas telomere (TRF1) signals could be detected in 13 of them (Figure 7B, top and middle panels). The lack of TRF1 staining in the other eight anaphases could reflect loss of the shelterin upon chromatin stretching. This result supports the hypothesis that unresolved replication intermediates from subtelomere/telomere regions interfere with chromosome segregation and lead to the formation of anaphase bridges. We also examined anaphases with lagging chromosomes and found that in all cases (20 out of 20) the lagging chromosome consisted of two paired chromatids (Figure 7B, bottom panels). In this case, we speculate that unreplicated regions in the short arm telomere, which is next to the centromere, prevent the separation of sister centromeres.
Figure 7.
Defective telomere replication causes chromosome missegregation. (A) Frequency of anaphase bridges and lagging chromosomes in wild-type MEFs untreated (–Aph) or treated (+Aph) with low doses of aphidicolin (0.5 μM 24 h). Examples of chromosome segregation defects induced by global inhibition of replication are shown on the right. (B) Examples of SA1-null anaphase cells with chromosome segregation problems stained with antibodies against TRF1 (red) and ACA (green). Cells were pre-extracted with detergent before fixation. The top and middle panels show the presence of telomeres but no centromeres at the chromatin bridges. The bottom panel shows a lagging chromosome containing two sister chromatids. Confocal microscopy was used to ensure that TRF1 signals were in the same focal plane as the DNA.
Taken all together, we show that the absence of telomere cohesion in cells lacking SA1 results in defective telomere replication. This generates fragile telomeres and is the cause of chromosome missegregation leading to mitotic catastrophe and aneuploidy.
Discussion
A mouse model to address cohesin functions in vivo
The importance of cohesin function in cell proliferation and differentiation is expanding rapidly (Merkenschlager, 2010; Dorsett, 2011). Mouse models appear as appropriate tools to unravel these different functions and their regulation both temporally (during embryo development and adult ageing) and spatially (in different tissues and organs). To date, mice deficient for genes encoding components of the cohesin complex have been generated for three meiosis-specific subunits, Smc1 β (Revenkova et al, 2004), Rec8 (Xu et al, 2005) and Rad21L (Herran et al, 2011) and also recently for Rad21 (Xu et al, 2010). The first three are viable but sterile whereas biallelic inactivation of Rad21 results in early embryonic lethality and only the radiosensitivity of Rad21 heterozygous animals was reported. A mouse with a conditional allele of the Rad21 locus has also been generated and used for depleting cohesin specifically in non-cycling thymocytes (Seitan et al, 2011). Thus, our study presents the first thorough characterization of a mouse model of a ubiquitously expressed cohesin subunit, SA1, for embryonic development and cancer. SA1-null embryos are embryonic lethal, which demonstrates that cohesin-SA1 performs a function that cannot be accomplished by cohesin-SA2. Inactivation of SA1 permits embryos to survive at least to E12.5 and, in some cases, even to E18.5. Decreased proliferation rates of the SA1-null cells due to metabolic stress resulting from aneuploidy (Williams et al, 2008), to reduced cell survival upon passage through mitosis (this study) or to transcriptional dysregulation of pro-proliferation genes like myc (Rubio et al, 2008; Rhodes et al, 2010; Remeseiro et al, 2012) likely contribute to the lethality of SA1-null embryos.
Cohesin-SA1 is required for efficient telomere replication
Cohesin complexes carrying SA1 and SA2 subunits are not functionally redundant. We have shown that cohesin-SA2 is more critical for centromeric cohesion and cohesin-SA1 has a specific role in telomere cohesion, consistent with previous results in HeLa cells (Canudas and Smith, 2009). Whereas both contribute to cohesion and prevent fragile site formation along chromosome arms, only cohesin-SA1 can perform this role at telomeres. We show mechanistic evidence of the deleterious effects of cohesion loss for efficient replication of telomeres, since downregulation of Sororin, a cohesin-interacting factor specifically required for cohesin to become cohesive, results also in telomere fragility. The repeated nature of telomeric sequences, the propensity of their G-rich strand to form G4 DNA structures, and the presence of the t-loop hinder the passage of replication forks (Gilson and Geli, 2007). Furthermore, stalled forks occurring at chromosome ends cannot be rescued by forks progressing in the opposite direction. Recent reports shed light on how the cell deals with this problem (Sfeir et al, 2009; Badie et al, 2010; Ye et al, 2010). Replicative stress specifically at telomeres increases dramatically in the absence of SA1, as evidenced by the high incidence of fragile telomeres and confirmed by the results of the single-molecule analyses (SMARD). We propose that cohesion mediated by cohesin-SA1 is an important component of the pathway that stabilizes arrested forks at telomeres and facilitates their restart and/or promotes HR-mediated repair of breaks generated upon fork collapse.
A new mechanism to generate aneuploidy when cohesin function is impaired
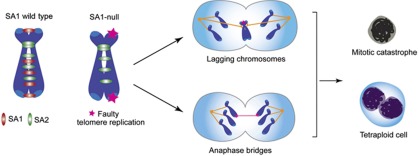
We have shown that SA1-null cells, despite their robust centromere cohesion, have defects in chromosome segregation that most likely arise from defective telomere replication (see model in Figure 8). Aberrant telomere structures resulting from incomplete replication might hinder proper orientation of the sister kinetochores in the acrocentric mouse chromosomes, thus increasing the chance of merotely (i.e., a single kinetochore attached to both spindle poles), which generates lagging chromatids in anaphase (Cimini et al, 2001; Salmon et al, 2005). A close analysis of defective anaphases in SA1-null cells shows that lagging chromosomes consist of two paired sister chromatids, not just one. We therefore favour the alternative possibility that unreplicated regions in subtelomere/telomere regions of the short arm prevent the separation of sister centromeres. If the unresolved replication intermediate happened in the long arm, it would generate instead a chromatin bridge containing telomere sequences, and we show evidence for these as well. Our hypothesis is consistent with recent reports showing that incompletely replicated regions or unresolved replication intermediates, or disruption of homology-directed repair, give rise to chromatin bridges and lagging chromosomes in anaphase (Torres-Rosell et al, 2007; Chan et al, 2009; Naim and Rosselli, 2009; Kawabata et al, 2011; Laulier et al, 2011; Lukas et al, 2011). The presence of these structures prevents proper cytokinesis and results in either mitotic catastrophe or tetraploidization, and the latter could drive aneuploidy in SA1-deficient cells. Ablation of telomere proteins can also lead to aneuploidy by additional mechanisms involving either chromosome fusion–breakage–bridge cycles following telomere de-protection (Smogorzewska et al, 2002) or bypass of mitosis upon persistent damage signalling in the uncapped telomeres (Davoli et al, 2010).
Figure 8.
The role of cohesin-SA1 in telomere cohesion and replication is essential for accurate chromosome segregation and to prevent aneuploidy. See text for details on the model.
Importantly, SA1 heterozygosity also leads to telomere fragility, chromosome segregation defects and aneuploidy. This aneuploidy is likely to contribute to tumourigenesis, as we show that SA1 heterozygous mice develop spontaneous tumours earlier than their wild-type littermates and with a distinct tumour spectrum. Until recently, little was known of the involvement of cohesin mutations in human cancer (Xu et al, 2011). Inactivation of SA2 has now been found in a diverse range of tumour types and it has been proposed that lack of SA2 function leads to aneuploidy due to PSSC in metaphase (Solomon et al, 2011), in agreement with previous reports (McGuinness et al, 2005; Canudas and Smith, 2009). We propose that inactivation of SA1 also generates aneuploidy, but through a completely different mechanism involving impaired telomere replication.
Materials and methods
Generation of SA1-knockout mouse model, MEFs isolation and cell culture
An ES cell line containing a SA1-Gene Trap allele (PO99A04) was obtained from the German Gene Trap Consortium (Schnutgen et al, 2005). Chimeric mice were generated by microinjection of ES clones into S129V host blastocysts, which were then implanted into pseudopregnant C57Bl/6J females, and germline transmission was assessed. Mice were housed in a pathogen-free animal facility following the animal care standards of the institution. The following primers were used for genotyping by PCR: F, 5′-GTGCTAGGATGACTCTGAAACTG-3; B32, 5′-CAAGGCGATTAAGTTGGGTAA CG-3′; R 5′-TGTGCTAGGCAGACAGTCTCC-3. Primary MEFs were isolated from E12.5 embryos resulting from intercrosses of SA1 heterozygous mice and cultured in DMEM/10% FBS.
Q-FISH, CO-FISH and interphase FISH
Metaphases for Q-FISH were prepared and hybridized as previously described (Martinez et al, 2009). Images were captured using Leica Q-FISH software and quantification of telomere intensities to determine telomere length was performed with TFL-Telo software. In the indicated cases, cells were cultured in 0.5 μM aphidicolin for 24 h. For CO-FISH, MEFs were cultured in the presence of 10 μM BrdU for 24 h to ensure complete replication and then CO-FISH was performed using first a fluorescein-labelled (TTAGGG)7 probe and then a Cy3-labelled (CCCTAA)7 probe (Applied Biosystems). FISH on interphase cells was done according to standard protocols, using probes from subtelomere and arm regions, which were labelled by nick-translation (Abbott Inc.) on BAC clones from a mouse library [BAC references: RP23-326G18 (telomere8), RP23-310L10 (arm8), RP23-71E10 (telomere10), RP23-453P21 (arm10)]. The percentage of cells with doublets was determined upon counting at least 100 cells per genotype and per experiment.
SMARD
For SMARD assay, asynchronously growing MEFs were sequentially labelled with 25 mM IdU (4 h) and 25 mM CldU (4 h). Cells were embedded in agarose plugs, lysed and DNA was digested with SwaI and fractionated by PFGE to select 50–150 kb fragments enriched in telomeric DNA as described (Norio and Schildkraut, 2001; Sfeir et al, 2009). DNA was stretched on microscope slides coated with 3-amino-propyltriethoxysilane (Sigma), denatured in alkali-denaturing buffer (0.1 N NaOH in 70% ethanol and 0.1% β-mercaptoethanol) for 12 min and fixed by addition of 0.5% glutaraldehyde for 5 min. Telomeric DNA was detected by hybridization with a Biotin-OO-(CCCTAA)4 PNA probe and Alexa Fluor 350-conjugated NeutrAvidin antibody (Molecular Probes) followed by biotinylated anti-avidin antibody (Vector). Halogenated nucleotides were detected with a mouse anti-IdU monoclonal antibody (BD), a rat anti-CldU monoclonal antibody (Accurate) and Alexa Fluor-conjugated secondary antibodies.
Chromosome spreads from MEFs and fetal livers
MEFs in culture were arrested in 0.1 μg/ml colcemide for 4–6 h, harvested by trypsinization, swollen in 75 mM KCl for 30 min at 37°C and fixed in methanol:acetic acid 3:1. Fetal livers from E14.5 embryos were minced with a scalpel and the cell suspension was incubated for 10 min at 37°C in 0.1 μg/ml colcemide in EDTA-containing buffer, further incubated in 75 mM KCl for 15 min and fixed in methanol:acetic acid 3:1. In both cases, the fixed suspension was dropped onto slides to obtain chromosome spreads that were stained and mounted with ProLong-Gold with DAPI (Invitrogen) and visualized using a Leica fluorescence microscope.
FACS
FACS analysis for DNA content was performed using propidium iodide staining, according to the standard procedures. For BrdU staining, cells were pulsed with 10 μM BrdU for 40 min, fixed and incubated with FITC-conjugated anti-BrdU antibody (BD Biosciences). Flow cytometry was performed using the FACS Canto II (Becton Dickinson) and data were analysed using FlowJo software (version 9.3.1).
Quantitative RT–PCR
The amount of SA1 and SA2 transcripts in MEFs, ES cells and different mouse tissues was determined by absolute quantitative RT–PCR. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and retrotranscribed with Superscript II (Invitrogen) using random hexamer primers. An Applied Biosystems 7900HT Fast qRT–PCR was used to determine the mRNA levels. The following primers were used: SA1 (F: 5′AGGCTTTCATGCTGCTCTGT3′ and R: 5′TCCATGCTTT GGTTTTCCTC3′), SA2 (F: 5′GGGGGAGGAACTGTCTTTCT3′ and R: 5′CCTTCAATGTCTTCAAAATCTGTG3′) and GAPDH as reference gene (F: 5′TGCACCACCAACTGCTTAGC3′ and R: 5′GAGG GGCCATCCACAGTCTTC3′).
Extract preparation, immunoblotting, ChIP-dot blot and immunofluorescence
For whole-cell extracts, cells were collected by trypsinization, washed once in cold PBS, resuspended in SDS–PAGE loading buffer and sonicated. For protein extracts from organs, a piece of tissue was pulverized in a mortar containing liquid nitrogen and lysed in RIPA buffer. Equal amounts of protein were run in either 7.5 or 12.5% Bis/Tris gels followed by western blotting. Chromatin fractionation was performed according to the protocol described by Mendez and Stillman (2000). For ChIP-dot blot, immunoprecipitates were dot-blotted into Hybond N+ membrane and hybridization was performed with a radiolabelled telomeric probe recognizing the TTAGGG repeats, and a centromeric probe annealing with the mouse major satellite (Benetti et al, 2007). For immunofluorescence, cells were cultured on polylysine-coated coverslips, fixed in 4% formaldehyde for 15 min at room temperature, permeabilized in 0.2% Triton-X100 for 5 min and subjected to antibody incubation. Images were taken using a Leica DM6000 microscope or with a confocal SP5-WLL (for Figure 7). Rabbit polyclonal sera against SA1, SA2, SMC1 and Sororin were obtained by using as immunogen either a synthetic peptide [SA1-C (CEDDSGFGMPMF), SA2 (CDPASIMDESVLGVSMF), SMC1 (CDLTKYPDANPNPNEQ)] or full-length recombinant protein (mSororin, transferred from IMAGE clone 30065848 into pDEST17) and affinity purified. Other custom made and commercial antibodies used in this study were as follows: Rad21 (Losada et al, 1998); TRF1 and Rap1 (Martinez et al, 2009, 2010); H3K9m3 (Millipore, 07-442); α-tubulin (Sigma, DM1A); GAPDH (Sigma, G8795); Histone 3 (Abcam, AB1791); Mek2 (BD, M24520); phalloidin-488 (Invitrogen); ACA (Antibodies Inc., 15-235).
RNA interference and rescue experiments
Interference of SA1, SA2, Sororin and SMC1 was performed with siGENOME SMARTpool siRNAs from Dharmacon (M-041989, M-057033, M-048366 and M-049483, respectively) at a final concentration of 100 nM and experiments were done 72–96 h after transfection. For interference of C2C12 cells and MEFs, DharmaFECT transfection reagent 1 (Dharmacon) and the Neon transfection system (Invitrogen) were used, respectively. Rescue experiments were performed by electroporating 106 cells with 12 μg of full-length SA1 cloned in pBABE-puro vector and telomere fragility was assessed 40 h later.
Recombinant protein expression
RNA obtained from MEFs was used for retrotranscription and subsequent PCR amplification of cDNA encoding SA1 and SA2 C-terminal regions (amino acids 1089–1258 and 992–1162, respectively). The resultant DNA fragments were then cloned into the EcoRI site of the pGEX-KG expression vector. Recombinant fusion proteins were then expressed in BL21-pLys strain and purified following standard methods. These proteins were used to quantitate the amount of SA1 and SA2 in MEFs (Supplementary Figure S1C).
Live-cell imaging
MEFs were transfected with H2B-mCherry expression vector using the Amaxa Nucleofector System and seeded onto chamber-slides. Time-lapse live-cell imaging was performed using the Delta Vision system (Applied Precision). Phase-contrast and fluorescent images were acquired every 5 min with a × 10 objective for 24 h. Image analysis was performed using ImageJ software.
Histology and immunohistochemistry
Embryos, normal tissues and tumour samples were fixed in 10% buffered formalin (Sigma) and embedded in paraffin using standard procedures. For histopathological studies, 3 μm sections were stained with haematoxylin and eosin (HE). Anti-BrdU (GE Healthcare), Ki67 (Master Diagnostica) and γH2AX (Millipore, 05-636) primary antibodies were used for immunohistochemical analysis, positive cells were visualized using 3,3-diaminobenzidine tetrahydrochloride plus (DAB+) as a chromogen and counterstaining was performed with haematoxylin.
Carcinogen treatments
For the induction of fibrosarcomas, 8-week-old mice received a single intramuscular injection of 1 mg of 3-MC diluted in 100 μl of sesame oil in one of the rear legs. Mice were observed on a daily basis until tumours of 1.5 cm in diameter developed in the injected leg, at which point the animals were sacrificed and the tumours were extracted for further analysis. Liver tumours were induced in 15-day-old male mice by intraperitoneal injection of 50 μg of DEN diluted in 100 μl of PBS. Tumour development was followed by computed tomography (CT). Animals were observed daily and sacrificed when they manifested signs of morbidity, in accordance with the Guidelines for Humane End Points for Animals used in biomedical research.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 software. A two-tailed Fisher's exact test was done in Figures 3C and D, 5B, 6C and 7A, and for the rest of statistical analysis a two-tailed Student's t-test was applied. Data are shown as mean±s.e.m (standard error of the mean); P<0.05 were considered significant and actual P-values are depicted in the figures.
Supplementary Material
Acknowledgments
We are grateful to I Barthelemy for her initial work on this project, M Rodríguez-Corsino for excellent technical assistance and B Ferreira, MC Martín and JC Cigudosa (Cytogenetics Unit, CNIO) for providing the BACs and invaluable help with FISH. We also acknowledge M Malumbres, A Martín-Pendás and M Soengas for helpful discussions and O Fernández-Capetillo and M Serrano for critically reading the manuscript. This research has been supported by the Spanish Ministry of Science and Innovation (SAF-2010-21517 and CSD2007-00015 Inesgen/FEDER to AL; ‘Ramón y Cajal’ grants for AC and PM), a La Caixa predoctoral fellowship for SR and NIH Grant GM045751 to CLS.
Author contributions: AL designed and supervised the study. SR designed, performed, analysed and interpreted most of the experiments with contributions from AC in Figures 1, 3 and 5; Supplementary Figures S1 and S4; from MC1 in Figure 1; and from PM and MB in Figure 4. SMARD was performed by SR in the laboratory of CLS with advise from WCD. MC4 performed the histopathological analyses. AL and SR wrote the manuscript with ideas and comments from the other authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, Blasco MA, Jonkers J, Tarsounas M (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Goodwin EH (2004) Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 32: 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA (2007) Suv4-20 h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S (2009) Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramolino L, Lee BC, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero JL, Zipori D (1997) SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene 195: 151–159 [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED (2001) Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T (2010) Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D (2007) Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 17: 157–162 [DOI] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D (2010) Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herran Y, Gutierrez-Caballero C, Sanchez-Martin M, Hernandez T, Viera A, Barbero JL, de Alava E, de Rooij DG, Suja JA, Llano E, Pendas AM (2011) The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30: 3091–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Fuchs J, Pichler P, Peters JM, Mechtler K (2010) Lesson from the stoichiometry determination of the cohesin complex: a short protease mediated elution increases the recovery from cross-linked antibody-conjugated beads. J Proteome Res 10: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N (2011) Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41: 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S (2010) Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA 107: 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulier C, Cheng A, Stark JM (2011) The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic Acids Res 39: 5935–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID (2009) Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 76: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253 [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA (2010) Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 12: 768–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M (2010) Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr Opin Genet Dev 20: 555–561 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M (2009) Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA (2005) XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet 37: 1063–1071 [DOI] [PubMed] [Google Scholar]
- Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11: 761–768 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A (2009) Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL (2001) Visualization of DNA replication on individual Epstein-Barr virus episomes. Science 294: 2361–2364 [DOI] [PubMed] [Google Scholar]
- Palidwor GA, Shcherbinin S, Huska MR, Rasko T, Stelzl U, Arumughan A, Foulle R, Porras P, Sanchez-Pulido L, Wanker EE, Andrade-Navarro MA (2009) Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput Biol 5: e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A (2012) A unique role of cohesin-SA1 in gene regulation and development. EMBO J 31: 2090–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R (2004) Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 6: 555–562 [DOI] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA (2010) Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol 344: 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera T, Losada A (2009) Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma 118: 223–233 [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Cimini D, Cameron LA, DeLuca JG (2005) Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci 360: 553–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, Chambon P, Wurst W, von Melchner H (2005) Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci USA 102: 7221–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M (2011) A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L (2007) Anaphase onset before complete DNA replication with intact checkpoint responses. Science 315: 1411–1415 [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Yanagida M (2006) Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell 17: 2287–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC (2004) Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep 5: 167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Balakrishnan K, Malaterre J, Beasley M, Yan Y, Essers J, Appeldoorn E, Tomaszewski JM, Vazquez M, Verschoor S, Lavin MF, Bertoncello I, Ramsay RG, McKay MJ (2010) Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS ONE 5: e12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ (2005) Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8: 949–961 [DOI] [PubMed] [Google Scholar]
- Xu H, Tomaszewski JM, McKay MJ (2011) Can corruption of chromosome cohesion create a conduit to cancer? Nat Rev Cancer 11: 199–210 [DOI] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, Verhoeyen E, Britton S, Calsou P, Salles B, Bizard A, Nadal M, Salvati E, Sabatier L, Wu Y, Biroccio A et al. (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142: 230–242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.