Abstract
The yeast splicing factor Cwc2 contacts several catalytically important RNA elements in the active spliceosome, suggesting that Cwc2 is involved in determining their spatial arrangement at the spliceosome's catalytic centre. We have determined the crystal structure of the Cwc2 functional core, revealing how a previously uncharacterized Torus domain, an RNA recognition motif (RRM) and a zinc finger (ZnF) are tightly integrated in a compact folding unit. The ZnF plays a pivotal role in the architecture of the whole assembly. UV-induced crosslinking of Cwc2–U6 snRNA allowed the identification by mass spectrometry of six RNA-contacting sites: four in or close to the RRM domain, one in the ZnF and one on a protruding element connecting the Torus and RRM domains. The three distinct regions contacting RNA are connected by a contiguous and conserved positively charged surface, suggesting an expanded interface for RNA accommodation. Cwc2 mutations confirmed that the connector element plays a crucial role in splicing. We conclude that Cwc2 acts as a multipartite RNA-binding platform to bring RNA elements of the spliceosome's catalytic centre into an active conformation.
Keywords: RRM, spliceosome, Torus domain, X-ray crystallography, zinc finger
Introduction
Nuclear pre-mRNA splicing is an essential step of gene expression in eukaryotes, whereby non-coding introns are excised from the pre-mRNA via two consecutive phosphoester transfer reactions. This process is catalysed by the spliceosome, a large and dynamic molecular machine which consists of the snRNPs U1, U2, U4/U6 and U5 and numerous proteins. An interesting design principle of the spliceosome is that the stepwise recruitment of the snRNPs to short conserved sequences at the 5′ and 3′ splice sites (SS) and the branchsite (BS) of the intron culminates in the assembly of a multi-megadalton ensemble, termed the B complex, which does not yet have an active site (Wahl et al, 2009). Subsequent activation of the spliceosome requires significant rearrangements that lead to the displacement of U1 and U4 snRNAs and the formation of the Bact complex. At the protein level, all U1 and U4/U6 snRNP proteins are also dissociated, while at the same time numerous proteins are stably integrated into the Bact complex (Fabrizio et al, 2009, see also below). The activated spliceosome undergoes additional rearrangements, driven by the RNA helicase Prp2, before step 1 of splicing occurs (Liu et al, 2007; Warkocki et al, 2009). During step 1, the BS adenosine attacks the 5′SS, generating the cleaved 5′ exon and intron-3′ exon lariat intermediates. The spliceosomal C complex is formed at this time and, following a further remodelling step, mediated by the RNA helicase Prp16, the second step of splicing is catalysed, leading to the ligation of the 5′ and 3′ exons and the excision of the intron (Will and Luhrmann, 2011).
During catalytic activation of the spliceosome, a complex RNA–RNA interaction network involving snRNAs and pre-mRNA is formed, and the resulting RNA structure is central in catalysing the two steps of splicing (Madhani and Guthrie, 1994). Thus, following dissociation of U1 and U4 snRNAs from the spliceosome, U6 snRNA rearranges and forms an internal stem–loop (ISL), which plays a central role in the catalysis of splicing (Fortner et al, 1994; Yean et al, 2000). U6 snRNA also forms additional base pairs with U2 RNA generating U2/U6 helices Ia and Ib, the later containing the invariant AGC triad of U6 that is essential for splicing (Fabrizio and Abelson, 1990; Hilliker and Staley, 2004). Finally, U6 snRNA via its conserved ACAGAGA sequence, also forms base pairs with the 5′ end of the intron (Kandels-Lewis and Seraphin, 1993). In addition, U5 snRNA initially contacts 5′ exon nucleotides (nts) near the 5′SS and also later during the splicing reaction, the 3′ exon (O’Keefe et al, 1996).
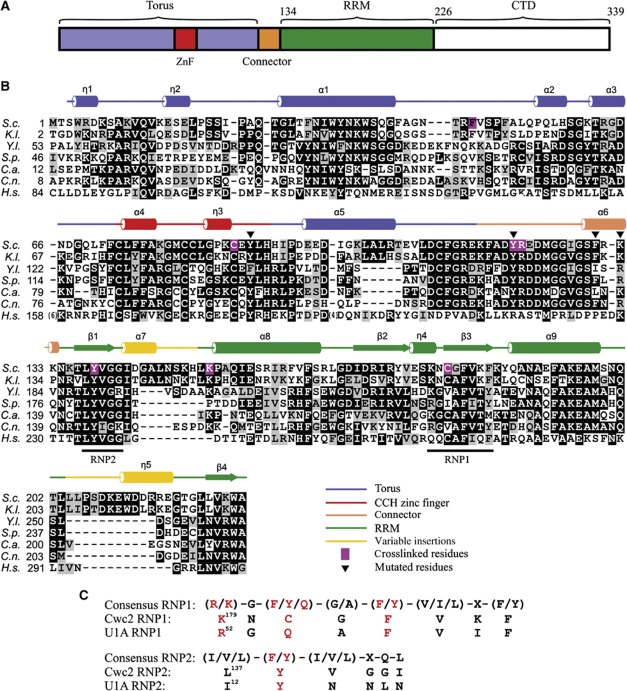
While it is clear that several of the aforementioned RNA structural elements play an important role in the catalysis of the splicing reaction, it is only poorly understood how these RNA elements are brought into a catalytically active tertiary conformation. There is evidence that spliceosomal proteins may play an important role in creating a catalytically active RNP core. For example, it was recently shown that a C-terminally located RNase H-like domain of Prp8 is situated at the heart of the catalytic core of the spliceosome (Pena et al, 2008; Ritchie et al, 2008; Yang et al, 2008). Moreover, during catalytic activation, numerous proteins become stably integrated into the spliceosome and they may also function in specifying an active catalytic RNA conformation. In yeast, these include a protein complex termed the ‘nineteen complex’ (NTC), that consists of eight core proteins including Prp19 (Tarn et al, 1994; Hogg et al, 2010) and an additional set of at least 12 proteins, which henceforth will be termed ‘NTC-related’ proteins, because several of them interact loosely with one or more of the NTC core proteins (Ohi and Gould, 2002). The NTC is required for promoting stable interactions of U5 and U6 snRNAs with the pre-mRNA during activation of the spliceosome (Chan et al, 2003; Chan and Cheng, 2005). Whether NTC proteins directly interact with the spliceosomal RNA network is not clear. Interestingly, several of the NTC-related proteins possess putative RNA-binding domains. Among these, the yeast Cwc2 protein is of particular interest in that it has a canonical RNA-binding domain (RNA recognition motif (RRM)) and a CCCH zinc finger (ZnF) of the CX7CX5CX3H type. Moreover, the likely human homologue of the yeast Cwc2 protein, Rbm22, also contains an evolutionary conserved RRM and ZnF domain. Cwc2 is essential for pre-mRNA splicing in vivo and was shown to contact U6 RNA during splicing in yeast extracts (McGrail et al, 2009). More recently, it was demonstrated that in purified catalytically active spliceosomes, Cwc2 can be crosslinked to several catalytically important RNA elements, including the U6-ISL, a region upstream of the ACAGAGA box, and the pre-mRNA intron near the 5′SS, placing Cwc2 at or near the spliceosome's catalytic centre. In the absence of Cwc2, a Bact-like complex could be assembled in vitro, but it was catalytically inactive (Rasche et al, 2012). Thus, by contacting multiple sites of the catalytic RNA–RNA interaction network, Cwc2 may assist in the formation of an active conformation of the spliceosome's catalytic centre. To gain insight into how Cwc2 might be capable of recognizing simultaneously several distinct RNA regions and possibly juxtapose these sites, we experimentally defined the folded core of Cwc2 that can functionally replace the full-length protein in an in vitro splicing assay. We determined its crystal structure, which revealed that Cwc2's ZnF, RRM and a previously uncharacterized Torus domain are integrated in a compact folding unit. We show by UV-mediated protein–RNA crosslinking and in vitro splicing assays that, via this unique architecture, Cwc2 acts as a multipartite RNA-binding platform.
Results
Experimental definition of a Cwc2 folding unit that contains the ZnF and the RRM domains
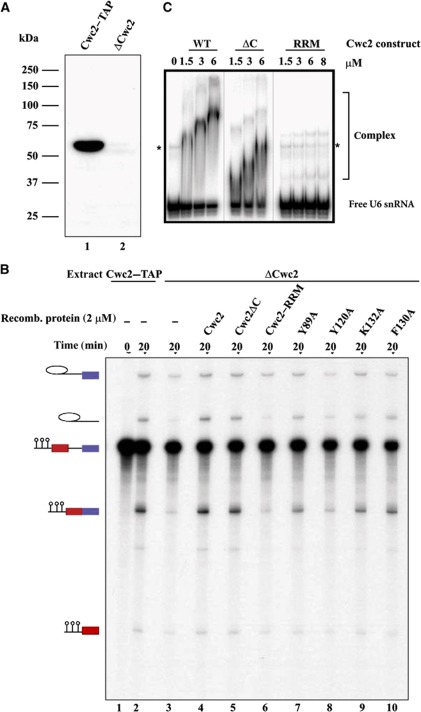
Purified, full-length Cwc2 from Saccharomyces cerevisiae, did not crystallize in our hands, possibly owing to the presence of a C-terminal, intrinsically disordered region. Limited proteolysis with the enzyme GluC resulted in complete digestion of this region and yielded a stable Cwc21–240 fragment (Supplementary Figure S1 in the Supplementary data available with this article online) that comprises the ZnF, the RRM domain, and two regions flanking the ZnF that have no predictable structural motifs.
Previously, yeast two-hybrid analysis showed that the C-terminal tail of Cwc2, comprising residues 230–339, interacts with the WD40 domain of the Prp19 protein, suggesting that the former is required for Cwc2 function in splicing (Ohi and Gould, 2002; Vander Kooi et al, 2010). To find out whether the Cwc21–240 fragment still functions like the full-length protein, we tested its activity in a standard yeast splicing extract depleted of the endogenous Cwc2 protein. Depletion was achieved by using a yeast strain that expresses Cwc2 tagged with the tandem affinity purification (TAP) tag at its C-terminus (Puig et al, 2001). Yeast extracts were incubated with IgG-Sepharose that binds the TAP-tag efficiently. Western blotting demonstrated that Cwc2 was removed to a large extent upon incubation of yeast extract with IgG-Sepharose (Figure 1A). Compared with the untreated splicing extract, the Cwc2-depleted extract (ΔCwc2) showed only very low splicing activity (Figure 1B, lanes 1–3), which is probably due to the residual amounts of Cwc2 in the depleted extract. These data are consistent with our previous finding that Cwc2 is essential for splicing in vitro (Rasche et al, 2012). For subsequent splicing complementation studies, we subcloned, expressed and purified a slightly shorter Cwc21–234 fragment in order to exclude the possibility of interference by traces of protease. Upon addition of Cwc21–234, as well as full-length protein to the ΔCwc2 extract, both steps of pre-mRNA splicing were restored (Figure 1B, lanes 3–5). Moreover, Cwc21–240 also retained the capacity to bind U6 snRNA in vitro with an affinity similar to that of the full-length Cwc2 protein, as revealed by an electrophoretic mobility shift assay (EMSA) (Figure 1C). Note that the decrease in electrophoretic mobility of the Cwc2–U6 RNP complex with increasing Cwc2 concentration may be due to oligomerization or changes in the structure and/or surface exposed charge of the complex, and was also observed in previous studies (McGrail et al, 2009; Lu et al, 2012). Together, these data indicate that the proteolytically resistant fragment represents the functional core of Cwc2 and that the C-terminal region 235–339 is not essential for splicing in vitro.
Figure 1.
Effects of Cwc2 mutagenesis on its splicing and RNA-binding activity. (A) Western blot analysis of yeast splicing extracts carrying TAP-tagged Cwc2 before depletion (lane 1) and after depletion of Cwc2 (lane 2). (B) Uniformly radiolabelled M3-actin pre-mRNA (0.4 nM) was incubated in 20% yeast whole-cell extract, which was either mock- (lanes 1 and 2) or Cwc2-depleted (lanes 3–10), under standard splicing conditions. Recombinant WT Cwc2 and mutant proteins indicated above each lane were then added to a final concentration of 2 μM (lanes 4–10). The splicing reactions were incubated at 23°C for the indicated time points. RNA was analysed on an 8% urea–polyacrylamide gel and visualized by autoradiography. The positions of the pre-mRNA, the splicing intermediates and products are indicated on the left. The intensities of the mRNA product signals were quantified using Quantity One software (Bio-Rad). The signal of the Cwc2–TAP extract (lane 2) was used as 100%, and relative to it the amount of mRNA produced was calculated. ΔCwc2 extract shows a residual level of mRNA of 16% (lane 3) compared with 109% for ΔCwc2 extract supplemented with WT Cwc2 (lane 4), 97% with Cwc2ΔC (lane 5), 16% with RRM (lane 6), 50% with Y89A (lane 7), 21% with Y120A (lane 8), 58% with K132A (lane 9) and 69% with F130A (lane 10). The mRNA level in the lane 3 was set as a background (remaining splicing activity of the Cwc2-depleted extract) and the restoration of splicing activity due to complementation with recombinant Cwc2 or its mutants was calculated as a difference between mRNA signals in the corresponding lane and lane 3. (C) Electrophoretic mobility shift analysis of the interaction of recombinant Cwc2 protein and its truncations with yeast U6 snRNA. Equal amounts of [32P]-5′-labelled U6 snRNA were incubated in the presence of increasing concentrations of purified full-length Cwc2 (wt), its stable GluC endoproteolytic fragment (ΔC) or its RRM. Resulting RNP complexes and free U6 RNA were resolved by native 6% PAGE. The identity and concentrations of the proteins used are indicated above the gel. Migration positions of RNA–protein complexes and unbound U6 snRNA is shown on the right. Asterisks mark a slower migrating band of U6 snRNA observed in protein-free samples.
Crystal structure determination and overall fold
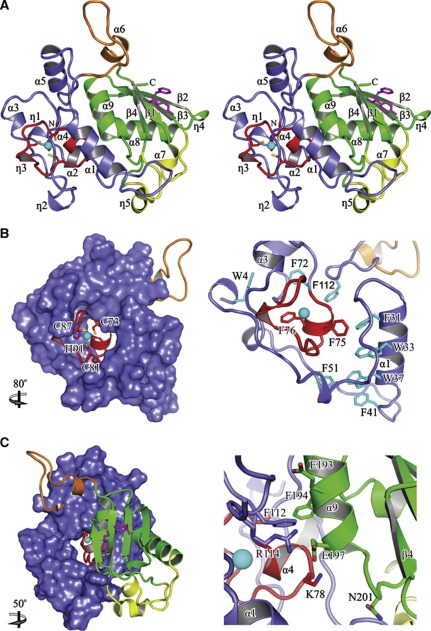
Cwc21–240 (hereafter designated Cwc2ΔC) produced by preparative proteolysis and purified to homogeneity by size-exclusion chromatography, reproducibly yielded well-diffracting crystals (Table I). The crystal structure was solved by single anomalous dispersion (SAD) with data collected around the zinc K-edge and refined against a 2.4 Å data set. The final model encompasses residues 3–226 and exhibits good stereochemistry (Table I). Consistent with the limited proteolysis, Cwc2ΔC exhibits a compact, globular structure that contains nine α-helices (α1–α9), five 310 helices (η1–η5) and four β-strands (β1–β4; Figure 2A).
Table 1. X-ray data collection and refinement statistics.
| Native | Native, Zn peak | |
|---|---|---|
| Data collection statistics a | ||
| Wavelength (Å) | 0.98 | 1.28 |
| Space group | P2(1)2(1)2(1) | P2(1)2(1)2(1) |
| Unit cell (Å) | a=58.8, b=79.8, c=113.9 | a=58.7, b=79.4, c=112.8 |
| Resolution (Å) | 47.2–2.4 (2.5–2.4) | 30–2.5 (3–2.5) |
| Unique reflections | 40 472 (4627) | 43 486 (14 867) |
| Redundancy | 5.5 | 6.8 |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| I/σ (I) | 16.3 (7.8) | 17.0 (10.1) |
| Rsym (%) | 8.6 (28.5) | 8.9 (19.7) |
| Refinement statistics | ||
| Resolution (Å) | 47.2–2.4 | |
| Reflections | 20 453 | |
| Test set (%) | 5 | |
| Rwork/Rfree | 22.3/25.5 | |
| Protein mol./res. | 2/427 | |
| Protein atoms | 3466 | |
| Water oxygens | 177 | |
| Mean B-factors (Å) | 16.36 | |
| Res. in preferred/allowed/disallowed regions (%) | 97.7/1.9/0.5 | |
| RMS deviations (bond length [Å]/bond angle [deg]) | 0.007/0.927 | |
| aValues for the highest resolution shell are in parentheses. | ||
Figure 2.
Structural overview. (A) Stereo ribbon plot of the Cwc2 structure. Secondary structure elements and termini are labelled. Red, ZnF domain; blue, Torus domain; green, RRM domain; yellow, variable insertions; orange, connector element. Aromatic residues from the two canonical RNP motifs are shown as magenta sticks. (B) Intimate association between the ZnF and the Torus domain. The residues coordinating the zinc atom are depicted as sticks and labelled (left). The close-up view highlights conserved aromatic residues that stabilize the Torus domain around the ZnF (right). (C) Interaction of the RRM domain with the ZnF and the Torus domain (left). Close-up view highlighting RRM contacts with the ZnF and Torus domains. Highly conserved residues are depicted as sticks and labelled (right). The relative orientations versus the position from (A) is indicated.
The two potential RNA-binding regions of Cwc2, the CCCH ZnF and the RRM domains are easily recognized in the crystal structure. The RRM domain (residues 134–226; Figure 3A), which represents about 40% of the structure, adopts the typical topology, with a four-stranded β-sheet (β1–β4) packed against two α-helices (α8, α9). In S. cerevisiae, the RRM domain of Cwc2 exhibits two insertions (positions 143–153 and 208–220), which have variable sizes and sequences among the orthologues and encompass the α7 and η5 helices, respectively (Figures 2A and 3B). RRM domains contain two well-conserved sequence motifs commonly referred to as RNP1 and RNP2. In known structures of RRM–RNA complexes, RNA molecules are bound with the 5′–3′ directionality along the RNP2–RNP1 axis; the binding involves two aromatic residues from the RNPs that undergo stacking interactions with the RNA bases (Maris et al, 2005; Clery et al, 2008). Both aromatic residues are present in the RRM domain of Cwc2: Y138 of RNP2 and F183 of RNP1, respectively. The other important aromatic residue of RNP1 in position 3 of the canonical sequence ((K/R)-G-(F/Y)-(G/A)-(F/Y)-(V/I/L)-X-(F/Y)), is replaced in S. cerevisiae Cwc2 by C181. However, a deviation from the consensus sequence at this position is not uncommon; for instance, the RRM of the human U1-A protein contains a Q residue at the third position of the RNP1 sequence (Figure 3C) (Oubridge et al, 1994). In the crystal structure of Cwc2, all of the residues commonly required for RNA binding by an RRM domain are exposed to the solvent, consistent with potential RNA-binding affinity of this Cwc2 domain (Figure 2A).
Figure 3.
Domain composition and sequence alignment of Cwc2. (A) Schematic depiction of domains and structural motifs in Cwc2. (B) Multiple sequence alignment of Cwc2 orthologues. S.c.—S. cerevisiae; K.l.—Kluyveromyces lactis; Y.l.—Yarrowia lipolytica; S.p.—Schizosaccharomyces pombe; C.a.—Candida albicans; C.n.—Cryptococcus neoformans; H.s.—Homo sapiens. Darker background corresponds to higher sequence conservation. Icons above indicate secondary structure elements from the Cwc2 crystal structure. Magenta background indicates the RNA-crosslinked sites. The triangles indicate mutated amino acids tested in splicing complementation assays. (C) Comparison of consensus RNP1 and RNP2 sequences (Maris et al, 2005) in Cwc2 and U1A, respectively. The amino-acid residues important for RNA binding are depicted in red. X represents any amino acid.
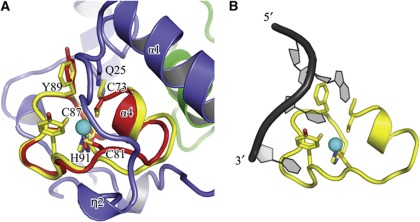
The ZnF domain adopts the common CCCH circular conformation, with the three cysteine residues (C73, C81, C87) and one histidine residue (H91) coordinating the zinc atom. The short α4 helix is located between the first and the second coordinating cysteines, while the η3 element is interposed between the second and the third cysteines. The Cwc2 ZnF adopts a fold very similar to those described in the atomic structures of TIS11d and MBNL1 (Hudson et al, 2004; Teplova and Patel, 2008) as shown by the structural superposition of Cwc2 upon MBNL1 and TIS11d, resulting in a Cα RMS pairwise deviation of 0.9 and 1.2 Å, respectively (Figure 4A). As described in detail below, the Cwc2 ZnF structure is surrounded by several α-helices and loops, which originate from its flanking regions.
Figure 4.
Comparison of the Cwc2 TlS11d ZnF. (A) Superposition of a CCCH-type ZnF domain of TIS11d on Cwc2. Cwc2 residues potentially involved in binding RNA and the four residues coordinated by zinc in the two proteins are shown and labelled. (B) Recognition of an AU-rich RNA element by the ZnF domain of TIS11d (pdb 1RGO) (Hudson et al, 2004), in the same orientation. The aromatic residues from TIS11d involved in stacking interactions with the bases are depicted as sticks.
Residues 116–133 adopt the conformation of a protruding loop with a short helix at the base (α6). This protrusion can be traced in only one of the two molecules from the asymmetric unit, possibly owing to intrinsic flexibility. The defined conformation is probably induced and stabilized by crystal contacts—like the salt bridge between E122, D123 from the protrusion and K224 from the β4 of a symmetry-related molecule—as well as by hydrophobic contacts (Supplementary Figure S2). While this manuscript was under review, the crystal structure of a slightly shorter Cwc2 fragment was reported (residues 1–237; Lu et al, 2012). The structure exhibits the same fold and topology, except for the flexible protruding loop. That is, residues 127–131 are not visible in the electron density while residues 116–126 are stabilized in a different conformation than the one observed in our structure.
The Cwc2 structure is dominated by a Torus domain that fastens the RRM and ZnF into a compact folding unit
A striking feature of the Cwc2 structure is a massive toroidal enclosure around the ZnF, formed by its two flanking appendages (3–72 and 92–115), like a ring on a finger (we refer to it hereinafter as the Torus domain; Figure 2B and C, left). The Torus domain has an outer diameter of 35–40 Å and encompasses four α-helices (α1–α3, α5) and two 310 helices (η1, η2), all connected by long loops (Figures 2A and 3B). Although they lack typical elements of secondary structure, these loops appear to be tightly stabilized as part of a compact fold, with virtually identical conformations in the two molecules of the asymmetric unit. The Torus domain is probably not an autonomously folding unit, as its conformation is established by the ZnF that provides several anchoring points along the two surrounding appendices, mainly through conserved hydrophobic residues (Figure 2B, right). Impairment of zinc coordination would probably result in the collapse of this entire circular architecture, in agreement with the lack of stable expression of Cwc2 in which one of the cysteines was mutated to a tyrosine (C73Y), a residue that does not coordinate zinc (McGrail et al, 2009). Notably, numerous aromatic residues are involved in the ZnF–Torus interface, which buries 1260 Å2 of protein surface, and in the inner reinforcement of the Torus domain. Thus, two of these (F75, F76) belong to the ZnF and eight (W4, F31, W33, W37, F41, F51, F72, F112) to the Torus domain (Figure 2B, right).
While the Torus domain engages in intimate interactions with the ZnF domain, at the same time it shares a large composite hydrophobic-polar interface (927 Å2) with the RRM domain (Figure 2C, left), mostly through contacts mediated by residues from the highly conserved α9 helix of the RRM. Among the most notable conserved and buried residues are F194, F112, R114, N201 and E193 (Figure 2C, right). The RRM domain is also in direct contact with the ZnF domain, which share an interface of 183 Å2. The highly conserved residues K78 of the ZnF and E197 from the RRM domain form a salt bridge (Figure 2C, right). In summary, the Torus domain represents the integrating element of the whole assembly, connecting tightly the ZnF and the RRM domain in one folding unit.
Previously reported atomic structures of CCCH ZnFs show them as individual domains in isolation or connected in tandem by structured linkers (Hudson et al, 2004; Teplova and Patel, 2008), but they have never been observed embedded in larger assemblies, such as the Torus domain in Cwc2. Thus, to our knowledge, the Cwc2 crystal structure is the first example of a structure in which a ZnF and an RRM domain are tightly interconnected by a Torus-like structural domain, thereby juxtaposing two distinct potential RNA-binding domains.
A positively charged connector element towers over a depression made up by the RRM and the Torus domains
At the sequence level, the C-terminal appendage of the Torus and the N-terminal part of the RRM domain are connected in Cwc2 by a linker (residues 116–133). In one of the molecules of the asymmetric unit, this region adopts the conformation of a loop with a short helix at its base (α6). When looking at the Cwc2 structure in the front view (Figure 2A), it is situated on top of the core structure of Cwc2. This structural element will hereinafter be called the connector element. It is interesting to note that the connector element comprises three conserved aromatic and three conserved positively charged residues, which would be consistent with the idea that the connector element might be involved in RNA recognition.
If one examines the electrostatic surface potential of the Cwc2 structure from the front, then three strongly positively charged regions become apparent; one on the connector, one on the RRM domain and one surrounding the ZnF (Figure 5A). In addition, these patches are interconnected by positively charged amino acids. If this front view is now rotated by 90° (Supplementary Figure S3), then it is clear that the Torus domain contributes to the front rim of a broad depression, whose bottom belongs to the RRM domain. The depression extends towards the ZnF and is carpeted with conserved, positively charged residues (see comparative Figure 5A and B). The side view (Supplementary Figure S3) also makes it clear that the rear rim of the depression is formed partly by amino-acid residues of the connector, and that the connector element towers over the depression. In summary, the structural and biophysical properties of the Cwc2 structure are consistent with the idea that Cwc2 is able to bind either a complex RNA structure or to recognize various RNA elements and to bring them into spatial proximity with one another.
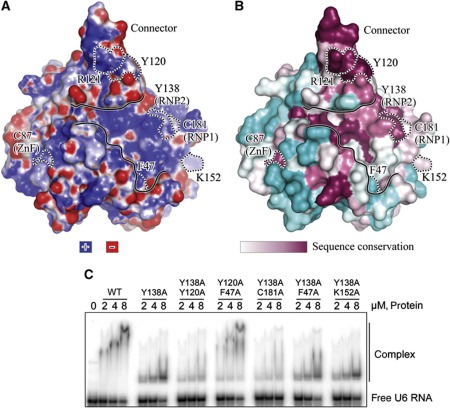
Figure 5.
Mapping of the RNA-crosslinked sites on the Cwc2 surface. (A) Electrostatic potential (B) surface conservation. Cyan—variable sequences. The molecule is shown in the same orientation as in Figure 2A. The residues crosslinked to RNA are outlined with dashed lines and labelled. The shallow depression is contoured with continuous lines. (C) Electrophoretic mobility shift analysis of the interaction of recombinant Cwc2 mutants with yeast U6 snRNA. Description as in Figure 1C. The amino acids mutated are shown above the gel.
Mapping of protein–RNA crosslinks and mutational analyses reveal that Cwc2 is an integrated multipartite RNA-binding unit
The accessibility of the ZnF and the RRM domains in the Cwc2 structure, together with the positively charged connector element and depression, suggests that Cwc2 may interact with RNA at several points on its surface. To gain initial insight into possible RNA contact sites of Cwc2, we performed UV-induced protein–RNA crosslinking, which relies on the UV-induced formation of covalent bonds between RNA bases and amino-acid residues when these are in close spatial proximity (zero-length crosslink; see Kramer et al, 2011 and references therein). Initially, we attempted to identify U6 snRNA crosslinking sites on Cwc2 obtained by UV irradiation of purified spliceosomes; however, these attempts failed due to insufficient amounts of spliceosomes that could be obtained. We therefore decided to use binary U6 snRNA–Cwc2 complexes formed in vitro, with the goal of mapping potential RNA contact sites in Cwc2 by mass spectrometry. Although Cwc2 interacts with several spliceosomal snRNAs in vitro (McGrail et al, 2009), we chose U6 snRNA for our crosslinking studies, because U6 is the only snRNA that is crosslinked to Cwc2 during splicing in yeast extracts or in purified catalytically active spliceosomes (McGrail et al, 2009; Rasche et al, 2012). Although U6 snRNA in isolation is not expected to adopt the conformation present in intact spliceosomes, UV irradiation of the binary U6 snRNA–Cwc2 complexes should reveal which regions of Cwc2 are in principle able to interact with RNA. Prior to crosslinking, we checked whether a stable and defined complex can be reconstituted between Cwc2 and U6 snRNA in solution. Thus, full-length Cwc2 was incubated with U6 snRNA at a molar ratio of 10:1 and then subjected to size-exclusion chromatography. The elution profile is dominated by a single peak that corresponds to a monodiperse complex with the apparent molecular weight of 260 kDa (Supplementary Figure S5). The elution volume of the complex is in agreement with the ones of the individual components. To drive the equilibrium towards complex formation, we incubated U6 snRNA with a three-fold higher excess of Cwc2 (30:1 ratio of protein to RNA) and subjected the mixture to UV irradiation. The complex was then hydrolysed with nucleases and endoproteinases, and titanium dioxide enrichment was subsequently performed. Crosslinked peptides were identified by the corresponding fragments obtained after higher energy collision-induced dissociation (HCD). The composition of the crosslinked RNA moiety was calculated from the difference between the mass of the crosslinked species and the calculated peptide mass.
We identified crosslinked peptides that encompassed residues W37–K61, F47–K61, G79–K101, C87–K101, F117–R131, T136–K149, H150–R159 and N180–R185 (Supplementary Table S1). Inspection of the fragment spectra of the peptide–RNA oligonucleotide conjugates also reveals the identity of the crosslinked amino acid, because the fragment ions containing this amino acid are shifted by the mass of the crosslinked RNA or fragments thereof (a nucleotide, a nucleoside or a base derived from the crosslinked RNA). We were able to narrow down the Cwc2-crosslinked sites to F47, C87, Y120/R121, K152 and C181 (Figure 3B; Supplementary Figures S7–S12). The crosslinked amino acid in the peptide encompassing T136–K149 could not be identified unambiguously. However, as we obtained sequence information (y-type and b-type fragment ions) of this particular peptide from amino-acid K149 (y1 ion) to V139 (y11 ion) and from T136 and L137 (a2 and b2 ion), but no fragment ion (b3, y12 and/or immonium ion) for Y138, this strongly suggests that Y138 is the actual crosslinked residue (Supplementary Figure S10).
The six residues of Cwc2 crosslinked to RNA are not clustered at a single site, but rather are sparsely distributed over a large and positively charged surface of the protein, although on the same side of the molecule (Figure 5A and B). Y138 and C181 are conserved residues located in the RNP2 and in the RNP1 motifs, respectively, while C87 is located in the ZnF (Figure 5B). This indicates that the two canonical RNA-binding domains, indeed function in RNA binding within the context of the Cwc2 protein. Crosslinked residues F47 and K152 are located on the same side as the RNP1 and RNP2 motifs and may thus be part of the RRM-based RNA-binding site of Cwc2. Noteworthy, while K152 is exposed to the solvent, the side chain of the crosslinked F47 is buried deeply within the Cwc2 core (Figure 6A, left). It therefore seems probable that the RNA crosslinked at this position contacts the backbone of F47, unless the side chain moves to the surface of Cwc2 when RNA is bound. Finally, the crosslinked residues Y120/R121 are located in the positively charged connector region, strongly supporting the idea that the connector element is also involved in Cwc2–U6 snRNA interactions. Thus, altogether, we have identified three distinct regions in the structure that are capable of contacting RNA, consistent with a possible role for Cwc2 in accommodating several RNA elements from the spliceosomal RNA–RNA network.
Figure 6.
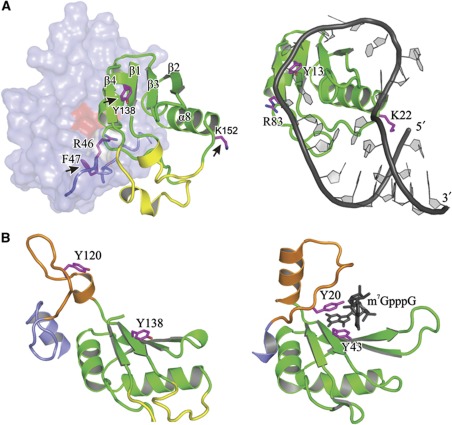
Notable similarities with other RRM domains. (A) Comparative view of Cwc2 (left) and the RRM domain of U1A in complex with its cognate RNA (right; Oubridge et al, 1994). Three U1A residues that contact its cognate RNA (right) and residues located in equivalent positions in Cwc2 (left) are depicted as magenta sticks. The Cwc2 residues crosslinked to RNA are marked with black arrows. The ZnF–Torus module is shadowed for the sake of clarity. (B) Comparative view of the connector-RRM (orange–green) region from Cwc2 (left) and the topologically equivalent region of CBP20 in complex with a cap analogue (black; pdb 1H2T, left). Other regions of the two protein molecules have been omitted for clarity sake. Aromatic residues of CBP20 that form the binding pocket of the cap (right) and Cwc2 residues that might cooperate in a similar manner (left) are depicted as magenta sticks.
Given that Cwc2 appears to bind RNA non-specifically (McGrail et al, 2009), we investigated whether all these regions recognize RNA indiscriminately or whether some of them possess a preference for particular RNA regions. Thus, we performed comparative crosslinking experiments with the binary complexes Cwc2–U6 snRNA versus Cwc2–U4 snRNA. As shown in Supplementary Table S2, the number of identified crosslinks between RNP1 (C181) and RNP2 (Y138) and both snRNA was comparable. Importantly, crosslinks involving the ZnF (C87), the connector element (Y120) and the Torus domain (F47) were much more frequently identified and validated (by MSMS) when Cwc2 crosslinked to U6 was analysed as compared with Cwc2 crosslinked to U4. This indicates a certain preference of these Cwc2 regions for U6 snRNA. To investigate which part of U6 snRNA interacts with which region of Cwc2, we also performed crosslinking with a binary Cwc2–U6-ISL (bases 59–90) complex. Interestingly, U6-ISL was crosslinked frequently to both RNP2 and RNP1, and to a lesser extent (compared with U6) to the ZnF and connector element, while no crosslinks were found for the Torus domain.
To substantiate the idea that Cwc2 acts as a multipartite RNA-binding protein, we have mutated the RNA-binding sites identified by crosslinking (Figure 5C), except for C87 whose structural role does not tolerate mutagenesis (McGrail et al, 2009), and the mutants were expressed and purified to homogeneity. Size-exclusion chromatography profiles demonstrated that all protein samples migrated as sharp single peaks, corresponding to the expected molecular weight. The purified mutants were assayed for RNA binding by EMSA. The RNA-binding affinity of Cwc2 for U6 snRNA was not affected by any of the single-point mutations tested (Figure 5C; Supplementary Figure S6). Interestingly, while mutation of Y138A in RNP1 did not change significantly RNA-binding affinity, it led to a significantly faster migrating RNP complex (compared with wild-type Cwc2) whose mobility did not change with increasing concentrations of protein (Figure 5C). This suggests that the conformation of the RNP complex formed with the Y138A mutant may be different due to changes in the interaction of the Cwc2 RRM with U6 snRNA. We next prepared double mutants that contained Y138A plus mutations in those residues found to crosslink RNA as well as a double mutant containing Y120/F47A (Figure 5C). A significant decrease in the amount of U6 snRNA shifted was observed with Y138A/Y120A and Y138A/C181A, demonstrating that Y120 from the connector element and C181 from RNP1 are important for RNA binding, and that there is an interplay between these two regions. Instead, no difference (compared with Y138A alone) was seen with Y138A/F47A and Y138A/K152A, indicating that F47 and K152 do not play a crucial role in RNA binding in a binary system. Furthermore, the Y120/F47A double mutant behaved like the wild-type Cwc2, indicating that these mutations in the connector loop and torus domain are not sufficient to alter RNA binding. Taken together, the mobility shift assays show that in addition to the RRM, the connector element contributes to RNA binding, supporting the idea that Cwc2 acts as a multipartite RNA-binding unit.
The ZnF and connector element are important for Cwc2 function in pre-mRNA splicing
To investigate the functional significance of selected structural elements of Cwc2 in splicing in vitro, point and truncation mutants of Cwc2 were used in complementation experiments with yeast splicing extracts depleted of their endogenous Cwc2, as described above (Figure 1B).
We first addressed whether the entire, Cwc2ΔC folding unit is required for splicing activity, or whether this can be achieved by using individual domains, such as those defined on the basis of the crystal structure. While we were unable to obtain soluble deletion mutants corresponding to the ZnF–Torus module, we could express a soluble fragment encompassing the C-terminal RRM domain (residues 124–234). Complementation assays showed that the single RRM domain could not restore pre-mRNA splicing activity (Figure 1B, lane 6). Furthermore, splicing complementation with full-length Cwc2 was not inhibited by addition of the RRM domain (unpublished data), raising the possibility that the single RRM domain is not able to bind RNA efficiently on its own. This idea was supported by an EMSA experiment, which showed that the isolated RRM domain does not bind U6 snRNA (Figure 1C). In sum, these data indicate that the RRM domain alone is not sufficient for Cwc2 function in splicing.
Although the Cwc2 structure and the crosslinking experiments show that the two RNPs motifs are exposed and are able to bind RNA, single-point mutations in amino acids Y138 and F183 (both of which are located in the RNP2 and RNP1 motifs of Cwc2, respectively) are not lethal (McGrail et al, 2009). Taken together, these results imply that the binding of RNA to the Cwc2–RRM is complex in nature and may involve cooperation with neighbouring regions of Cwc2 (see below and Discussion). Therefore, we did not attempt a more exhaustive mutational analysis of the RRM domain, and chose instead to examine the functional significance of the other two regions of Cwc2 that were found to contact RNA.
First, we mutated residue Y89 of the ZnF to alanine, on the basis of the following considerations: in RNA–protein co-crystals of MBNL1 and TIS11d, there are respectively one and two aromatic amino acids in the ZnF structures that stack against nucleotide bases of the RNA (Hudson et al, 2004; Teplova and Patel, 2008). While one of the corresponding aromatic residues is replaced in Cwc2 by a leucine (L83), the other one is conserved (Y89). Addition of the Cwc2–Y89A mutant to the Cwc2-depleted extract resulted in some restoration of splicing, but only to a low level (34% of the activity obtained with the wild-type Cwc2 protein; Figure 1B, lane 7). This result supports the importance of residue Y89 and, accordingly, of the entire ZnF for the function of Cwc2.
To test the function of the connector element, we generated several single-point mutants. Substitution of Y120 by alanine essentially abolished the ability of Cwc2 to restore splicing in the depleted extract (Figure 1B, lane 8). The replacement of residue K132 or F130 in the connector element of Cwc2 with alanine, likewise led to a significant loss of splicing activity (mRNA production levels of ca. 42 and 53%, respectively, compared with complementation with the wild-type protein) (Figure 1B, lanes 9 and 10). These results show that the connector element is important for the function of Cwc2 in splicing in vitro.
Discussion
An unusual toroidal domain acting as a scaffold for the ZnF, RRM and connector in Cwc2
Cwc2 belongs to the group of so-called NTC-related proteins and is stably integrated into the spliceosome during its activation. Cwc2 is essential for the viability of S. cerevisiae and is required for the catalytic activation of the spliceosome. The numerous contact points between Cwc2 and the spliceosomal RNA–RNA interaction network probably provide a means for Cwc2 to assist in the attainment of the active conformation of the catalytic centre (Rasche et al, 2012). In this work, we have determined the crystal structure of Cwc21−240 (Cwc2ΔC), which includes the ZnF and RRM domain (Figure 3A), to a resolution of 2.4 Å. We were able to show that the Cwc2ΔC fragment can replace endogenous Cwc2 in a splicing complementation assay in vitro, indicating that Cwc2ΔC retains the basic functions of the full-length protein.
The crystal structure of Cwc2ΔC reveals a new design principle for an RNA-binding protein. That is, three separate RNA-binding modules (ZnF, RRM and the intervening connector element) are organized into a compact globular structure by a toroidal domain that acts as a scaffold. This so-called Torus domain is composed of the flanking regions of the ZnF, around which they wrap like a ring on a finger. At the same time, the Torus domain is closely packed against the ‘back’ side of the RRM domain, resulting in a compact structure in which the potential RNA-binding sites of the ZnF and RRM domains are located on opposite sides. The RRM domain alone (residues 124–234) binds RNA much less efficiently than the entire Cwc2ΔC, indicating the importance of the tight association with the Torus and the ZnF domains in creating a functional unit. Accordingly, the RRM domain does not restore activity to a Cwc2-depleted splicing mixture (Figure 1B), and does not inhibit splicing complementation by competing with full-length Cwc2 in such an assay (Rasche, unpublished data). These observations do not conflict with the observation that Cwc2124–339 bound RNA efficiently in vitro (McGrail et al, 2009), as it is probable that the C-terminus of Cwc2 (residues 235–339) also stabilizes the structure of the RRM domain. In addition to a scaffolding role, the Torus domain frames the broad and positively charged depression on the Cwc2ΔC surface, which may accommodate RNA elements (Figure 5A). Thus, the front rim of the depression is formed by the α1 helix and loop located between α1 and α2 helices.
The Torus domain does not resemble any other domain reported so far and its structure suggests that it has co-evolved as an expansion of the ZnF domain. Although the entire ZnF–Torus module might be typical for Cwc2, similar design principles of domains that have acquired structured expansions in order to meet requirements for new functions have been previously reported in other proteins. For instance, the C-terminal Jab-MPN domain of the spliceosomal protein Prp8 has acquired several insertions and appendages that form a well-structured external layer that acts as a protein–protein interaction platform (Pena et al, 2007).
The human homologue of Cwc2, namely RBM22, also contacts the U6 snRNA and pre-mRNA intron within isolated spliceosomes at nucleotides corresponding to those contacted by its yeast counterpart (Rasche et al, 2012). Sequence comparison of RBM22 and Cwc2 shows that the ZnF and the RRM are well conserved (47 and 35% identity), while the Torus and the connector element exhibit poor sequence conservation (Figure 3B). Although only the 3D structure of RBM22 will show to which extent its folded core resembles that of Cwc2, the high conservation of several residues located at the interfaces between the RRM, ZnF and Torus suggests that the inner architecture of RBM22 is built up in a similar manner. Indeed, in the NMR-derived structure of the isolated RRM domain of hRBM22 (pdb 2YTC), there are three highly conserved RRM residues that in Cwc2 contact the ZnF–Torus module (E288, E292, N296 from hRBM22; Supplementary Figure S13). Moreover, comparison of the ZnF sequences of the two proteins shows that the residue pair K78–E195, which forms a salt bridge between the ZnF and the RRM in Cwc2, is conserved as K170–E292 in RBM22 (Supplementary Figure S13). Nevertheless, it should be noted that hRBM22 has a much longer N-terminal region that may also cooperate with its ‘Cwc2-like’ core in recognizing the catalytic RNA elements of the human spliceosome.
Evidence that Cwc2 is a multipartite RNA-binding protein
It was recently shown by crosslinking and chemical structure probing that in isolated, catalytically active spliceosomes Cwc2 simultaneously contacts several elements of the catalytic RNA network, including the intron as well as U6 snRNA in the ISL and the vicinity of the ACAGAGA box (Rasche et al, 2012). It is not yet known which regions of Cwc2 interact with these various elements of the spliceosome. Therefore, we UV-irradiated binary complexes of Cwc2 with U6 snRNA or U4 snRNA and detected the crosslinked positions by MS, in order to identify domains of Cwc2 with affinity for RNA. We were able to map a total of six sites that were crosslinked with U6 snRNA. They are located in the ZnF (C87), the connector element (Y120–R121), the RRM (K152, C181 from RNP1 and Y138 from RNP2) and in the Torus domain (F47) adjacent to the RNA-binding side of the RRM domain. These data show that not only the RRM domain, but also the ZnF and the connector element are able to contact RNA. Although Cwc2 does not show binding specificity towards a particular snRNA in isolation (McGrail et al, 2009), our comparative crosslinking experiments (Supplementary Table S2), suggest that the connector element and ZnF have a stronger preference for U6 snRNA compared with U4 snRNA. In contrast, the RRM domain was crosslinked with similar intensities to both U4 and U6 snRNA.
Cwc2 contacts several bases in two distinct U6 snRNA regions in the spliceosome (Rasche et al, 2012). We thus asked whether these regions are differentially recognized in binary Cwc2–U6 snRNA complexes by the ZnF, connector element or the RRM. Crosslinking experiments with U6 snRNA and U6-ISL showed that the RRM domain is crosslinked to the two molecules with equal intensity, while the ZnF, the connector element and the Torus are not crosslinked to the U6-ISL. This indicates that in binary complexes, different Cwc2 sites can discriminate between distinct regions of U6 snRNA. Thus, the RRM binds the ISL, while the ZnF and the connector element contact other U6 snRNA regions. However, whether U6 snRNA recognition by Cwc2 in the spliceosome occurs in a similar manner is currently not clear.
Single-point mutations in the crosslinked residues of Cwc2 did not affect RNA binding as assayed by EMSA, probably due to multiple contacts of Cwc2 with RNA. However, double mutations in crosslinked residues from the RRM domain (Y138, C181) and connector element (Y120) revealed that these regions are important for RNA binding in binary complexes. Consistent with our findings, Lu et al. have recently shown that RNA binding is not affected by single-point mutations within the connector element but is completely abolished by a quadruple mutant (Lu et al, 2012). Similarly, a triple mutation in the RNP1 and RNP2 motifs leads to complete abolishment of RNA binding (Lu et al, 2012). Together, the crosslinking experiments and mobility shift assays show that Cwc2 is a multipartite RNA-binding protein, with a structure and biophysical properties that enable it to organize several RNA elements. Furthermore, as the RNA-binding sites are also connected by positively charged regions, such as the surface depression described above (see Figure 5A and Supplementary Figure S3), Cwc2 appears to be able to accommodate a complex RNA structure. The structure of Cwc2 thus provides strong support for the hypothesis, based upon our earlier structure–function investigations (Rasche et al, 2012), that Cwc2 induces or promotes an active conformation of the spliceosomal RNA network through multiple contacts with catalytically important RNA elements.
Importance of the ZnF and connector element for pre-mRNA splicing
Guided by the crystal structure and the UV crosslinking results, we performed mutational analyses in order to assay whether, in addition to the RRM domain, the ZnF and the connector element are important for the function of Cwc2 in pre-mRNA splicing. Using an in vitro complementation splicing assay, we demonstrated that a Y89A point mutation in the ZnF inhibits Cwc2 splicing activity significantly (ca. 65%), while a Y120A point mutation in the connector element abolishes activity almost completely. Although our assay does not provide direct evidence that the reduction in splicing activity is due to inhibition of an RNA–Cwc2 interaction by the point mutations, this seems the most plausible interpretation. In support of this idea, aromatic residues of other ZnF domains corresponding to Y89 in Cwc2 are known to take part in RNA binding (Hudson et al, 2004; Teplova and Patel, 2008). Furthermore, Y120 of the connector element belongs to the Y120–R121 region that was crosslinked to U6 snRNA and its mutation together with Y138A reduced RNA binding as shown by our EMSA experiments (Figure 5C).
The architecture of Cwc2 and possible implications for RNA recognition
The crystal structure and RNA-binding studies presented in this work indicate that Cwc2 possesses a multipartite RNA-binding platform with a unique composite architecture, probably as an adaptation to the particular spatial configuration of the cognate RNA elements from the catalytic centre of the spliceosome. Despite the novelty of this assembly, Cwc2 has evolved from wide spread RNA-binding domains like the ZnF and the RRM.
A structural comparison of Cwc2 domains with the known structures from the Protein Data Bank offers additional insight into the function of this protein. As shown in Figure 4A, the ZnF structure of the CCCH type present in Cwc2 resembles closely that from MBNL1 and TIS11d. The atomic structures of ZnFs of MBNL1 and TIS11d have been determined in complex with RNA, and in both cases nitrogen atoms from the bases are hydrogen bonded to the sulphur atom of the third cysteine coordinated with the zinc atom (Hudson et al, 2004; Teplova and Patel, 2008), in full agreement with the RNA crosslink observed for the equivalent residue (C87) of Cwc2. In both complexes, the major contribution to RNA recognition is made by stacking interactions undergone by two aromatic residues from TIS11d and by an arginine and an aromatic residue in MBNL1 (Figure 4B). One of these residues is conserved as Y89 in Cwc2 (Figures 3 and 4A) and Y89A mutation leads to reduction in splicing activity, suggesting that Y89 is involved in RNA binding. Superposition of MBNL1 complexed with RNA upon that of Cwc2 leads to major steric interference of the RNA backbone with the Torus domain. Instead, RNA recognition similar to that by TIS11d is in principle possible only if the interaction between Y89 and Q25 from the Torus domain would be disrupted in order to allow stacking with the RNA base (Figure 4A).
In addition to canonical RRM residues, two other U6 snRNA-crosslinked residues—F47 and K152—are located on the same side as the RNP1 and RNP2 motifs of the RRM and may strengthen RNA binding. This possibility is illustrated when one compares the co-crystal structures of the RRMs of the human U1A protein in complex with stem–loop II of U1 snRNA (Oubridge et al, 1994). In this case, the non-canonical RNA-binding residue, R83 of U1A contacts one base from the loop of the U1 snRNA hairpin, while K22 from the same protein contacts the loop and the double-helical RNA stem (Figure 6A, right). Notably, structural superposition of this structure with Cwc2 shows that K22 has a position equivalent to that of K152, a Cwc2 residue that has been crosslinked to RNA. Moreover, the side chain of R46, which precedes the crosslinked F47 of the Torus domain in Cwc2, occupies a position equivalent to that of the guanidinium group of R83 in the U1A protein (Figure 6A). Although K152 and F47 do not play an important role in RNA binding based on EMSA assays, their crosslinking to the U6 snRNA suggests that the binding tract of U6-ISL on Cwc2 may follow the one observed for the intramolecular stem–loop II of U1 snRNA when complexed with the U1A protein.
The connector element is a flexible and highly conserved RNA-binding stretch that connects the Torus and RRM domains (116–133; Figure 3A). It is interesting to note that a similar functional and topological association has been described within the nuclear cap-binding complex (CBC; Mazza et al, 2002). The co-crystal structure of CBC and a cap analogue shows the methylated guanosine of the cap sandwiched between two aromatic residues of the CBP20 subunit. One aromatic residue belongs to the RNP2 motif of the RRM domain, while the second, Y20, belongs to the N-terminal extension of the RRM domain. Similar to the Cwc2 connector element, the N-terminal extension of CBP20 exhibits intrinsic flexibility and in the presence of the cap becomes structured as a loop, which contains the crucial Y20, followed by a short helix (Figure 6B, right). Notably, the functionally important Y120 residue appears to be located in a similar position in the Cwc2 connector (Figure 6B, left), suggesting a possible mode of recognizing an RNA base in cooperation with the RRM domain. The connector element of Cwc2 might be in a position to act as a molecular switch in modulating RNA contacts during the different spliceosomal transitions. For instance, a protein ligand might lock the connector into a conformation that either cannot accommodate RNA at all, or can accommodate it, but in a different manner. Such a ligand could be other spliceosomal proteins (for instance Prp8, NTC or NTC-related proteins) that are remodelled or exchanged during the spliceosomal transitions. This scenario would explain the change in the interaction pattern between Cwc2 and U6 snRNA in purified Bact and C complexes (Rasche et al, 2012). As Cwc2 interacts with proteins of the NTC (Ohi and Gould, 2002), it may play a central part in transmitting forces between spliceosomal proteins and the catalytic RNA network that lead to rearrangements in spliceosome structure essential for its activation.
Taken together, our data indicate that Cwc2 is a compact multipartite RNA scaffold with a conformation suitable to bridge and accommodate catalytically important RNA elements and thereby induce an active configuration of the catalytic centre of the spliceosome.
Materials and methods
Details of the cloning, mutagenesis, protein expression, purification, limited proteolysis, EMSAs and the crystallographic procedures are in the Supplementary data.
Mass spectrometric analysis of UV-induced crosslinking sites
UV crosslinking and crosslink enrichment were performed according to established protocols (Luo et al, 2008; Kramer et al, 2011). In all, 100 μg of Cwc2 and 3 μg of U6 snRNA were incubated in 100 μl buffer (20 mM HEPES-NaOH pH 7.5, 100 mM NaCl, 1 mM DTT) on ice for 30 min for complex formation prior to crosslinking at 254 nm for 2 and 10 min (crosslinking apparatus built in-house, equipped with four 8 W lamps, G8T5; Sankyo Denki, Japan). The complex was hydrolysed by RNases A and T1 (Ambion, Applied Biosystems, Darmstadt, Germany) and protease trypsin (Promega, Mannheim, Germany) in the presence of 1 M urea, desalted via C18 material and enriched by titanium dioxide chromatography (columns prepared in-house; C18 material from Dr Maisch GmbH, Ammerbuch, Germany; TiO2 material from FL Sciences, Tokyo, Japan). Nano-LC-ESI-MS was performed on an Orbitrap Velos with HCD as fragmentation method. Spectra were evaluated and annotated manually.
Immunodepletion of Cwc2
For depletion of TAP-tagged Cwc2, yeast extract in AGK buffer (20 mM HEPES-KOH pH 7.9, 200 mM KCl, 1.5 mM MgCl2, 10% glycerol) was incubated either with IgG-Sepharose, or (for mock depletion) with Protein A Sepharose (PAS), for 2 h at 4°C. After incubation, the Sepharose beads were sedimented by brief centrifugation and the supernatant (depleted extract) was dialysed against 20 mM HEPES-KOH pH 7.9, 50 mM KCl, 20% (v/v) glycerol, 0.2 mM EDTA pH 8.0, 0.5 mM DTT, 0.5 mM PMSF and 2 mM benzamidine and then analysed by western blotting.
Data deposition
The coordinates and structure factors have been deposited in the Protein Data Bank with PDB code 3TP2.
Supplementary Material
Acknowledgments
We thank Julius Nitsche and Mirjam Sommer for technical assistance, Monika Raabe for mass spectrometric analysis, the teams of the beamline PXII (SLS, Villigen, Switzerland) for support during diffraction data collection and Cindy Will for critical reading of the manuscript. This study was supported by the Max Planck Society.
Author contributions: JS performed the cloning, mutagenesis, expression, purification and limited proteolysis; KK and HU performed the crosslinking and mass spectrometric analyses; NR and PF performed the splicing complementation assay; OD performed the mobility shift assay; JS and VP performed the crystallographic analyses; JS, RL and VP wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Chan SP, Cheng SC (2005) The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199 [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Clery A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–298 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J (1990) Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250: 404–409 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R (2009) The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- Fortner DM, Troy RG, Brow DA (1994) A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev 8: 221–233 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP (2004) Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg R, McGrail JC, O’Keefe RT (2010) The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem Soc Trans 38: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264 [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S, Seraphin B (1993) Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039 [DOI] [PubMed] [Google Scholar]
- Kramer K, Hummel P, Hsiao H, Luo X, Wahl M, Urlaub H (2011) Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int J Mass Spectrom 304: 184–194 [Google Scholar]
- Liu YC, Chen HC, Wu NY, Cheng SC (2007) A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Lu G, Yan C, Wang L, Li W, Yin P (2012) Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition. Biochem J 441: 591–597 [DOI] [PubMed] [Google Scholar]
- Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC (2008) Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol Cell 32: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1994) Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet 28: 1–26 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S (2002) Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J 21: 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail JC, Krause A, O’Keefe RT (2009) The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res 37: 4205–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe RT, Norman C, Newman AJ (1996) The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Gould KL (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K (1994) Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438 [DOI] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC (2007) Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC (2008) Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Rasche N, Dybkov O, Schmitzová J, Akyildiz B, Fabrizio P, Lührmann R (2012) Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J 31: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM (2008) Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Patel DJ (2008) Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol 15: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi CW, Ren L, Xu P, Ohi MD, Gould KL, Chazin WJ (2010) The Prp19 WD40 domain contains a conserved protein interaction region essential for its function. Structure 18: 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwalder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R (2009) Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2011) Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: pii: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R (2008) Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.