Abstract
Vertebrates have two cohesin complexes that consist of Smc1, Smc3, Rad21/Scc1 and either SA1 or SA2, but their functional specificity is unclear. Mouse embryos lacking SA1 show developmental delay and die before birth. Comparison of the genome-wide distribution of cohesin in wild-type and SA1-null cells reveals that SA1 is largely responsible for cohesin accumulation at promoters and at sites bound by the insulator protein CTCF. As a consequence, ablation of SA1 alters transcription of genes involved in biological processes related to Cornelia de Lange syndrome (CdLS), a genetic disorder linked to dysfunction of cohesin. We show that the presence of cohesin-SA1 at the promoter of myc and of protocadherin genes positively regulates their expression, a task that cannot be assumed by cohesin-SA2. Lack of SA1 also alters cohesin-binding pattern along some gene clusters and leads to dysregulation of genes within. We hypothesize that impaired cohesin-SA1 function in gene expression underlies the molecular aetiology of CdLS.
Keywords: CdLS, ChIP-sequencing, embryonic development, mouse model, transcription
Introduction
In addition to its role in sister chromatid cohesion, essential for accurate chromosome segregation and postreplicative DNA repair (Losada and Hirano, 2005; Peters et al, 2008; Nasmyth and Haering, 2009), cohesin contributes to transcriptional regulation (Rollins et al, 2004; Dorsett et al, 2005; Horsfield et al, 2007; Hallson et al, 2008; Pauli et al, 2008, 2010; Schaaf et al, 2009; Lin et al, 2011), DNA replication (Guillou et al, 2010), V(D)J recombination (Degner et al, 2009) and T-cell receptor rearrangement (Seitan et al, 2011). Chromosome conformation capture (3-C) studies and other experimental evidences suggest that cohesin performs these functions by promoting the formation of chromatin loops in collaboration with additional factors (Hadjur et al, 2009; Mishiro et al, 2009; Nativio et al, 2009; Hou et al, 2010; Chien et al, 2011; Seitan et al, 2011). One such factor is CTCF, which colocalizes with cohesin at many sites along the human and mouse genomes (Parelho et al, 2008; Rubio et al, 2008; Wendt et al, 2008). Cohesin is also found at non-CTCF-binding sites occupied by tissue-specific transcription factors (TFs), and possibly contributes to set up tissue-specific transcriptional programs (Schmidt et al, 2010). In murine embryonic stem cells, cohesin and the transcriptional coactivator Mediator facilitate DNA looping between the enhancers and promoters of some genes required to maintain pluripotency (Kagey et al, 2010).
The involvement of cohesin in all these different processes that are critical for cell proliferation and differentiation makes it difficult to determine which is most relevant for cohesinopathies, human syndromes caused by mutations in proteins related to cohesin (Bose and Gerton, 2010). The most prevalent cohesinopathy described to date is the Cornelia de Lange syndrome (CdLS), which is caused by heterozygous mutations in the cohesin loader Nipbl (Krantz et al, 2004; Tonkin et al, 2004) or, in a reduced number of cases, in the cohesin subunits Smc1 or Smc3 (Musio et al, 2006; Deardorff et al, 2007). CdLS is characterized by mental retardation, reduced body size, dysmorphic face, upper limb defects and several additional organ abnormalities (Liu and Krantz, 2009). Cells from patients rarely show cohesion defects and instead, microarray studies reveal altered patterns of gene expression (Castronovo et al, 2009; Liu et al, 2009). Nipbl-heterozygous mice exhibit many features of CdLS and display modest but significant transcriptional changes in mouse embryonic fibroblasts (MEFs) and embryonic brains (Kawauchi et al, 2009). Whether the wide range of defects observed in CdLS patients are elicited by expression changes in a few critical genes or by the sum of multiple small alterations is unclear (Muto et al, 2011). Moreover, the exact role of cohesin in CdLS pathogenesis is still far from understood.
Importantly, somatic vertebrate cells have two distinct versions of cohesin that consist of Smc1, Smc3, Rad21/Scc1 and either SA1 or SA2 (Losada et al, 2000; Sumara et al, 2000). The two SA proteins show 75% sequence identity along their central region and only differ in short regions at both ends of each protein. Cohesin-SA1 and cohesin-SA2 present similar cell-cycle regulation in terms of loading and dissociation from chromatin (Losada et al, 2000) and are found in all mouse tissues (Remeseiro et al, 2012). However, they are not functionally equivalent. Mice homozygous for a gene-trap allele that abolishes SA1 expression are embryonic lethal (Remeseiro et al, 2012). We have shown that cohesin-SA1 plays a specific role in telomere cohesion that is essential for efficient telomere replication. In contrast, cohesion at centromeres, which is critical for chromosome segregation, relies on cohesin-SA2 (Canudas and Smith, 2009; Solomon et al, 2011). Despite their robust centromeric cohesion, chromosome segregation is defective in SA1-deficient cells and often leads to aneuploidy, increasing the incidence of spontaneous tumours in heterozygous animals (Remeseiro et al, 2012).
To further understand the functional specificity of cohesin-SA1 and cohesin-SA2, we have compared their genomic distributions. We report, for the first time, the genome-wide binding sites of four different cohesin subunits in both wild-type and SA1-null mouse cells. We show that cohesin-SA1 is enriched at promoters and, more importantly, that it determines the distribution of cohesin. In the absence of SA1, cohesin is significantly less abundant at gene promoters and relocates to genomic positions featured by low cohesin occupancy and absence of CTCF. As a result, gene expression is altered, affecting genes involved in developmental pathways relevant to CdLS.
Results
SA1 is essential for embryonic development
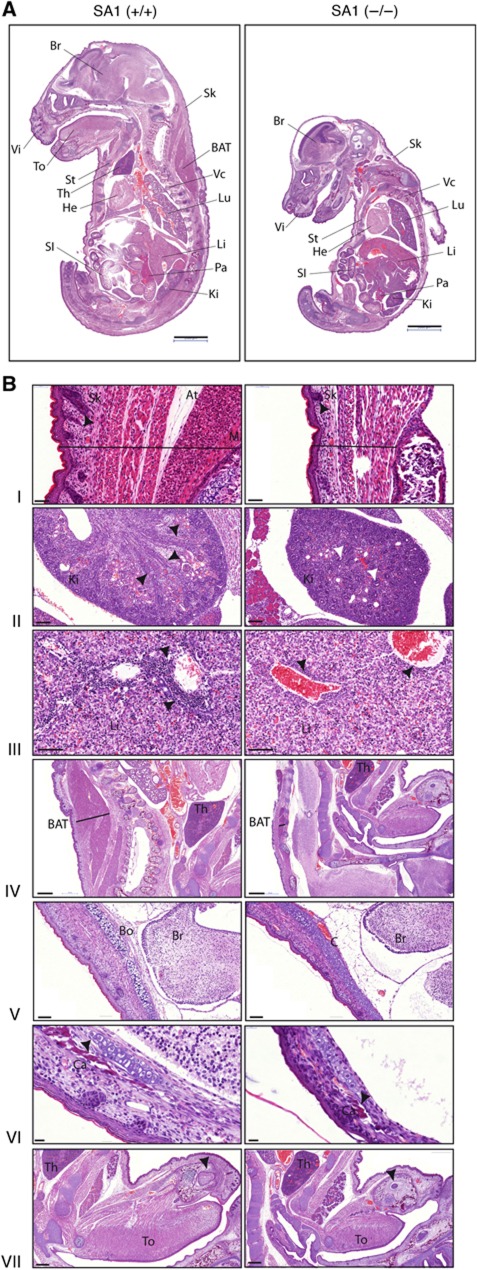
We recently generated a genetically modified mouse model lacking the expression of Stag1 gene encoding the SA1 cohesin subunit (Remeseiro et al, 2012). Viability of SA1-null embryos strongly decreases by E12.5, but some exceptionally survive to E18.5 (see Supplementary Table S1 in Remeseiro et al, 2012). Those late embryos present a clear growth delay and general hypoplasia (Figure 1A). Their skin is much thinner, with less hair follicles and a reduced muscle layer compared with their wild-type littermates (Figure 1B, panel I). The developmental delay is also observed in organs such as kidney and liver (Figure 1B, panels II and III, respectively). Interestingly, SA1-null embryos present some features characteristic of CdLS in addition to the reduced body size. These include impairment of lipid metabolism, as illustrated by a dramatically diminished interscapular layer of brown adipose tissue (BAT) (Figure 1B, panel IV), and severe abnormalities in skeletal and bone development, featured by delayed ossification and defective calcium deposition (Figure 1B, panels V–VII). Both cohesin-SA1 and cohesin-SA2 are ubiquitously expressed in wild-type E17.5 embryos and, importantly, SA2 expression is not altered in SA1-null embryonic tissues, including those described above (Supplementary Figure S1A). Protein and mRNA levels of other cohesin subunits remain invariable in the absence of SA1 (shown for brain in Supplementary Figure S1B and C). Moreover, the amount of total cohesin bound to chromatin does not change in MEFs lacking SA1 (Figure 4G in Remeseiro et al, 2012). Thus, it is unlikely that the inability of cohesin-SA2 to compensate the cohesin-SA1 deficiency has merely a quantitative basis. Instead, cohesin-SA1 must perform specific function(s) in embryonic development that cannot be assumed by cohesin-SA2.
Figure 1.
SA1-null embryos present growth delay. Histological analysis of HE-stained tissue sections from E17.5 embryos shows general hypoplasia and developmental delay in SA1-null embryos. (A) SA1-null embryos present a much reduced body size compared with their wild-type littermates. The following organs/tissues are indicated: BAT, brown adipose tissue; Br, brain; He, heart; Ki, kidney; Li, liver; Lu, lung; Pa, pancreas; SI, small intestine; Sk, skin; St, sternum; Th, thymus; To, tongue; Vc, vertebral column; Vi, vibrissa. Scale bars, 2 mm. (B) Detailed sections of different tissues showing either developmental delay or CdLS features. (I) Thinner skin (black line) in SA1-null embryos with less hair follicles (arrowheads), reduced adipose tissue (At) and thinner muscle layer (M). Scale bars, 50 μm. (II) Kidney sections showing collecting ducts already formed in wild-type (black arrowheads) and not radially arranged in SA1-null embryos. In the latter, glomeruli are dispersed all over as primitive glomeruli at earlier stages (white arrowheads). Scale bars, 100 μm. (III) Decreased number of haematopoietic precursors in livers from SA1-null embryos compared with wild-type (arrowheads). Scale bars, 100 μm. (IV) Thickness of the interscapular layer of BAT is dramatically diminished in SA1-null embryos (black line). Scale bars, 500 μm. (V) Delayed intramembranous ossification in the cranium in the absence of SA1. Notice the presence of cartilage (C) in SA1-null embryos instead of bone (Bo) already formed in the wild-type. Scale bars, 100 μm. (VI) Defective bone calcification (Ca) in the cranium from SA1-null embryos. Scale bars, 20 μm. (VII) Delay in dentition in SA1-null embryos. Arrowheads in both sections indicate tooth buds. Scale bars, 500 μm.
Genome-wide distribution of cohesin-SA1 and cohesin-SA2
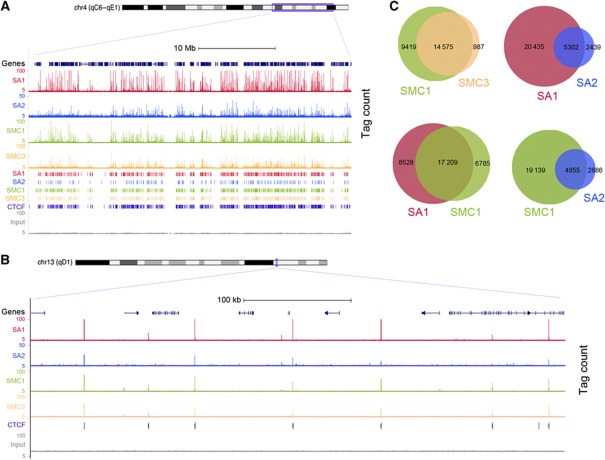
In order to assess if a differential distribution of the two cohesin complexes contributes to their functional specificity, we performed chromatin immunoprecipitation followed by massive parallel DNA sequencing (ChIP-seq) in primary MEFs (E12.5) using SA1, SA2, SMC1 and SMC3-specific antibodies (Supplementary Figure S2A and B). Genome-wide analysis of the sequenced tags using MACS peak detection algorithm (see Materials and methods) defined 25 737 binding sites for SA1, 7741 for SA2, 23 994 for SMC1 and 15 546 for SMC3, with a cutoff P-value of 10−5 and FDR<0.1 (Figure 2A and B; ChIP-seq data summary in Supplementary Tables S1 and S2). A subset of these regions was further validated by quantitative PCR analysis (ChIP-qPCR; Supplementary Figure S2C, D and E). As expected for proteins forming a complex, a large overlap was observed between the binding sites obtained with SMC1 and SMC3 antibodies, as well as between those seen with SMC1 and either SA1 or SA2 antibodies (Figure 2C). Lack of a more complete overlap is likely attributable to slight differences in the accessibility of the antibodies to the complex depending on its chromatin environment. Since cohesin has been reported to colocalize with the insulator protein CTCF, we compared all our cohesin data sets with a data set of CTCF-binding sites from a murine cell line available online (ENCODE/Stanford/Yale data set). Most positions detected with any given cohesin antibody correspond to a CTCF-binding site, with the SA2 data set showing the lowest overlap (63%; Supplementary Figure S3A). Thus, our ChIP-seq data, obtained with four different antibodies, provide an accurate map of cohesin-binding sites along the mouse genome.
Figure 2.
Genome-wide distribution of cohesin-SA1 and cohesin-SA2 in the mouse genome. (A) Representative genomic distribution of SA1, SA2, SMC1 and SMC3 in a region of mouse chromosome 4. Tag counts (upper) and called peak tracks (lower) are shown. A track with CTCF-binding sites (from ENCODE/Stanford/Yale data set) and the input are also depicted. Higher magnification of a region of chromosome 13 is shown in (B). (C) Venn diagrams showing the overlapping between the binding sites identified in wild-type MEFs by ChIP-seq with the indicated antibodies.
Cohesin distribution and enrichment at promoters depend on SA1
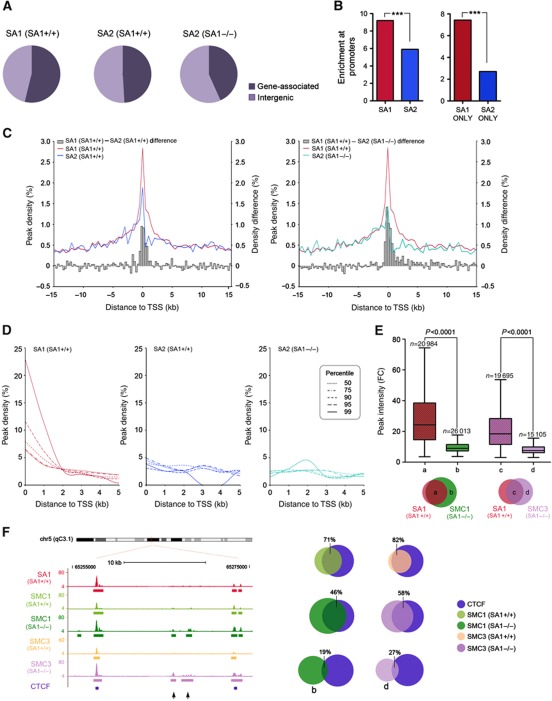
Cohesin-SA1 and cohesin-SA2 binding sites are distributed among gene-associated and intergenic regions to a similar extent in wild-type MEFs (Figure 3A, left and middle pie charts). Importantly, cohesin-SA1 is highly enriched at regions 1-kilobase (kb) upstream transcription start sites (TSS) considering that they represent only 0.62% of the mouse genome (Figure 3B, left). The enrichment is significantly less noticeable for cohesin-SA2. This difference is even more remarkable if we compare enrichment at TSS of cohesin-binding sites that are exclusive for SA1 or SA2 (Figure 3B, right). In addition, the frequency of binding at TSS is higher for cohesin-SA1 than for cohesin-SA2 (Figure 3C, left). We next asked whether the absence of SA1 affects cohesin distribution. Indeed, ChIP-seq analysis of SA1-null MEFs with the SA2 antibody resulted in 6349 peaks (P-value of 10−5, FDR<0.1) from which only 45% overlap with the SA2 peaks found in wild-type MEFs (Supplementary Figure S3B; Supplementary Tables S1 and S2). The new positions occupied by SA2 correspond to binding sites preferentially located at intergenic regions (Figure 3A, pie chart on the right). The frequency of SA2 binding at TSS is further reduced in SA1-null cells, which suggests that SA2 is unable to replace SA1 particularly at these genomic positions (Figure 3C, right). Interestingly, we observed a gradual increase in frequency of binding at TSS as the cohesin-bound regions lengthen only in the case of cohesin-SA1, but not for cohesin-SA2 in either wild-type or SA1-null cells (Figure 3D). This indicates that genomic regions containing the highest density of cohesin-SA1 molecules are mainly gene promoters.
Figure 3.
Cohesin distribution depends on SA1. (A) Distribution of SA1 and SA2 in SA1+/+ (wild-type) cells and of SA2 in SA1−/− cells represented as percentage of peaks detected at gene-associated regions (including 1 kb upstream TSS, Gene Body and 1 kb downstream TTS) and intergenic regions. Notice that SA2 relocates towards intergenic regions in the absence of SA1. (B) Left: The percentage of SA1 and SA2 binding sites located in regions 1 kb upstream TSS in wild-type cells was normalized against the frequency of these regions in the genome and displayed as fold enrichment. Right: Same analysis performed for positions that are exclusive for SA1 or SA2 (shadowed bars). ***P<0.0001. (C) Cohesin distribution around TSS (±15 kb) defined as peak density (%). The histogram represents the difference in peak density between SA1 and SA2. (D) Frequency of binding at TSS in percentiles of peak length distribution (P50, P75, P90, P95 and P99). (E) Peak intensity (measured as fold change) for SMC1 and SMC3 peaks identified in SA1-null MEFs that overlap (groups ‘a’ and ‘c’) or not (groups ‘b’ and ‘d’) with SA1 peaks, as indicated in the Venn diagrams below the graph. Notice that cohesin positions in groups ‘a’ and ‘c’ present significantly higher occupancy (medians=24.4 and 18.5, respectively) than those in groups ‘b’ and ‘d’ (medians=9.2 and 7.6, respectively). See Supplementary Figure S3B for the number of peaks in each group. (F) Left: Representative image showing the redistribution of SMC1 and SMC3 in SA1-null cells towards low occupancy sites (arrowheads). Right: Venn diagrams showing the overlapping between CTCF-binding sites and the cohesin-binding sites indicated, including those defined as groups ‘b’ and ‘d’ in (E). More detailed information regarding the peak number within each group is shown in Supplementary Figure S3A and B.
To further characterize the redistribution of cohesin-SA2 in the absence of SA1, we performed ChIP-seq with SMC1 and SMC3 antibodies in SA1-null MEFs. The number of cohesin positions detected with these antibodies in the SA1-null MEFs (46 997 and 34 800, respectively) is higher than the number detected with the SA2 antibody, most likely due to a more limited sensibility of the latter (Supplementary Tables S1 and S2). The fact that the number of peaks doubles in both cases with respect to wild-type MEFs suggests a major redistribution of cohesin in the absence of SA1 (Supplementary Figure S3C). Of all SMC1-binding sites identified in SA1-null MEFs, 45% correspond to cohesin-SA1 positions in wild-type cells (group ‘a’ in Figure 3E). The remaining 55% (group ‘b’) show significantly lower peak intensity than those in group ‘a’, and the same is true for the SMC3-binding sites in SA1-null MEFs (groups ‘c’ and ‘d’ in Figure 3E; examples are shown in Figure 3F, left). Binding to some of these regions with lower cohesin occupancy was validated by ChIP-qPCR (Supplementary Figure S2C, lower panels). These positions have an additional feature. Whereas most sites bound by SMC1 and SMC3 in wild-type MEFs are also CTCF sites (71 and 82% respectively, see charts in Figure 3F, right), the overlap decreases to 46 and 58% in SA1-null cells, mostly due to the modest overlapping of the new positions with CTCF (groups ‘b’ and ‘d’, 19 and 27%, respectively). These results indicate that in the absence of SA1, cohesin spreads to CTCF-negative positions in which the complex shows significantly weaker enrichment compared with the positions defined in the wild-type cells. Taken all together, we conclude that cohesin accumulation at promoters and CTCF sites depends largely on the presence of SA1. This points to a unique role of cohesin-SA1 in transcriptional regulation.
Transcriptome changes in SA1-null cells affect CdLS-related functions
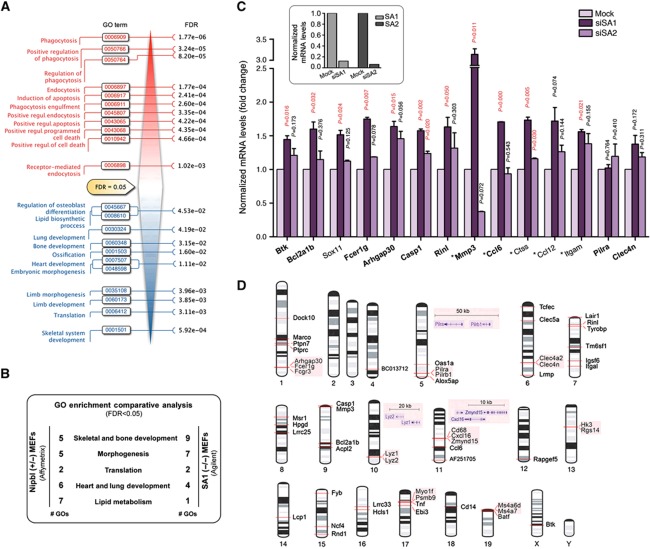
To obtain a global view of expression changes in the absence of SA1, we analysed the transcriptomes of SA1-null and wild-type MEFs using microarrays (Supplementary Table S3). An enrichment analysis of Gene Ontology (GO) terms revealed profound functional differences between both expression profiles. Transcriptional changes in the SA1-null cells led to upregulation of GO-defined processes related to phagocytosis, endocytosis and apoptosis. In contrast, processes associated with abnormalities observed in CdLS patients, such as limb and skeletal system morphogenesis, heart and lung development and lipid metabolism, were downregulated (Figure 4A; Supplementary Table S4). Moreover, we found a significant overlap (GO Comparative Analysis, FDR<0.05) between the transcriptional changes linked to the loss of cohesin-SA1 and those reported for Nipbl-heterozygous MEFs (Kawauchi et al, 2009) (Figure 4B; Supplementary Table S5). A stringent statistical analysis (FDR<0.15) identified 55 differentially expressed genes (DEGs), many of them significantly upregulated in the SA1-null MEFs (Supplementary Table S6). Validation of microarray data was performed by qPCR for a subset of 15 genes (Supplementary Figure S4). To determine if this transcriptional regulation is SA1 specific, we knocked down either SA1 or SA2 in MEFs by siRNA. Transient downregulation of SA1 significantly reduced the expression of 11 out of 14 genes selected from those found to be regulated by SA1 in the microarray analysis (Figure 4C). In contrast, reduction of SA2 expression only affected two of the tested genes, supporting the specificity of cohesin-SA1 in this regulation. Since SA1 is present in the vicinity of the TSS in only six of the DEGs (Supplementary Table S6), it is likely that cohesin-SA1 affects gene expression by additional mechanisms that do not require its presence at promoters. According to recent literature, cohesin promotes the proper topological organization of gene expression domains or gene clusters. Consistent with this role for cohesin-SA1, we observed that 18 out of 55 DEGs are located in close proximity of one another (<800 kb; Figure 4D). Finally, cohesin may indirectly affect the expression of a gene by regulating a factor required for its transcription (Horsfield et al, 2007). As described in the next sections, we have found evidence of the importance of cohesin-SA1 in these different modes of regulation of gene expression by cohesin.
Figure 4.
Gene expression changes in the absence of SA1. (A) GO analysis (FDR<0.05) reveals biological processes upregulated (in red) and downregulated (in blue) in SA1-null MEFs compared with wild-type. FDRs for each of the enriched GO terms are indicated. (B) GO Comparative Analysis (FDR<0.05) between transcriptomes from SA1-null and Nipbl-heterozygous MEFs. Common GO terms were grouped in five big biological processes: skeletal and bone development, morphogenesis, translation, heart and lung development and lipid metabolism. The number of GO terms belonging to each group is shown (# GOs). (C) Expression of a set of SA1-regulated genes that includes 10 DEGs (shown in bold) and additional genes (Sox11 and skin-related genes, marked with an asterisk) was measured in wild-type MEFs after transfection with SA1 or SA2 siRNAs (results come from triplicate qPCR reactions from two independent experiments). The upper inset shows the efficiency of siRNAs. (D) Chromosomal location of the 55 DEGs (FDR<0.15). Notice that 18 of them are located in close proximity (shadowed).
Cohesin-SA1 positively regulates c-myc expression
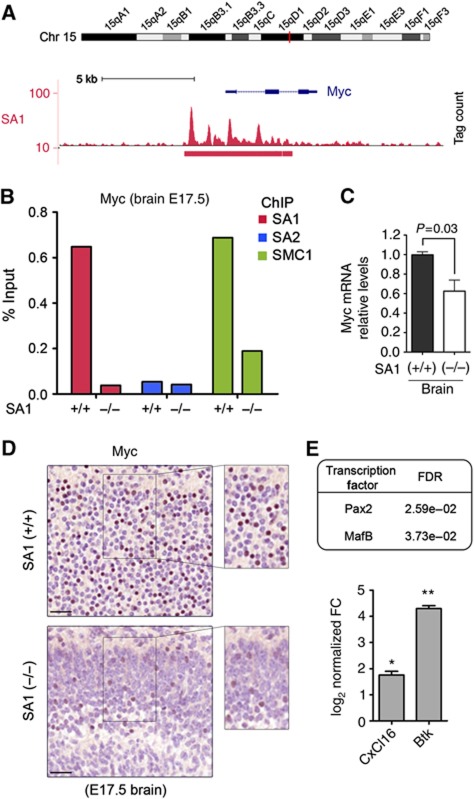
Myc is a major modulator of cell proliferation, growth and differentiation whose transcription is regulated by cohesin in different organisms (Stedman et al, 2008; Rhodes et al, 2010). Mutant mice expressing reduced levels of Myc have decreased body mass owing to multiorgan hypoplasia (Trumpp et al, 2001). Our ChIP-seq data show that the c-myc gene constitutes one of the most prominent SA1-binding regions in the mouse genome (Figure 5A). Quantitative qPCR analyses in MEFs validate this result and show that SMC1 levels in this region are severely reduced in the SA1-null cells (Supplementary Figure S2D). Expression of myc target genes is significantly altered in these cells (FDR=0.06; Supplementary Figure S5), supporting the relevance of cohesin-SA1 in positively regulating c-myc. More importantly, ChIP-qPCR performed in brain tissue obtained from wild-type and SA1-null E17.5 embryos indicates that also in vivo SA2 does not efficiently replace SA1 (Figure 5B). Myc mRNA and protein levels are reduced in brains from SA1-null embryos, as shown by qPCR and immunostaining (Figure 5C and D, respectively). It is likely that decreased cell proliferation rates due to transcriptional downregulation of c-myc contributes to the lethality of SA1-null embryos. We asked whether other TFs might be regulated by cohesin-SA1. Indeed, gene set analysis revealed that among the transcriptionally altered genes, there is a statistically significant enrichment in genes with binding sites for Pax2 and MafB (Figure 5E, top), two TFs involved in differentiation and development (Cordes and Barsh, 1994; Mansouri et al, 1996) whose encoding genes contain cohesin-SA1 at their promoters. Consistently, two of the DEGs are MafB downstream targets (Figure 5E, bottom) and MafB itself is upregulated in SA1-null MEFs (Supplementary Table S3). Therefore, part of the expression changes associated to cohesin-SA1 loss can be secondary to the regulation of genes encoding TFs.
Figure 5.
Cohesin-SA1 regulates myc expression. (A) SA1-binding region at myc gene (5350 bp) is the widest in the mouse genome (the median is 531 bp). (B) Validation by ChIP-qPCR of SA1, SA2 and SMC1 binding at myc promoter in wild-type (n=12) and SA1-null (n=9) E17.5 brains. (C) Myc mRNA levels are significantly reduced in SA1-null brains from E17.5 embryos. Three embryonic brains per genotype were used. (D) Immunohistochemistry on E17.5 brains showing reduced Myc protein levels in the cortex of SA1-null embryos. Notice the different cortical structure between wild-type and SA1-null brains. Scale bars, 40 μm. (E) Table showing two TFs whose target genes are dysregulated in SA1-null cells. Expression levels of two of those target genes (CxCl16 and Btk) were estimated from three independent qPCR reactions of two clones per genotype. Values are represented as log2 of fold change (FC) versus wild-type. **P<0.01, *P<0.05.
Cohesin-SA1 regulates the expression of gene clusters involved in skin functions
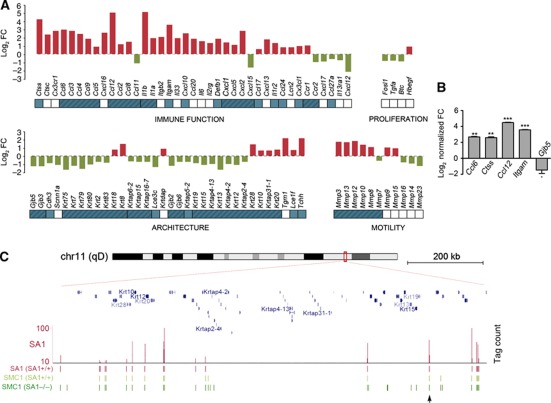
Among DEGs not having cohesin at TSS there are three genes involved in skin development and function (Ccl6, Cxcl16 and Mmp3). Since SA1-null embryos show defects in skin formation (Figure 1B, panel I), we decided to take a closer look at expression changes caused by the absence of SA1 in genes with skin-related functions including immunity, proliferation, skin architecture and motility. Gene Set Enrichment Analysis (GSEA) revealed a highly significant enrichment in those genes in SA1-null MEFs (FDR<10−3; Supplementary Figure S6A). Only 15 out of 88 (17%) skin-related genes whose transcription changes have SA1-binding sites up to 5 kb from their TSS (Supplementary Table S7). Strikingly, 63 of the genes (72%) are located in clusters (labelled in blue in Figure 6A), such as the Ccl and Cxcl clusters involved in skin immune function or the keratin clusters involved in skin structure. Analysis of mRNA levels further confirmed the SA1-specific transcriptional changes for a subset of genes implicated in the immune response (Figure 6B; see also genes labelled with an asterisk in Figure 4C). As shown by ChIP-seq for the keratin cluster in chromosome 11 and for the Cxcl cluster in chromosome 5, cohesin distribution within the cluster is considerably altered in SA1-null cells, as judged by the appearance of multiple ‘new’ sites with low cohesin occupancy (Figure 6C; Supplementary Figure S6B). At the same time, there is much less cohesin at SA1-binding sites, as exemplified by the reduced binding of SMC1 and SMC3 to a site located in the vicinity of Krt15/19 genes in SA1-null cells (indicated with an arrowhead in Figure 6C; qPCR data in Supplementary Figure S2C, upper panels). We therefore hypothesize that cohesin-SA1 plays an architectural role in the organization of these gene clusters that is essential for regulation of their gene expression.
Figure 6.
SA1 regulates clustered genes involved in skin development. (A) Transcriptional changes detected in genes involved in skin development and function in SA1-null cells. Bars represent the log2 FC in SA1-null compared with wild-type cells obtained from microarray analysis. Genes in clusters are depicted in blue and genes belonging to the same cluster are grouped (blue shadow). (B) Validation by RT–qPCR of transcriptional changes in some of the genes shown in (A) (from three independent qPCR reactions of two clones per genotype). ***P<0.001, **P<0.01, *P<0.05. (C) Detail of SA1 (SA1+/+), SMC1 (SA1+/+) and SMC1 (SA1−/−) binding sites at the Keratin cluster located on chromosome 11. Arrowhead points to a cohesin-SA1-binding site that is validated by ChIP-qPCR in Supplementary Figure S2C.
Cohesin-SA1 regulates the expression of Protocadherins in the brain
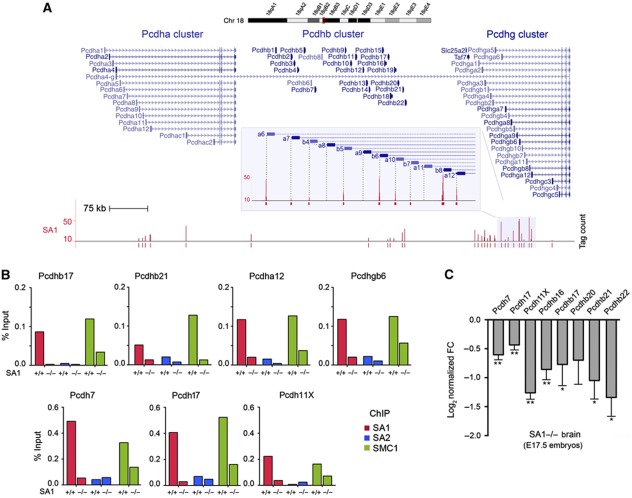
A detailed analysis of the transcription data revealed the downregulation of many members of the protocadherin (Pcdh) gene family in SA1-null cells. Most, but not all, Pcdh genes are present in three consecutive clusters (Pcdh a, b and g) at mouse chromosome 18. The expression of the Pcdh genes in these clusters is regulated by multiple promoters and alternative cis splicing (Yagi, 2008). Our ChIP-seq data identified SA1-binding sites located precisely at most of the multiple TSS of the clustered Pcdh genes (Figure 7A) and also at non-clustered Pcdh genes (e.g., Pcdh7; validation shown in Supplementary Figure S2E). In vivo, ChIP-qPCR performed in the brains from wild-type and SA1-null E17.5 embryos showed on the one hand, the prevalence of SA1 over SA2 at the TSS of several of these genes, and on the other, that SA2 does not efficiently replace SA1 in the SA1-null brains, since SMC1 occupancy in these regions is much reduced (Figure 7B). Given that protocadherins are essential for central nervous system development (Morishita and Yagi, 2007), the contribution of cohesin-SA1 to their transcriptional regulation may be particularly critical in the brain. Indeed, we observed a highly significant downregulation of 7 out of 8 Pcdh genes in the brains of E17.5 SA1-null embryos compared with wild-type (Figure 7C). We had chosen these genes because they had also been found downregulated in the brains of Nipbl-heterozygous mice, leading the authors to propose that reduced levels of protocadherins in the brain might contribute to the mental retardation observed in CdLS patients (Kawauchi et al, 2009). Importantly, we here show that regulation of Pcdh gene expression in the brain relies on the presence of cohesin-SA1 at their TSS.
Figure 7.
SA1 regulates the expression of protocadherins in mouse brain. (A) Detail of SA1-binding sites at Pcdh clusters located in chromosome 18. Notice the position of SA1 at multiple TSS. (B) SA1, SA2 and SMC1 binding at the TSS of four clustered and three non-clustered Pcdh genes was validated in vivo in wild-type (n=12) and SA1-null (n=9) E17.5 brains. (C) Significant downregulation of Pcdh genes in the brains from E17.5 SA1-null embryos (three embryos per genotype and three independent qPCR reactions per condition). Values are represented as log2 of FC versus wild-type. **P<0.01, *P<0.05, Pcdhb20 P-value=0.13.
Discussion
Vertebrates have two distinct cohesin complexes that contain either SA1 or SA2 (Losada et al, 2000; Sumara et al, 2000), but most studies assume that both function in a similar way and/or that cohesin-SA2 is more relevant because it is more abundant in somatic cells (Xiao et al, 2011). However, ablation of SA1 results in embryonic lethality in mice and late SA1-null mouse embryos present a clear developmental delay (Remeseiro et al, 2012; this study). Thus, cohesin-SA1 and cohesin-SA2 are not functionally equivalent. On one hand, we have shown that cohesin-SA1 is responsible for telomere cohesion whereas cohesin-SA2 is critical for centromere cohesion. On the other hand, we report here that cohesin-SA1 has an important role in gene regulation that cannot be assumed by cohesin-SA2.
Differences between SA1 and SA2
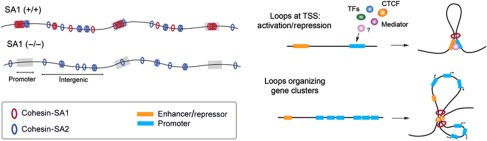
We have analysed the genome-wide distributions of SA1 and SA2 in MEFs and found that cohesin-SA1 has a greater propensity for localizing at gene promoters and gene bodies, while cohesin-SA2 prefers intergenic regions. Moreover, in the absence of SA1, cohesin-SA2 (the only cohesin present) redistributes to intergenic regions and fails to accumulate at promoters and other sites nearby genes that could be important for transcriptional regulation, such as sites also bound by CTCF (see model in Figure 8, left). The molecular mechanisms by which cohesin-SA1 regulates gene expression are likely related to the ability of the complex to bring together two DNA sequences not only in trans (the sister chromatids), but also in cis, thereby facilitating DNA looping (Hadjur et al, 2009; Mishiro et al, 2009; Nativio et al, 2009; Hou et al, 2010; Chien et al, 2011). These loops may dictate gene expression by promoting or preventing communication between enhancers and promoters in different configurations. One would require the presence of cohesin at the promoter (model in Figure 8, upper right) whereas in other cases, cohesin would be located away from the promoter and would mediate the organization of a gene cluster (Figure 8, lower right). The transcriptional changes observed in SA1-null cells and tissues indicate that cohesin-SA2 cannot fulfil the function of cohesin-SA1. We envision that SA1 and SA2 must confer distinct properties to cohesin. The mechanism by which cohesin associates with chromatin, namely by topological embrace, allows cohesin sliding along DNA (Lengronne et al, 2004; Ocampo-Hafalla and Uhlmann, 2011). Cohesin-SA1 could be less prone to sliding than cohesin-SA2 or, in other words, more prone to occupy a fixed genomic position. Cohesion, probably the most important function of cohesin-SA2, does not require localization of cohesin at precise sites, whereas regulation of transcription does. The molecular mechanisms underlying the potential differences in mobility between cohesin-SA1 and cohesin-SA2 remain to be elucidated. One possibility is that the interaction of SA1 and SA2 with Wapl and Pds5, two cohesion factors that modulate the association of cohesin with chromatin throughout the cell cycle, has distinct characteristics (Losada et al, 2005; Gandhi et al, 2006; Gause et al, 2010; Nishiyama et al, 2010). Another is the preferential interaction of SA1 with proteins present at defined sites such as CTCF, Mediator or Polycomb (Misulovin et al, 2008; Parelho et al, 2008; Rubio et al, 2008; Wendt et al, 2008; Nativio et al, 2009; Kagey et al, 2010; Strubbe et al, 2011; Xiao et al, 2011). The identification of proteins that specifically interact with either SA1 or SA2 is one goal of our future work.
Figure 8.
A model for the differential distribution of cohesin-SA1 and cohesin-SA2 and its implications in transcription. Left: Different dynamics and localization of cohesin-SA1 and cohesin-SA2. Cohesin-SA1 is enriched at gene promoters to a much larger extent than cohesin-SA2. In SA1-null cells, cohesin-SA2 fails to accumulate at gene promoters and relocates to intergenic positions. Right: Proposed mechanisms for cohesin-SA1 in regulation of gene expression. Cohesin-SA1 present at gene promoters (upper panel) or in the proximity of genes organized in clusters (lower panel) is required for the formation of loops that arrange the chromatin for gene transcription. The insulator protein CTCF as well as different TFs are likely involved in cohesin-SA1 recruitment to specific genomic positions.
Cohesin-SA1 and CdLS
The role of cohesin in transcriptional regulation has become particularly relevant for human health after identification of mutations in the cohesin loader Nipbl and in cohesin subunits as a major cause of CdLS and the subsequent finding that cells from CdLS patients do not show overt cohesion defects (Liu and Krantz, 2009; Dorsett, 2011). A mouse heterozygous for a gene-trap mutation in Nipbl recapitulates some of the pathologies observed in CdLS patients including small size, craniofacial anomalies, heart defects, impaired hearing, delayed bone maturation, reduced body fat and behavioural disturbances (Kawauchi et al, 2009). Mouse lacking either Pds5A or Pds5B also exhibit CdLS-like features (Zhang et al, 2007, 2009). SA1-heterozygous mice do not show apparent CdLS phenotypes, but SA1-null embryos that survive to late stages of embryogenesis (E17.5–18.5) have clear developmental delay, impaired lipid accumulation and delayed ossification, features that resemble CdLS. Limb or heart defects have not been clearly observed in the limited number of embryos examined. In mice, reduction of Nipbl mRNA levels to 70% is sufficient to elicit CdLS phenotypes without causing apparent cohesion defects (Kawauchi et al, 2009). In Drosophila, reduction of Nipped-B expression to 30% of wild-type levels reduces the stable binding of cohesin SA subunit by the same percentage (Gause et al, 2010). How the decrease in Nipbl levels affects loading of cohesin-SA1 and cohesin-SA2 on chromatin in mammalian cells remains to be addressed. One scenario is that it affects more acutely the loading of cohesin-SA1 either because it is less abundant (Losada et al, 2000; Holzmann et al, 2010; Remeseiro et al, 2012) or because being more frequently found within genes than cohesin-SA2, cohesin-SA1 may be released from chromatin to allow passage of the transcriptional machinery and thus needs to be constantly reloaded by Nipbl. Alternatively, reduction of Nipbl levels may decrease the amount of both cohesins on chromatin but while this barely affects cohesion, it affects gene transcription (Schaaf et al, 2009; Heidinger-Pauli et al, 2010). Although we cannot rule out the possibility that cohesin-SA2 affects also gene expression, comparative microarray analysis of human glioblastoma cells expressing or not SA2 shows no evidence for such a role (Solomon et al, 2011), consistent with our results in MEFs after downregulation of SA2 by siRNA.
In summary, we have found that cohesin-SA1 plays a unique role in transcriptional regulation that is essential for embryonic development and could underlie the aetiology of CdLS. Although the analysis of SA1-null embryonic fibroblasts combining transcriptional profiling and genome-wide distribution data by ChIP-seq is an important first step, we are aware of the necessity of employing a similar strategy in specific tissues or developmental/differentiation stages in order to understand the contribution of cohesin-SA1 to the establishment of their transcriptomes.
Materials and methods
Identification of SA1, SA2, SMC1 and SMC3 binding sites by ChIP-sequencing
ChIP was performed in SA1-null and wild-type MEFs with custom-made rabbit polyclonal antibodies against SA1, SA2 (described in Remeseiro et al, 2012), SMC1 and SMC3 (raised against the C-terminal peptides CEMAKDFVEDDTTHG and CDLTKYPDANPNPNEQ, respectively), as described (Cuadrado et al, 2010) with some modifications. Cells were cross-linked by addition of 1% formaldehyde for 10 min at room temperature, harvested and frozen in liquid nitrogen. Cells were then resuspended in lysis buffer (2 × 107 cells/ml) and sonicated on Covaris system (shearing time 30 min, 20% duty cycle, intensity 10, 200 cycles per burst and 30 s per cycle) in a volume of 2 ml. From 4 to 15 ng of immunoprecipitated chromatin (as quantitated by fluorometry) were electrophoresed on agarose gel and independent sample-specific fractions of 100–200 bp were taken. These samples were processed into sequencing libraries and analysed according to Illumina's ‘ChIP-Sequencing Sample Prep Guide’ (part #11257047 Rev. A), with the exception that gel extraction was replaced with Agencourt AMPure XP (Beckman Coulter) bead purification. Adapter-ligated library was completed by limited-cycle PCR with Illumina PE primers (14 cycles). DNA libraries were applied to an Illumina flow cell for cluster generation and sequenced on the Illumina Genome Analyzer IIx (GAIIx). Image analysis was performed with Illumina Real Time Analysis software (RTA1.8). Sequence alignment to the reference genome (NCBIm37/mm9, April 2007) was made with Illumina's ELANDv2 algorithm on its ‘eland_extended’ mode from within CASAVA-1.7 package, using default settings. Only reads with a unique alignment in the reference genome were used for the peak detection, which was performed using MACS v1.4 setting a FDR<0.1 and a P-value cutoff of 10−5 (Zhang et al, 2008). All comparisons were done using the input tracks as control, and each one of the data sets as treatment, using the following naming convention: SA1 (SA1+/+), SA2 (SA1+/+), SMC1 (SA1+/+), SMC3 (SA1+/+), SA1 (SA1−/−), SA2 (SA1−/−), SMC1 (SA1−/−) and SMC3 (SA1−/−). In the case of SA1 (SA1+/+) data set, the 176 peaks obtained from SA1 ChIP in SA1−/− cells were cleared. CTCF peaks were obtained from the UCSC Genome Browser track ‘MEL CTCF D Pk’, part of the ENCODE/Stanford/Yale data set of TF-binding sites (TFBS) in mouse. Genomic interval overlaps and signal distributions were obtained using BEDTools v2.12 (Quinlan and Hall, 2010), PeakAnalyzer v1.3 (Salmon-Divon et al, 2010) and custom UNIX shell scripting. All the statistical tests and correlations were calculated using R functions (http://cran.r-project.org).
ChIP-qPCR in embryonic brain
The brains from E17.5 embryos were extracted and minced in cold PBS with protease inhibitors cocktail (Roche #11873580001). Small tissue pieces (<1 mm) were then cross-linked for 20 min RT in fixing solution (1% formaldehyde, 50 mM HEPES-KOH, 100 mM NaCl, 1 mM EDTA and 0.5 mM EGTA). Cross-linking was stopped by adding 1/20 volume of 2.5 M Glycine for 5 min at RT. After two washes in PBS, tissues were resuspended in lysis buffer and processed for further ChIP analysis as described in the above section.
Validation of ChIP-seq results
ChIP-seq results were validated by ChIP-qPCR on immunoprecipitated chromatin from two clones per genotype. Chromosome coordinates of the validated peaks and the corresponding primers are listed in Supplementary Table S8. The relative amount of each amplified fragment was normalized with respect to the amplification obtained from input DNA and represented as percentage of 1 μg of input DNA.
Gene expression analysis
Total RNA from three wild-type and three SA1-null MEF clones was analysed by two-colour hybridization on Whole Mouse Genome DNA microarrays (G4122F; Agilent), and images were quantified with Agilent Feature Extraction Software (v. 10.1.1). DEGs between SA1-null and wild-type MEFs were obtained by limma (Smyth GK; Bioconductor project; http://www.bioconductor.org). FDR adjustment was employed to account for multiple testing. Raw data from gene expression microarray experiments in Nipbl+/− MEFs were kindly provided by the authors (Kawauchi et al, 2009). In this case, Affymetrix Murine 430A 2.0 were normalized using Robust Multiarray Average (RMA) algorithm available in Bioconductor's affy package and limma package was used to obtain DEGs.
Functional analysis for GO terms
GO analyses at Biological Process, Cellular Component and Molecular Function were performed using FatiScan tool available at Babelomics suite (http://www.babelomics.org). To this end, genes were ranked based on limma's moderated t statistic and GO enrichment was evaluated by segmentation test. GO terms showing FDR <0.05 were considered statistically significant.
Gene set analysis of Myc targets and skin-related genes
GSEA (Subramanian et al, 2005) was employed to evaluate the enrichment of custom gene sets in our microarrays experiments. Myc targets were obtained from literature (Chen et al, 2008; Kim et al, 2008, 2010; Sridharan et al, 2009; Smith et al, 2010) whereas skin gene set was built from Nagarajan et al (2010) and references therein. GSEA was run using gene expression values ranked by limma moderated t statistic. After Kolmogorov–Smirnoff testing, those gene sets showing FDR <0.1, were considered enriched between SA1-null and wild-type MEFs.
Enrichment analysis for target genes of TFs
GSEA for Jaspar TFBS was done using Fatiscan tool available at Babelomics platform (http://www.babelomics.org). Genes were ranked by limma moderated t statistic. TFBS showing FDR <0.05 were considered enriched between SA1-null and wild-type MEFs.
mRNA isolation and quantitative real-time PCR (qRT–PCR) analysis
Total RNA was isolated from MEFs using RNeasy Kit (Qiagen) and cDNA was synthesized with SuperScript™ II reverse transcriptase (Invitrogen) using random hexamer primers. An Applied Biosystems 7900HT Fast qRT–PCR was used to determine mRNA levels. GAPDH was used for normalization. Primers used for mRNA amplification are described in Supplementary Table S9.
RNA interference, immunoprecipitation and immunoblotting
Interference of SA1 and SA2 was performed with siGENOME SMARTpool siRNAs from Dharmacon (M-041989 and M-057033, respectively) at a final concentration of 100 nM and using DharmaFECT transfection reagent 1 in the case of C2C12 cells and the Neon transfection system (Invitrogen) in the case of MEFs. ChIP with SA1- and SA2-specific antibodies was performed 72–96 h after transfection. Immunoprecipitation was carried out with Nuclear Complex Co-IP Kit (Active Motif, 54001) from cell extracts according to the manufacturer's instructions, with SA1, SA2, SMC1 and SMC3 specific antibodies. Whole-cell extracts were prepared by lysing and sonicating a cell pellet in SDS–PAGE loading buffer and equal amounts of protein were run in 7.5% Bis/Tris gels followed by western blotting.
Histology and immunohistochemistry
E17.5 embryos were fixed in 10% buffered formalin (Sigma) and embedded in paraffin using standard procedures. In all, 3 μm sections were stained with haematoxylin and eosin (HE) and subjected to histopathological analysis. Anti-myc (Santa Cruz, sc-764), anti-SA1 and anti-SA2 were used for immunohistochemical analysis of 3 μm sections. Positive cells were visualized using 3,3-diaminobenzidine tetrahydrochloride plus (DAB+) as a chromogen, and counterstaining was performed with haematoxylin.
Statistical analysis
GraphPad Prism 5 software was used to calculate two-tailed χ2 test (with Yates’ correction) in Figure 3B, Mann–Whitney U-test in Figure 3E and two-tailed Student's t-test in Figures 4C, 5C, 5E, 6B and 7C, Supplementary Figures S1C and S4. Limma package was used for GO, GSEA and gene expression analysis to obtain DEGs.
Data access
Microarray and ChIP-sequencing data from this study have been submitted to GEO database and have been approved with the following Series reference: GSE32320 (GSE32234 and GSE32319 SubSeries correspond to microarray and ChIP-seq data, respectively).
Supplementary Material
Acknowledgments
We are very grateful to M Cañamero (Pathology Unit, CNIO) for histopathological analyses and to O Domínguez (Genomics Unit, CNIO) for help and advise on ChIP-seq. We also thank M Serrano and O Fernández-Capetillo for critically reading the manuscript. Research in our laboratory is supported by the Spanish Ministry of Science and Innovation (SAF-2010-21517 and CSD2007-00015 to AL; Ramón y Cajal grant for AC). SR is the recipient of a ‘La Caixa’ predoctoral fellowship.
Author contributions: AL designed and supervised the study. SR and AC designed, performed, analysed and interpreted all the experiments. DGP and GGL performed and interpreted the ChIP-seq and microarray data analysis, respectively. AL, SR and AC wrote the manuscript with ideas and comments from GGL and DGP.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bose T, Gerton JL (2010) Cohesinopathies, gene expression, and chromatin organization. J Cell Biol 189: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S (2009) Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castronovo P, Gervasini C, Cereda A, Masciadri M, Milani D, Russo S, Selicorni A, Larizza L (2009) Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res 17: 763–771 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR Jr, Calof AL, Lander AD, Groudine MT, Yokomori K (2011) Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. J Biol Chem 286: 17870–17878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS (1994) The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79: 1025–1034 [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Munoz-Canoves P, Nebreda AR (2010) Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J 29: 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID (2007) Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ (2009) Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 182: 44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K (2005) Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D (2010) Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, Sinclair DA, Honda BM (2008) The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci USA 105: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D (2010) Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol 20: 957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Fuchs J, Pichler P, Peters JM, Mechtler K (2010) Lesson from the stoichiometry determination of the cohesin complex: a short protease mediated elution increases the recovery from cross-linked antibody-conjugated beads. J Proteome Res 10: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS (2007) Cohesin-dependent regulation of Runx genes. Development 134: 2639–2649 [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A (2010) Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA 107: 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, Yokomori K, Hallgrimsson B, Lander AD (2009) Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet 5: e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH (2010) A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M et al. (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36: 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Jin H, Liu X, Hampton K, Yu HG (2011) Scc2 regulates gene expression by recruiting cohesin to the chromosome as a transcriptional activator during yeast meiosis. Mol Biol Cell 22: 1985–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID (2009) Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 76: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID (2009) Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol 7: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Hallonet M, Gruss P (1996) Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol 8: 851–857 [DOI] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M (2009) Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, Macarthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D (2008) Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Yagi T (2007) Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol 19: 584–592 [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L (2006) X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 38: 528–530 [DOI] [PubMed] [Google Scholar]
- Muto A, Calof AL, Lander AD, Schilling TF (2011) Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol 9: e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, Chin SS, Wang D, Liu S, Sinha S, Garrett-Sinha LA (2010) Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J Cell Sci 123: 3566–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A (2009) Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Ocampo-Hafalla MT, Uhlmann F (2011) Cohesin loading and sliding. J Cell Sci 124: 685–691 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K (2008) Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 14: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, van Steensel B, Nasmyth K (2010) A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Curr Biol 20: 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A (2012) Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA (2010) Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol 344: 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D (2004) Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24: 3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P (2010) PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D (2009) Regulation of the Drosophila enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One 4: e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT (2010) A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M (2011) A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S (2010) Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R (2011) Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci USA 108: 5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Smith M, Eichhorn P, Jones S, Imamwerdi B, Lindsay S, Jackson M, Wang TJ, Ireland M, Burn J, Krantz ID, Carr P, Strachan T (2004) A giant novel gene undergoing extensive alternative splicing is severed by a Cornelia de Lange-associated translocation breakpoint at 3q26.3. Hum Genet 115: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM (2001) c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414: 768–773 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G (2011) Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 31: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T (2008) Clustered protocadherin family. Dev Growth Differ 50(Suppl 1): S131–S140 [DOI] [PubMed] [Google Scholar]
- Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Kulkarni S, Deardorff MA, Uzielli ML, Dorsett D, Beebe DC, Jay PY, Heuckeroth RO, Krantz I, Milbrandt J (2009) Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One 4: e5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J (2007) Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134: 3191–3201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.