Abstract
The skin epidermis contains different appendages such as the hair follicle and the sebaceous glands. Recent studies demonstrated that several types of stem cells (SCs) exist in different niches within the epidermis and maintain discrete epidermal compartments, but the exact contribution of each SC populations under physiological conditions is still unclear. In addition, the precise mechanisms controlling the balance between proliferation and differentiation of epidermal SC still remain elusive. Recent studies provide new insights into these important questions by showing the contribution of hair follicle SC to the sebaceous lineage and the importance of chromatin modifications and micro-RNAs (miRs) in regulating epidermal SCs renewal and differentiation. In this review, we will discuss the importance of these papers to our understanding of the mechanisms that control epidermal SC functions.
Keywords: epigenetics, keratinocytes, lineage tracing, micro-RNA, stem cells
Introduction
The skin epidermis is a stratified epithelium that constitutes a barrier protecting the animals against microorganisms, external stress and water loss. To ensure these vital functions, the skin epidermis must replace the cells that are naturally lost and must be able to repair the tissue following injuries. The skin epidermis contains different appendages such as the hair follicle, the sweat gland and the sebaceous gland (SG), which ensure protection, camouflage and thermoregulation. Different stem cells (SCs) present in the skin epidermis regulated the maintenance and repair of the different part of the skin epidermis during physiological conditions (Blanpain et al, 2007).
During embryonic development, epidermis originates from a single layer of ectodermal cells that will undergo a stratification process allowing the formation of the different layers of differentiated cells required to fulfil its barrier function (basal, spinous and granular layers). The basal layer cells express high level of basal integrins, keratin 5 (K5) and K14, while the spinous layers express K1 and K10 and the granular layers accomplish the terminal differentiation process and expressed filaggrin and loricrin. P63, a gene of the p53 family, is expressed in basal cells of all stratified epithelia and is thought to represent a master regulator of the stratification process (Blanpain and Fuchs, 2007). In the absence of p63, epidermis indeed fails to form a stratified epithelium (Mills et al, 1999; Yang et al, 1999). Different theories have been proposed to account for the critical p63 function during epidermal morphogenesis. p63 has been proposed to be required for the initiation of the stratification programme (Mills et al, 1999), for epidermal SC renewal (Yang et al, 1999; Senoo et al, 2007) or both processes (Truong et al, 2006).
HF morphogenesis begins during embryonic development through a series of epithelial-to-mesenchymal interactions and it is complete by postnatal day 8 in mice. After reaching a critical size, the lower part of the HF degenerates in a process called catagen. The permanent portion of the HF called the bulge region enters a resting stage (telogen) before entering in a new growing stage (anagen) (Paus et al, 1999). Throughout the life of the animals, HF will alternate cycle of growth and degeneration called the hair cycle. The cyclic HF regeneration is mediated by the presence of multipotent SC located in the bulge region. The first defined characteristic of bulge SC that has been identified is their relative quiescence, which can be visualized by their ability to retain nucleotide analogues during pulse chase experiments and called for this reason labelled retaining cells or LRCs (Cotsarelis et al, 1990; Morris and Potten, 1999). Bulge SC have been purified based on their slow cycling property (Tumbar et al, 2004) or the expression of several markers including CD34/CD49f (α6 integrin) (Trempus et al, 2003; Blanpain et al, 2004), keratin 15 (Morris et al, 2004), Lgr5 (Jaks et al, 2008). While bulge SC are able to differentiate into all epidermal lineage upon transplantation into immunodeficient mice, adult bulge SC lineage tracing experiments demonstrated that bulge SCs participate to the homeostasis of all HF cells below the SGs but not contribute to SG and IFE under physiological conditions (Morris et al, 2004; Levy et al, 2005). However, during pathological conditions such as during wounding, bulge SC are rapidly mobilized and participate into the repair of the IFE (Ito et al, 2005; Jaks et al, 2008; Nowak et al, 2008).
The continuous replacement of terminally differentiated cells that are shed from the skin surface is mediated by the presence of multiple small epidermal proliferation units (EPUs) scattered all along the IFE that contain one SCs or progenitor cells (Ghazizadeh and Taichman, 2005). Upon division, these SC can undergo three fates: most of the divisions are asymmetric, giving rise to one SC and one committed cell, the remaining divisions are symmetric (renewal or differentiation). Based on clonal analysis and mathematical modelling, it has been recently proposed that the homeostasis of the IFE can be maintained by the presence of only one type of progenitors that stochastically choose between these three different fates with a fixed probability (Clayton et al, 2007; Doupe et al, 2010).
Recent studies demonstrated that even more progenitors and SC exist in the epidermis and contribute to the maintenance of the other epidermal compartments such as the isthmus, the junctional zone, the infundibulum (Nijhof et al, 2006; Jensen et al, 2008; Jensen et al, 2009; Raymond et al, 2010; Snippert et al, 2010). But it is not clear what is the precise contribution of each SC populations under physiological conditions and how the compartmentalization of the epidermis is orchestrated. In addition, while different signalling pathways regulating epidermal SC functions have been identified, the precise mechanisms controlling the balance between proliferation and differentiation of epidermal SC still remain elusive. Especially, the impact of chromatin modifications and micro-RNAs (miRs) expression on epidermal SCs renewal and differentiation is also still poorly understood.
Recently, a flurry of papers published in the EMBO Journal provides new insights into these outstanding questions (Chikh et al, 2011; Driskell et al, 2011; Mejetta et al, 2011; Petersson et al, 2011). In this review, we discuss the importance of these papers to the understanding of the heterogeneity of skin SCs, and the role of miR and histone modifications on epidermal SC functions.
Contribution of bulge SC to the sebaceous gland lineage
SG is a part of the pilosebaceous unit that through holocrine differentiation of the sebocytes secretes the sebum, which maintain the waterproof nature of the fur. The important cellular turnover within the SG is thought to be fuelled by the presence of SCs. Until recently, very little was known about the cellular and molecular mechanisms that control SG morphogenesis and homeostasis. Lineage tracing performed during epidermal development demonstrate that SG originates from bipotent embryonic progenitors common for both HF and SG lineages that expressed Shh and Sox9 at one time of their specification (Levy et al, 2005; Nowak et al, 2008). Lineage tracing of adult bulge SC was performed using different inducible CRE expressed under the regulatory region of K15, Lgr5 and K19, which are expressed specifically in bulge SC during telogen (Morris et al, 2004; Ito et al, 2005; Levy et al, 2005; Youssef et al, 2010). K19CREER and Lgr5CREER are knockin mice, in which the CREER is inserted into the endogenous locus of these genes (Barker et al, 2007; Means et al, 2008), whereas K15CREPR are transgenic mice made by random integration of fragment of the K15 promoter (Morris et al, 2004). In mice expressing one of these inducible CRE together with a reporter gene (e.g., Rosa/YFP or Rosa/LacZ), tamoxifen (TAM) or RU486 administration induces the activity of the inducible CRE, allowing the excision of a stop cassette leading to the reporter gene expression in bulge SC and all their future progeny. These experiments showed that only rarely were SG cells labelled using these different bulge specific inducible CRE, suggesting that the SG is mainly maintained independently of bulge SC during physiological condition. The transcriptional repressor Blimp1 is expressed during morphogenesis in the upper HF adjacent to the emergence of the newly formed SGs. Blimp1 expressing cells encompass progenitors committed to SG lineage as demonstrated by lineage tracing using Blimp1CRE mice. The persistence of Blimp1 expressing cells in adult HF suggested that the SG could be maintained by unipotent progenitors during adult homeostasis. In the absence of Blimp1, increased bulge SC proliferation is observed, possibly through a derepression of c-Myc mediated by Blimp1 (Horsley et al, 2006). Two independent groups have shown that cells located in the upper follicle called the upper isthmus (UI), expressing high level of MTS24 or low level of Sca1, are highly clonogenic in vitro and multipotent in vivo, suggesting that this region of the HF might contain a subset of SCs that sustain SG homeostasis (Nijhof et al, 2006; Jensen et al, 2008). Another population of cells located at the junction zone (JZ) between UI and IFE, expressing Lrig1 and enriched for Blimp1 transcript, contain multipotent SC as demonstrated by transplantation assays (Jensen et al, 2009). Finally, a recent study using Lgr6CREER, which is expressed in the UI, demonstrated the presence of unipotent SG SCs responsible for the long-term labelling of the SG and the surrounding UI HF (Snippert et al, 2010). It is still unclear whether SGs are only maintained by Lgr6+ SCs, a substet of which expressed MTS24 and possibly Blimp1, or that several population of SCs, including HF SCs, are mobilized and contribute to the SG turnover during adult life.
In a recent study, Petersson et al (2011) used new bulge SC lineage tracing to show that a subpopulation of bulge SCs may contribute to the homeostasis of SG. Using new K15CreERT2 transgenic mice crossed with the Rosa26–YFP or Rosa–LacZ reporter mice, the authors performed lineage tracing of bulge SCs. TAM administration labels cells preferentially in the bulge region (90% of LacZ+ cells) while the remaining labelled cells are already localized in the SG and its duct. Clonal analysis showed that a subset of YFP+ cells apparently coming from the bulge migrates to the UI and the SG 5 days later, as suggested by the increased number of the SG labelled. These K15CREER-labelled cells then locally self-renew and continue to contribute to SGs homeostasis up to 6 months after TAM administration.
K15CREER-labelled cells also renew and expand overtime within the bulge region, consistent with the notion that some of the bulge SCs undergo symmetric self-renewing division within their niche, as previously suggested (Waghmare et al, 2008; Zhang et al, 2009, 2010). The absence of contiguous trail of YFP+ cells between the bulge and the SGs suggests that the K15-derived SG progenitors arising from the bulge are not bipotent at the clonal level, and leave the bulge niche once committed to the SG lineage. The apparent contribution of bulge SC to the SG homeostasis is not limited to a particular stage of the hair cycle, although the flux of bulge SC to the SG seems to increase during the activation stage of bulge SC that occurs during the initiation of HF regeneration.
The migration of bulge SC to the SG has been substantiated by two different experimental approaches. The level of expression of SG progenitor markers (Lgr6, Lrig1 and Blimp1) in YFP-labelled cells increases with time after TAM administration, consistent with a progressive enrichment of SG progenitors within the progeny of K15CREER-derived cells. The authors also developed a new method to perform ex vivo live cell imaging of whole mount preparation of the skin epidermis in order to visualize the migration of bulge cells in real time. This elegant approach allowed the authors to follow the fate of individual bulge SC during 22 h. As expected, most of the labelled bulge SC remained quiescent during the observation but a fraction of those cells divide and migrate locally, downward but also upward towards the UI and SG (Figure 1). However, the whole mount preparation could not be imaged long enough to visualize the whole process of bulge SC migration to the SG and their subsequent contribution to the sebocyte differentiation.
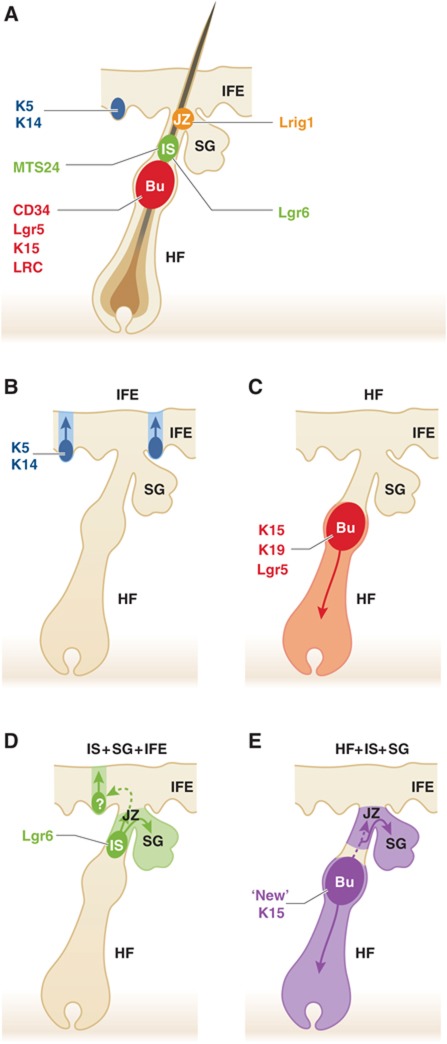
Figure 1.
Bulge SC contribution to the sebaceous lineage. (A) Scheme highlighting the different epidermal SC populations found in the skin epidermis: the IFE SC (shown in blue), the bulge SC (Bu in red), the isthmus SC (Is in green) and the junctional SC (JZ in yellow). (B) Lineage tracing experiments have shown that IFE is maintained independently of the HF by the presence of number small unit of proliferation. (C) Lineage tracing of bulge SC using K15CREPR, Lgr5CREER and K19CREER demonstrate their ability to contribute to the homeostasis of the hair follicle but not the SG or the IFE. (D) Lgr6 lineage tracing show the long-term renewal of Lgr6+ isthmus cells and their contribution to the sebaceous lineage. (E) The recent study of Petersson and colleagues using a new K15CreER shows the potential contribution of bulge SCs to the SG homeostasis.
It has been previously demonstrated that overexpression of a mutant of Lef1 that cannot bind β-catenin (K14ΔNLef1) leads to development of ectopic SGs, suggesting that β-catenin/Lef1 signalling regulates SG differentiation (Merrill et al, 2001; Niemann et al, 2002, 2007). To determine the cellular origin of the de novo SGs in K14ΔNLef1, the authors performed lineage tracing experiments using K15CreERT2/Rosa–YFP in K14ΔNLef1 mice. The presence of YFP-labelled cells in de novo SG above and below the bulge demonstrates that these lesions arise from the bulge SC that progressively expressed markers of the SG progenitors (Lgr6, Lrig1, Blimp1 and Plet1) ectopically. Interestingly, while de novo SG arise from cells below and above the bulge region in K14ΔNLef1, de novo SGs arise only in their normal niche when ΔNLef1 is preferentially expressed in the bulge region (K15ΔNLef1 transgenic mice).
These new results are very interesting and intriguing. Indeed, while it has been previously reported that bulge SC can contribute to the SG lineage using K15CrePR and Lgr5CreER lineage tracing, but the contribution of the SG was reported to be limited to only rare follicles (Morris et al, 2004; Jaks et al, 2008). One possibility is that these new CREER mice target a subpopulation of the upper bulge more prone to give rise to the SG lineages than the previously described bulge specific inducible CRE (Morris et al, 2004; Levy et al, 2005; Jaks et al, 2008; Youssef et al, 2010) or that these new K15CREER also targeted a rare subpopulation of SG progenitors known to participate to both isthmus and SG maintenance such as Lgr6+ cells (Snippert et al, 2010), Lrig1+ cells (Jensen et al, 2009) or MTS24+ cells (Nijhof et al, 2006). The long-term labelling of SG by resident progenitors observed using Lgr6CREER (Snippert et al, 2010) or K15CreER in this study (Petersson et al, 2011), demonstrates that the SG contain resident progenitor that locally self-renew and that not all SGs are constantly renewed through a continuous flux of bulge SC. It will be important to determine what mechanisms regulate the asymmetric cell fate decision of bulge SCs during the specification of SG progenitors and what regulate the flux of bulge SC towards the SG.
Regulation of p63 expression by an autoregulatory loop involving miRs
p63, a member of the p53 family of protooncogenes, has been identified as a key regulator of stratified epithelium development. Mice lacking p63 are born alive but lack all major stratified epithelia including the epidermis (Mills et al, 1999; Yang et al, 1999). p63 has several isoforms depending on the transcription site and alternative splicing (Yang et al, 1998). ΔNp63 is the major isoform express in the epidermis and is expressed mainly in the basal layer of the epidermis (Koster et al, 2004; Laurikkala et al, 2006). Subsequent studies suggest that p63 can regulate renewal and differentiation of the epidermal progenitors (Truong et al, 2006; Senoo et al, 2007) but the mechanisms regulating p63 expression in epidermal cells remain elusive.
miRNAs provide an additional layer of complexity to the transcriptional regulatory switch of p63 during epidermal differentiation. miR-203 is upregulated during epidermal stratification suprabasally at E15.5 and was shown to directly represses p63 expression in the suprabasal differentiated cells to promote differentiation and stratification of the epidermis (Lena et al, 2008; Yi et al, 2008).
In a recent study published in the EMBO Journal, Chikh et al (2011) showed that iASPP regulates p63 expression in the epidermis through the regulation of miRNA expression. Apoptosis stimulating proteins of p53 (ASSP) are a family of proteins that regulates p53-mediated apoptosis. ASPP1, ASPP2 potentiate the pro-apoptotic function of p53, while the third member iASPP inhibits this function (Bergamaschi et al, 2003). iASPP is mostly expressed in epithelial cells from several tissues including skin (Herron et al, 2005) and is frequently mutated in human cancers (Bergamaschi et al, 2003). Structural studies predicted that iASPP preferentially binds to p63, suggesting that iAPSS could be involved in the regulation of p63 expression in the skin (Robinson et al, 2008).
In this new study, the authors showed that iASPP is expressed in the early developing mouse epidermis and colocalizes with p63 in the nucleus of basal epidermal cells at E15.5 and thereafter. The same pattern of iASPP expression is found in human epidermis. Stimulation of human keratinocyte differentiation decreases both ΔNp63 and iASPP expression, suggesting that iASPP might be involved in p63 downregulation. The authors performed ChIP experiments to demonstrate that p63 directly binds to iASPP promoter in vivo. Overexpression of the two p63 isoforms stimulates iASPP expression, while their knockdown by siRNA downregulates iASPP, thus functionally demonstrating that p63 regulates iASPP in vitro. Reciprocally, downregulation of iASPP decreases expression of TAp63 and ΔNp63 at protein level, demonstrating that iASPP and p63 form a positive autoregulatory feedback loop. Intriguingly, iASPP knockdown does not regulate p63 mRNA expression, suggesting that iASPP may regulate the expression of p63 protein through a posttranscriptional or a postranslational mechanism. By profiling miR expression after iASPP KD, the authors found that miR-574-3p and miR-720, two miRs predicted to target p63, were upregulated following iASPP silencing. Antagomirs against these miRs restored the expression of p63 in the context of iASPP knockdown, suggesting that iASPP represses miR-574-3p and miR-720 expression, which in turn promote p63 expression.
To determine the role of iASPP in keratinocytes, the authors used transcriptional profiling by microarray of iASPP-depleted cells. This experiment showed that iASPP is involved in the regulation of several members of the desmosomal complexes, tight junction and gap junction components, and a decrease in β1 integrin expression. Cells depleted for iASPP acquire a phenotype of differentiated cells with reduced proliferation. Human skin equivalent using organotypic culture of iASPP-depleted keratinocytes are thicker, less proliferative and present premature differentiation characterized by the expression of K1, involucrin and loricrin-positive cells within the basal compartment. This phenotype was partially rescued by specific antagomirs against miR-574-3p and miR-720, suggesting that iASPP promotes self-renewal and inhibits differentiation by repressing miR-574-3p and miR-720 expression (Figure 2).
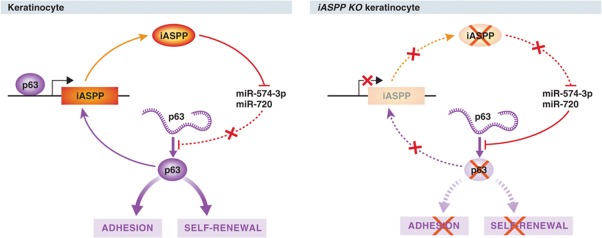
Figure 2.
iASPP regulates keratinocyte renewal and differentiation by regulating miR expression. Under physiological conditions, p63 regulates iASPP expression, which in turn, represses miR-574-3p and miR-720. Upon iASPP knockdown in human keratinocytes, these miRs are upregulated and inhibit p63 expression, leading to defect in keratinocyte adhesion, renewal and differentiation (Chikh et al, 2011).
This study shows that p63, a major regulator of epidermal morphogenesis and differentiation, is indirectly regulated by iASPP through the modulation of miR expression. While miR-574-3p and miR-720 regulate p63 expression, iASPP KD does not exactly phenocopy p63 KD (Carroll et al, 2006; Truong et al, 2006). Indeed, while iASPP-depleted keratinocytes show a decrease of β1 integrin and an increase of α3 integrin expression without modifications of β4 integrin; p63 knockdown induces a strong decrease in the expression of β1, β4 and α3 integrins at mRNA level (Carroll et al, 2006). In addition, iASPP KD impairs cell proliferation similarly as p63 KD, but also induces a premature differentiation of skin equivalent, which is inhibited upon p63 silencing (Truong et al, 2006), suggesting that iASPP may regulate other signalling pathways in addition of p63. Future studies will be required to clarify the discrepancy between iASPP and p63 KD, and determine what could be the other targets regulated by miR-574-3p and miR-720.
Role of the polycomb repressor complex in the regulation of epidermal SCs
The polycomb group proteins (PcG) constitute a chromatin remodelling complex referred to as Polycomb Repressor Complexes (PRC). PRC2 is composed of Ezh2 (or Ezh1), Eed, suz12 and RBAP46/48 (also known as Rbbp7/4) (Margueron and Reinberg, 2011). This complex is recruited to chromatin and Ezh2 mediates the trimethylation of histone H3 on Lysine 27 (H3K27me3). This histone mark induces recruitment of PRC1, which participates to repress adjacent locus. PRC1 complexes are more variable and are composed of Ring1a/b together with Bmi1, Cbx and PH proteins that mediate the monoubiquitination of H2A at Lys119, which induces gene silencing (Simon and Kingston, 2009).
Deletion of PRC2 complexes subunit leads to the upregulation of lineage commitment genes normally silenced in embryonic SCs that lead to early embryonic lethalilty (O’Carroll et al, 2001; Voncken et al, 2003). In the skin, loss of Ezh2 impaired proliferation and induced the premature differentiation of the epidermis during embryonic development but this phenotype disappeared postnatally (Ezhkova et al, 2009). Combined deletion of Ezh1 and Ezh2 leads to hair loss and hyperproliferation of IFE cells (Ezhkova et al, 2011). In HF lineages, Ezh1/Ezh2 cKO induces upregulation of Ink4b/Ink4a/Arf and subsequent overexpression of p16/p19 (Ezhkova et al, 2011). Since Ink4b/Ink4a/Arf shRNA restore HF cells self-renewal in vitro, these results suggest that Ezh1/Ezh2 regulate HF SCs proliferation mostly by repressing Ink4b/Ink4a/Arf locus, similarly to Bmi1, another PRC2 subunit (Jacobs et al, 1999; Cordisco et al, 2010). In addition, Jmjd3 histone demethylase, which is involved in H3K27me3 demethylation, has been shown to be upregulated following keratinocyte differentiation and its overexpression in basal epidermal cells induces premature differentiation. This effect is associated to the derepression of differentiation-related genes such as krt1 and S100A8 and ectopic expression of krt1, krt10, filaggrin and involucrin. Inhibition of Jmjd3 demethylase activity by point mutation blocked its promoting effect on differentiation, suggesting that jmjd3 stimulates epidermal differentiation by the demethylation of H3 at several loci (Sen et al, 2008). Jarid2, a jumonji/JmjC domain-containing protein, has been shown to help the recruitment of PRC2 to the promoter of repressed genes in ESC (Peng et al, 2009; Shen et al, 2009; Landeira et al, 2010). However, conflicting results have been published concerning the role of Jarid2 in regulating the methyltransferase activity of PRC2 (Herz and Shilatifard, 2010; Landeira and Fisher, 2011). Little is known about the mechanisms that contribute to the recruitment of PRC2 complex to the promoter of repressed genes in adult tissue.
A recent study by Mejetta et al (2011) published in EMBO Journal showed that Jarid2 regulates epidermal progenitor proliferation and differentiation in the mouse skin epidermis. Moreover, Jarid2 also recruits PRC1 to the promoters of lineage-specific genes in embryonic SCs (Landeira et al, 2010). Conditional deletion of Jarid2 in epidermal cells during embryonic development induced no initial defect of proliferation and differentiation. However, after birth, Jarid2 null epidermis is characterized by an increased number of differentiated cells as well as a decrease in the proliferation of basal epidermal progenitors. Jarid2 null keratinocytes present fewer proliferative colonies than control keratincoytes and present characteristics of terminally differentiated keratinocytes in vitro. While HF morphogenesis is apparently unaffected in the absence of Jarid2, there is a delay in the initiation of HF regeneration, despite the similar number of adult HF bulge SC. HFs present a delay in anagen re-entry, but they eventually resume hair cycle and regenerate new hairs. In addition, Jarid2 null HFs are less responsive to TPA-induced bulge SC proliferation, and their epidermis presented increased differentiation upon TPA treatment.
The authors then showed by co-immunoprecipitation that Jarid2 interacts with PRC2 components Ezh2 and Suz12 in mouse keratinocytes, as it does in ESC. By performing ChIP experiments, they showed that Jarid2 localized to the promoter of several epidermal and non-epidermal target genes of the PRC2 complex previously shown to be occupied by Ezh2 in the skin epidermis (Ezhkova et al, 2009). In the absence of Jarid2, there is a small but significant decrease in the occupancy of these target genes by Suz12 as well as a decrease in the level of H3 trimethylation at these loci. These results are consistent with the notion that Jarid2 helps the recruitment of PRC2 and positively regulates the methyl transferase activity of the PRC2 complex at these loci. Accordingly, these PRC2 target genes including genes involved in keratinocyte differentiation, as well as p16, a gene that negatively regulates cell-cycle progression were upregulated in the absence of Jarid2 in newborn skin epidermis. In contrast, in the adult epidermis, the absence of Jarid2 has no effect on the expression of differentiated genes but still present a decrease in the recruitment of Suz12 at the Ink4a promoter, leading to an increase of p16 expression (Figure 3).
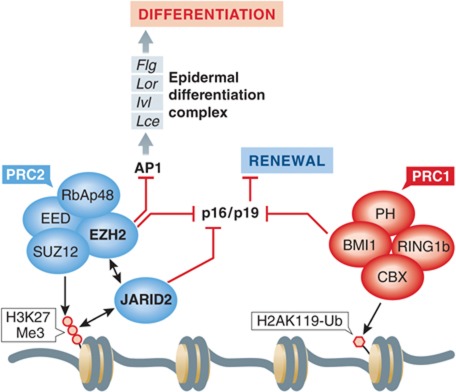
Figure 3.
Jarid2 regulates epidermal SC proliferation and differentiation. Jarid2 modulates the recruitment of the PRC2 complex and the trimethylation of lysine 27 Histone H3 (H3K27Me3). This epigenetic mark represses the Arf4 locus, leading to repression of p16/p19. In Jarid2 cKO mice (K14Cre∷Jarid2fl/fl), trimethylation of H3K27 is decreased and subsequently, p16 is upregulated leading to a delay in bulge SC activation and increased IFE differentiation. This effect could be related, at least partially, by a decrease in the recruitment of Ezh2, which that is involved in repressing of p16 and epidermal differentiation complex encompassing Filaggrin (Flg), Loricrin (Lor), Involucrin (Ivl) and late cornified envelope (Lce) genes in an AP1-dependent manner (Ezhkova et al, 2009). In the absence of Jarid2, cultured keratinocytes present a decrease in their clonogenic potential, suggesting that Jarid2 controls epidermal SC renewal.
These results highlight the complexity of the regulation of epidermal SCs by histone modification. While Jarid2 and Ezh2 null epidermis present some similarities (e.g., increase of p16), there are clear differences in the timing and the extent of their phenotypes (Ezhkova et al, 2009; Mejetta et al, 2011). These apparent discrepancies between the two models suggest that Jarid2 and Ezh2 functions have only partially overlapping functions and other factors are probably involved in the regulation of gene expression mediated by these two proteins. Consistent with this possibility, it has been shown that Ezh2 regulates differentiation by inhibiting AP1 (c-Jun/c-Fos) recruitment to the promoter of genes involved in epidermal differentiation (Ezhkova et al, 2009). It will be interesting to determine whether the difference between Ezh2 and Jarid2 functions is at least partially related to their ability to modulate AP1 function. Moreover, the modest reduction in the level of H3K27me3 in the absence of Jarid2 suggests that some functions of Jarid2 in the skin epidermis may be not related to its ability to stimulate the histone methylation but potentially serves to recruit other proteins that mediate distinct functions. Furthermore, the work of Ezhkova et al (2011) had shown that despite the complete absence of H3K27me3 marks in Ezh1/2 null keratinocytes, only few genes bound by PRC2 are actually upregulated in the absence of Ezh1/2, suggesting that the repression of differentiation genes is not only mediated by the trimethylation of H3 at K27.
The notion that the recruitment of specific chromatin-associated protein can function independently of their catalytic activity is reinforced by a recent study that showed that Cbx4, a PRC1-associated protein, controls proliferation and differentiation of human epidermal SCs by distinct mechanisms (Luis et al, 2011). Surprisingly, both gain and loss of function for cbx4 inhibit the clonogenicity of epidermal SC in vitro. While Cbx4 overexpression inhibits cell proliferation without modifying differentiation, Cbx4 KD promotes differentiation. Overexpression of a chromodomain mutant of Cbx4 that can no longer interact with PRC1 induces senescence by upregulating Ink4a locus (p16) and p57 without modifying differentiation, while overexpression of a Cbx4 SUMO-U3 ligase activity mutant stimulates differentiation.
The histone H4 monomethyltransferase Setd8 is required for epidermal SC maintenance
Setd8 is the sole enzyme responsible for the monomethylation of histone H4 at lysine 20 (H4K20me1) (Xiao et al, 2005). This methylation of histone H4 has been reported to be essential for cell-cycle progression in vitro (Jorgensen et al, 2007; Tardat et al, 2007) and its deletion is lethal in flies and during early embryonic development in mice (Nishioka et al, 2002; Karachentsev et al, 2005; Oda et al, 2009). Deletion of Setd8 in ESC induces DNA damage response, genomic instability and cell-cycle arrest (Oda et al, 2009). While in vitro studies suggest a critical role of Setd8 in cell-cycle progression in vitro and early embryonic development, little is known about the role of Setd8 during the later stage of embryogenesis and adult renewing tissue. In the epidermis, it has been previously shown that Myc regulated the methylation of histone H4 (H4K20me1) by an HDAC-dependent mechanism, and this chromatin modifications was associated with differentiation (Frye et al, 2007). A recent study of Driskell et al (2011) published in the EMBO Journal demonstrated the essential role of Setd8 in skin development. In epidermis, H4K20me1 is found in basal undifferentiated cells of IFE, SG and anagen hair follicle and is mostly excluded from cells in S-phase, although rare cells positive for BrdU and H4K20me1 is seen in HF-committed cells. While no antibodies recognized specifically Setd8 in the epidermis, reporter mouse expressing β-galactosidase under the regulatory region of the Setd8 locus (Huen et al, 2008) indicated that the strongest setd8 expression in the epidermis is found in the developing epidermis, adult SG and HF matrix cells. Lower and patchy expression of the Setd8 reporter activity is seen in the adult IFE.
The authors first investigated the role of Setd8 during morphogenesis using conditional deletion of Setd8 using the K14CRE mice, which is expressed in the early stage of mouse epidermal development (around E12). Mice deficient for Setd8 are born alive but they lack limbs and skin and die immediately after birth, a phenotype reminiscent of the p63 null mice (Mills et al, 1999; Yang et al, 1999). Indeed, p63 was severely reduced in Setd8 null epidermis, which is composed of a single layer of K8 expressing cells that failed to renew and stratify and which are progressively lost from the skin surface, identically to p63 null mice.
To determine the role of Setd8 in adult epidermis, the authors performed the conditional deletion of Setd8 using the K14CREER. Setd8 deletion in adult epidermis results in decrease of IFE progenitors proliferation, increase in progenitor apoptosis and decrease in the expression of progenitor markers including α6 integrin and p63, as well as a decrease in the expression of differentiation markers. Concomitant expression of YFP and deletion of Setd8 demonstrated that Setd8-deficient cells are progressively lost, which induced a wounding-like stimulus that simulated the non-deleted Setd8 HF cells (due to the poor efficiency of HF targeting by K14CREER) to replace the Setd8 YFP+deficient IFE cells. Similarly, homeostasis of the SG was also severely affected following the conditional deletion of Setd8 in the adult epidermis that can also recover through the repopulation of the SG by Setd8 HF SCs.
c-Myc overexpression stimulates epidermal cell proliferation and differentiation (Waikel et al, 1999, 2001; Arnold and Watt, 2001), as well as monomethylation of H4K20me1 (Frye et al, 2007). Driskell and colleagues have now demonstrated that that Myc directly binds to the promoter of Setd8 and stimulates its expression. Deletion of Setd8 rescues the increase in cell proliferation and differentiation associated with c-Myc overexpression, suggesting that Setd8 is required to mediate some Myc-associated functions.
The authors showed that p53 expression was majorly increased upon Setd8 deletion in the epidermis. Combined deletion of p53 and Setd8, rescues the proliferation defect, p63 expression and the differentiation defect seen in Setd8 null epidermis in vivo, and overexpression of p63 rescues the proliferation defect of Setd8 null keratinocytes in vitro. The authors proposed a model by which Setd8 represses p53 and stimulates p63 to allow epidermal renewal and differentiation (Figure 4). An interesting parallel with these findings is the recent study reporting that epidermal deletion of HDAC1/2, which normally remove the histone acetylation marks important for gene repression, also induces a phenotype reminiscent of the p63 null mice, presenting a major defect in epidermal differentiation (LeBoeuf et al, 2010). In contrast to Setd8 deficiency, which leads to a decrease of p63 expression, the absence of HDACA1/2 did not modify p63 expression but impairs its ability to repress gene expression including p16 (Su et al, 2009; LeBoeuf et al, 2010), which in turn induces cell-cycle arrest. In addition, HDAC1/2 deficiency also lead to increased p53 acetylation, increasing its transcriptional activity leading to p21 overexpression, which also concurs to inhibit epidermal cell proliferation (LeBoeuf et al, 2010).
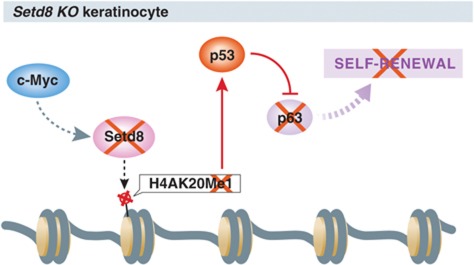
Figure 4.
The Histone monomethyltrasferase Setd8 is required for human keratinocytes self-renewal. The recent study of Driskell and colleagues showed that Setd8 KO leads to the loss of H4K20Me1 imprint in keratinocytes and impairment of self-renewal due to the loss of p63 (Driskell et al, 2011). Intriguingly, loss of H4K20Me1 leads to the stabilization of p53 and deletion of p53 rescues the phenotype of Sted8 cKO mice by restoring the level of p63. It is still unclear what exactly regulates Setd8 expression in keratinocytes albeit it is known that it is a target gene of c-Myc. Moreover, the mechanism of stabilization of p53 in the absence of the H4K20Me1 imprint is still unknown.
It is still not clear why exactly p53 is massively increased upon loss of Setd8. As DNA damage is one of the major trigger of p53 stabilization, it would be interesting to determine whether Setd8 prevent DNA damage and consequently p53 activation? Or does H4K20me1 methylation regulate p53 activation? Do other histone modifications regulate directly and/or indirectly p53 and p63 expression?
Conclusions and perspectives
These recent studies published in the EMBO Journal stress the complexity of the regulation of the skin epidermis maintenance. Recent lineage tracing experiments has demonstrated the heterogeneity of bulge SC and suggested that bulge may directly participate into SG homeostasis during epidermal homeostasis (Petersson et al, 2011). More studies will be required to define more precisely the mechanisms that regulate the balance between asymmetric and symmetric cell division within the bulge SC, the mechanisms that trigger SG commitment of bulge SC and that regulate their migration towards the SG as well as the mechanisms that prevent migration of bulge SC towards the IFE in the absence of wounding.
miRs appeared as important regulators of the main signalling pathways that regulated epidermal identity, proliferation and differentiation. It will be important to determine in the future whether similar mechanisms are also impaired in skin cancers in which epithelial cells are characterized by increased proliferation and defect of differentiation. It will be also crucial to determine more precisely what regulate the expression of these different miRs in the skin epidermis.
Many studies have now demonstrated the crucial role of epigenetic and histone modifications in the regulation of SC proliferation and differentiation (Frye et al, 2007; Ezhkova et al, 2009, 2011; Sen et al, 2010; Driskell et al, 2011; Mejetta et al, 2011). While the deletion of the different members of the PRC2 complex in the skin epidermis lead to similar but also distinct phenotypes (Ezhkova et al, 2009, 2011; Mejetta et al, 2011), they all lead to the repression of Ink4a locus encoding p16, preventing cellular senescence as it was initially discovered for Bmi1 deficiency in fibroblasts (Jacobs et al, 1999) as well as in haematopoietic and leukaemic SCs (Lessard et al, 1999). Clearly, more studies are needed to fully understand the functions of epigenetic modifications in the regulation of skin SCs. Does DNA methylation also play an important function in regulating proliferation and differentiation at it was recently suggested in human epidermal SC (Sen et al, 2010) and how are DNA modifications connected to histone modifications? How are these different epigenetic marks established and dynamically regulated in the different stages of SC ontogeny and activation? How exactly do they control gene expression and DNA damage? The answers to these questions and the role of these processes during diseases such as in cancer will constitute important challenges for future research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnold I, Watt FM (2001) c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol 11: 558–568 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, O’Neil NJ, Trigiante G, Crook T, Hsieh JK, O’Connor DJ, Zhong S, Campargue I, Tomlinson ML, Kuwabara PE, Lu X (2003) iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet 33: 162–167 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E (2007) p63: revving up epithelial stem-cell potential. Nat Cell Biol 9: 731–733 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Horsley V, Fuchs E (2007) Epithelial stem cells: turning over new leaves. Cell 128: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E (2004) Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118: 635–648 [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 8: 551–561 [DOI] [PubMed] [Google Scholar]
- Chikh A, Matin RN, Senatore V, Hufbauer M, Lavery D, Raimondi C, Ostano P, Mello-Grand M, Ghimenti C, Bahta A, Khalaf S, Akgul B, Braun KM, Chiorino G, Philpott MP, Harwood CA, Bergamaschi D (2011) iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J 30: 4261–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH (2007) A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189 [DOI] [PubMed] [Google Scholar]
- Cordisco S, Maurelli R, Bondanza S, Stefanini M, Zambruno G, Guerra L, Dellambra E (2010) Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J Invest Dermatol 130: 1048–1062 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM (1990) Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329–1337 [DOI] [PubMed] [Google Scholar]
- Doupe DP, Klein AM, Simons BD, Jones PH (2010) The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell 18: 317–323 [DOI] [PubMed] [Google Scholar]
- Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M (2011) The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J 31: 616–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM (2007) Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One 2: e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S, Taichman LB (2005) Organization of stem cells and their progeny in human epidermis. J Invest Dermatol 124: 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron BJ, Rao C, Liu S, Laprade L, Richardson JA, Olivieri E, Semsarian C, Millar SE, Stubbs L, Beier DR (2005) A mutation in NFkB interacting protein 1 results in cardiomyopathy and abnormal skin development in wa3 mice. Hum Mol Genet 14: 667–677 [DOI] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A (2010) The JARID2-PRC2 duality. Genes Dev 24: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, O’Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E (2006) Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 126: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J (2008) Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem 283: 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G (2005) Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291–1299 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM (2009) Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen UB, Yan X, Triel C, Woo SH, Christensen R, Owens DM (2008) A distinct population of clonogenic and multipotent murine follicular keratinocytes residing in the upper isthmus. J Cell Sci 121: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R (2005) PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18: 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Fisher AG (2011) Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol 21: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L et al. (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I (2006) p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133: 1553–1563 [DOI] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE (2010) Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell 19: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E (2008) miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ 15: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G (1999) Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev 13: 2691–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA (2005) Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 9: 855–861 [DOI] [PubMed] [Google Scholar]
- Luis NM, Morey L, Mejetta S, Pascual G, Janich P, Kuebler B, Roma G, Nascimento E, Frye M, Di Croce L, Benitah SA (2011) Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9: 233–246 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means AL, Xu Y, Zhao A, Ray KC, Gu G (2008) A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 46: 318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejetta S, Morey L, Pascual G, Kuebler B, Mysliwiec MR, Lee Y, Shiekhattar R, Di Croce L, Benitah SA (2011) Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J 30: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E (2001) Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 15: 1688–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G (2004) Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Potten CS (1999) Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J Invest Dermatol 112: 470–475 [DOI] [PubMed] [Google Scholar]
- Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM (2002) Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 129: 95–109 [DOI] [PubMed] [Google Scholar]
- Niemann C, Owens DM, Schettina P, Watt FM (2007) Dual role of inactivating Lef1 mutations in epidermis: tumor promotion and specification of tumor type. Cancer Res 67: 2916–2921 [DOI] [PubMed] [Google Scholar]
- Nijhof JG, Braun KM, Giangreco A, van Pelt C, Kawamoto H, Boyd RL, Willemze R, Mullenders LH, Watt FM, de Gruijl FR, van Ewijk W (2006) The cell-surface marker MTS24 identifies a novel population of follicular keratinocytes with characteristics of progenitor cells. Development 133: 3027–3037 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E (2008) Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D (2009) Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Muller-Rover S, Van Der Veen C, Maurer M, Eichmuller S, Ling G, Hofmann U, Foitzik K, Mecklenburg L, Handjiski B (1999) A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J Invest Dermatol 113: 523–532 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson M, Brylka H, Kraus A, John S, Rappl G, Schettina P, Niemann C (2011) TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J 30: 3004–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K, Richter A, Kreft M, Frijns E, Janssen H, Slijper M, Praetzel-Wunder S, Langbein L, Sonnenberg A (2010) Expression of the orphan protein Plet-1 during trichilemmal differentiation of anagen hair follicles. J Invest Dermatol 130: 1500–1513 [DOI] [PubMed] [Google Scholar]
- Robinson RA, Lu X, Jones EY, Siebold C (2008) Biochemical and structural studies of ASPP proteins reveal differential binding to p53, p63, and p73. Structure 16: 259–268 [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA (2010) DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA (2008) Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F (2007) p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536 [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgard R, Clevers H (2010) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER (2009) Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J 28: 1904–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW (2003) Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol 120: 501–511 [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E (2004) Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M (2003) Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA 100: 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T (2008) Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J 27: 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR (2001) Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet 28: 165–168 [DOI] [PubMed] [Google Scholar]
- Waikel RL, Wang XJ, Roop DR (1999) Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene 18: 4870–4878 [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR (2005) Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev 19: 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E (2008) A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, Sotiropoulou PA, Blanpain C (2010) Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol 12: 299–305 [DOI] [PubMed] [Google Scholar]
- Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T (2009) Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, White BS, Shalloway DI, Tumbar T (2010) Stem cell dynamics in mouse hair follicles: a story from cell division counting and single cell lineage tracing. Cell Cycle 9: 1504–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]