Abstract
Mitochondria are dynamic subcellular organelles that convert nutrient intermediates into readily available energy equivalents. Optimal mitochondrial function is ensured by a highly evolved quality control system, coordinated by protein machinery that regulates a process of continual fusion and fission. In this work, we provide in vivo evidence that the ATP-independent metalloprotease OMA1 plays an essential role in the proteolytic inactivation of the dynamin-related GTPase OPA1 (optic atrophy 1). We also show that OMA1 deficiency causes a profound perturbation of the mitochondrial fusion–fission equilibrium that has important implications for metabolic homeostasis. Thus, ablation of OMA1 in mice results in marked transcriptional changes in genes of lipid and glucose metabolic pathways and substantial alterations in circulating blood parameters. Additionally, Oma1-mutant mice exhibit an increase in body weight due to increased adipose mass, hepatic steatosis, decreased energy expenditure and impaired thermogenenesis. These alterations are especially significant under metabolic stress conditions, indicating that an intact OMA1-OPA1 system is essential for developing the appropriate adaptive response to different metabolic stressors such as a high-fat diet or cold-shock. This study provides the first description of an unexpected role in energy metabolism for the metalloprotease OMA1 and reinforces the importance of mitochondrial quality control for normal metabolic function.
Keywords: ageing, apoptosis, degradome, metabolism, mitochondrial dynamics
Introduction
Mitochondria are fundamental and highly dynamic organelles of eukaryotic cells that play critical roles in energy generation, control of intermediate metabolism, homeostasis of intracellular calcium and regulation of apoptosis events (Mammucari and Rizzuto, 2010; Wallace et al, 2010). Additionally, mitochondria are also the primary source of endogenous reactive oxygen species (Orrenius et al, 2007). Due to these multiple functions, it is not surprising that mitochondrial alterations have been associated with many pathological conditions including cancer, neurodegenerative disorders and cardiovascular diseases (Green and Kroemer, 2004; Wallace, 2005; Chen and Chan, 2009; Fulda et al, 2010). To maintain their functional activities, mitochondria have developed a quality control mechanism, in which several proteolytic enzymes play important roles (Tatsuta and Langer, 2008). These mitochondrial proteases, together with some mitochondrial chaperones, monitor the folding and assembly of mitochondrial proteins and selectively degrade misfolded and non-assembled polypeptides from the organelle (Voos, 2009). Among the proteolytic enzymes implicated in the mitochondrial quality control system, there are several ATP-dependent proteases such as Yme1L and m-AAA proteases located in the inner membrane, or ClpP and Lonp1 that carry out their functions in the matrix (Koppen and Langer, 2007). The functional relevance of these mitochondrial enzymes is underscored by the finding that deficiencies in m-AAA proteases SPG7 and AFG3L2 are responsible for important neurodegenerative diseases such as hereditary spastic paraplegia or a dominant form of spinocerebellar ataxia (Casari et al, 1998; Di Bella et al, 2010).
In addition to the key role of mitochondrial proteases in the quality control system characteristic of these organelles, these enzymes may also regulate the activity of some mitochondrial proteins through their ability to perform highly specific reactions of proteolytic processing, which contribute to the maturation or inactivation of these substrates. This may be the case of OMA1, a proteolytic enzyme located in the inner mitochondrial membrane, which was first identified in yeast as a protease with overlapping activities with m-AAA proteases (Kaser et al, 2003). OMA1 is a zinc metalloprotease of the M48 family, which exhibits multiple transmembrane domains and significant amino-acid sequence similarity with FACE1/ZMPSTE24, a protease involved in the processing of prelamin-A and whose mutations cause premature ageing disorders (Navarro et al, 2005; Varela et al, 2005, 2008). Yeast OMA1 seems to play a role in mitochondrial quality control through a mechanism similar to that of m-AAA proteases, although in an ATP-independent manner. This activity has been proposed to be part of a salvage system of quality control, reminiscent of that performed in Escherichia coli by HtpX, a stress protease with overlapping activities with the AAA protease FtsH (Shimohata et al, 2002). Recently, a series of in vitro studies have permitted the conclusion that OMA1, in collaboration with m-AAA protease isoenzymes, contributes to the proteolytic processing of OPA1 (optic atrophy 1), a dynamin-related GTPase involved in mitochondrial inner membrane fusion as well as in the regulation of mitochondrial morphology and in the protection of cells from apoptosis (Ehses et al, 2009; Head et al, 2009).
To further characterize the in vivo role of this mitochondrial metalloprotease, we have generated mutant mice deficient in OMA1. These mice are viable and fertile, but exhibit a marked obesity with metabolic alterations, reduced energy expenditure and altered thermogenic response. Furthermore, we have found that OMA1 plays an essential and non-redundant role in the in vivo proteolytic inactivation of the GTPase OPA1. Finally, we describe studies of mitochondrial activity and function in brown fat tissue and primary adipocytes deficient in OMA1. On the basis of these findings, we propose that OMA1 participates in mitochondrial quality control regulating mitochondrial dynamics, and its absence induces mitochondrial dysfunctions, which have an important impact on mouse metabolic homeostasis.
Results
Generation of mice deficient in OMA1
To evaluate the in vivo roles of OMA1 mitochondrial protease, we engineered a targeting vector to generate a null allele of Oma1. This vector was designed to replace exon 2 of the Oma1 gene with a neo cassette (Supplementary Figure S1A). The linearized construct was electroporated into G4 embryonic stem cells and after homologous recombination, we obtained seven positive targeted clones that were used to generate chimeric mice. These mice were then bred to C57BL/6 mice to generate heterozygous mice. After intercrossing these heterozygous mice, we generated Oma1−/− animals at the expected Mendelian ratio. Homozygosity for the mutation was confirmed by Southern blot and PCR analysis (Supplementary Figure S1B and C).
Despite the Oma1 deficiency, these mutant mice developed normally, with males and females being fertile, and their survival rates indistinguishable from those of their wild-type littermates (Supplementary Figure S2A). During the time period we studied the animals, we did not observe any physical indicators of neurological abnormalities. Moreover, histopathological analysis of the brain, specifically the hippocampus, from 18-month-old mice did not reveal any neuronal loss, nor increment in ubiquitin staining in Oma1-deficient mice compared with their littermates controls. Finally, all samples analysed for both genotypes displayed changes consistent with normal ageing (Supplementary Figure S2B). These findings demonstrate that OMA1 is dispensable for embryonic and adult mouse development, as well as for normal growth and fertility, possibly due to functional redundancy with other peptidases present in the inner membrane of mitochondria.
Diet-induced obesity in Oma1−/− mice
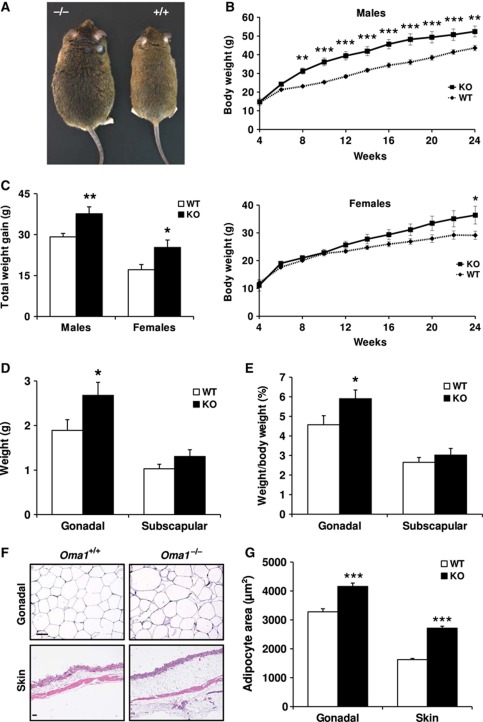
In the course of phenotypic characterization of Oma1-deficient mice, we noticed a significant increase in the body weight of Oma1−/− mice as compared with their wild-type littermates kept on standard chow (Supplementary Figure S2C). This observation encouraged us to evaluate a putative role for OMA1 in adipose tissue biology. Accordingly, wild-type and Oma1 knockout mice were fed a high-fat diet and total body weights were determined for 24 weeks. We started our study with 4-week-old mice, which displayed similar weights and adipose deposit composition in males and females of each genotype (Supplementary Figure S2D). At 8 weeks, we observed significant weight differences in Oma1−/− males when compared with controls, and these marked changes continued until the end of the experiment. In the case of Oma1−/− females, there was also an increase of weight when compared with the corresponding controls, but lower than in male mice (Figure 1A and B). Statistical analysis revealed that the gain of weight observed at the end of the experiment was significantly higher in both male and female mice deficient in Oma1 than in their littermate controls (Figure 1C).
Figure 1.
Increase of body weight and fat content in Oma1−/− mice after diet-induced obesity. (A) Photograph of representative Oma1+/+ and Oma1−/− mice after high-fat diet induced obesity. (B) Body weight curves of males and females of Oma1+/+ (♦) and Oma1−/− (▪) mice (n=8–12). (C) Lean body mass in the same animals. (D) Gonadal and subscapular fat mass and (E) gonadal and subscapular fat mass as a percentage of total body weight of the same animals at the end of the experiment. (F) H&E sections of gonadal WAT and skin of Oma1+/+ and Oma1−/− mice. Original magnifications WAT: × 200; skin: × 40. Scale bar: 60 μm. (G) Mean of adipocyte area in gonadal WAT and skin. Results are mean±s.e.m. (n=6–12). *P<0.05; **P<0.01; ***P<0.001.
To determine whether the increase of body weight observed in Oma1-deficient mice was due to an increase in fat content, gonadal and subscapular fat pad deposits were separately weighted. We only detected an increase of gonadal fat pads weight and their weight normalized to total body weight in Oma1-null mice (Figure 1D and E), whereas subscapular fat pads and their weight relative to total body weight did not reach statistical significant differences. Histological analysis of white adipose tissue (WAT) from gonadal and subcutaneous deposits showed a marked adipocyte hypertrophy in Oma1−/− mice (Figure 1F), which was confirmed by morphometric measurement of adipocyte area (Figure 1G). Analysis of brown adipose tissue (BAT) of Oma1-deficient mice did not show any significant alteration in fat content (Supplementary Figure S3A), indicating that the obesity-induced phenotype was only related to WAT hypertrophy. Interestingly, those Oma1−/− mice exhibiting the most severe obesity phenotype often presented granuloma-like lesions in the intestinal mesenteric fat. Further histological analysis of these granulomatous lesions revealed the presence of fat necrosis foci containing foamy histiocytes and cholesterol crystals (Supplementary Figure S3B). Similar lesions have been described previously as crown-like structures, which appear to manifest as a result of increased inflammation and adipocyte cell death (Cinti et al, 2005). All these findings suggest that the absence of OMA1 protease leads to profound alterations in WAT homeostasis after nutritional challenges.
Metabolic changes in Oma1−/− mice
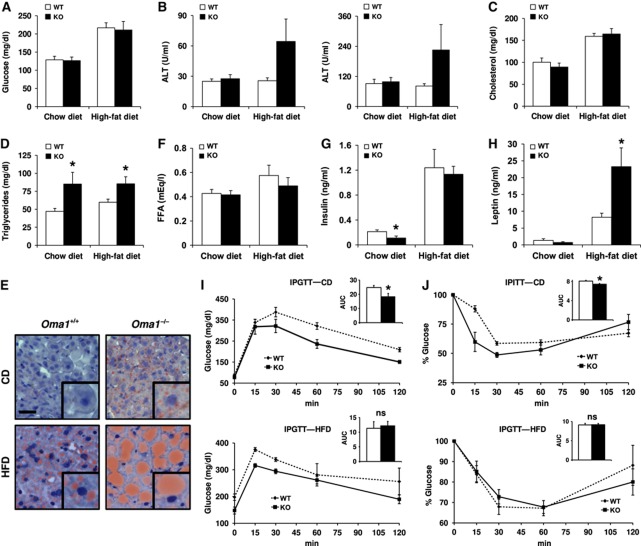
To further evaluate the phenotypic alterations observed in Oma1−/− mice, we performed an analysis of biochemical parameters and metabolic enzymes in the liver of these mutant mice under both standard and high-fat diet. First, and because in many animal models of obesity blood glucose tends to be above normal due to obesity-induced insulin resistance, we measured blood glucose levels in Oma1−/− adult mice, but we did not observe any differences with control mice in both standard and high-fat diet conditions (Figure 2A). Furthermore, analysis of liver parameters did not reveal significant differences in levels of hepatic transaminases under standard chow diet. However, under high-fat diet, we observed an increment in the levels of both Ala- and Asp-transaminases (Figure 2B). Total levels of cholesterol in mutant mice did not exhibit significant differences with those of control animals under chow and high-fat diet (Figure 2C). Nevertheless, levels of triglycerides were significantly higher in Oma1−/− mice compared with littermate controls, in both standard and high-fat diet (Figure 2D). Consistent with this, Oil Red O staining revealed a marked steatosis in the liver of Oma1−/− mice maintained on a normal chow diet, which increased significantly under a high-fat diet (Figure 2E). Levels of free fatty acids were comparable between genotypes on both diets (Figure 2F). Interestingly, analysis of insulin and leptin plasma levels showed different patterns under standard and high-fat diet. Thus, under standard rodent chow, both insulin and leptin plasma levels were reduced in mutant mice. However, under high-fat diet, insulin levels did not show differences despite the increment levels in control and mutant mice. Furthermore, leptin levels were significant enhanced as expected in an obese mouse model (Figure 2G and H).
Figure 2.
Lipid metabolism and glucose tolerance alterations in Oma1−/− mice under standard chow and high-fat diet. Analysis of serum and plasma parameters of Oma1+/+ and Oma1−/− mice (12–20 weeks old, n=6–12 for each group). Mice were fed on standard chow and high-fat diet, and analysis was determined after an overnight fast. (A) Blood glucose levels in Oma1+/+ and Oma1−/− mice. (B) Serum levels of alanine aminotransaminase (ALT) and aspartate aminotransaminase (AST), (C) cholesterol and (D) triglycerides. (E) Oil Red O staining of histological sections of liver from Oma1+/+ and Oma1−/− mice under chow diet (CD) and high-fat diet (HFD). Scale bar: 20 μm. Levels of (F) free fatty acids, (G) insulin and (H) leptin in plasma of Oma1+/+ and Oma1−/− mice. (I) IPGTT and (J) IPITT in wild-type and Oma1−/− male mice (8–12 weeks old, n=6–8). AUC, area under the curve (arbitrary units). Bars represent mean values±s.e.m. ns denotes no significant difference. *P<0.05.
The absence of hyperglycaemia in Oma1-deficient mice, together with their low insulin levels under standard chow diet, suggested the possibility of an increased insulin sensitivity of Oma1−/− mice. Accordingly, intraperitoneal glucose tolerance tests (IPGTTs) revealed that Oma1−/− mice showed improved glucose clearance compared with control mice (Figure 2I). Intraperitoneal insulin tolerance tests (IPITTs) confirmed that Oma1−/− mice presented increased insulin sensitivity (Figure 2J), which is consistent with their low insulin levels. Remarkably, these advantages in glucose metabolism were lost when Oma1-deficient mice were fed a high-fat diet. Thus, the metabolic tests in mutant mice showed a decrease in glucose tolerance and the same insulin insensitivity than control mice (Figure 2I and J, lower panels). Insulin tolerance was also analysed at 8 weeks, with animals displaying similar insulin responses to those observed at 12 weeks (data not shown). In addition, obese Oma1−/− mice showed the same higher levels of insulin than controls (Figure 2F), which explains the observed insulin insensitivity.
To further investigate these metabolic alterations observed in Oma1-deficient mice, we performed oligonucleotide-based microarrays to analyse transcriptional changes in adipose tissue from Oma1−/− mice kept under high-fat diet. As can be seen in Supplementary Table S1, we found a marked up-regulation in the expression of genes encoding proteins related with lipid metabolism and transport such as fatty-acid binding protein 1 (FABP1) and several apolipoproteins and cytochromes. Further, bioinformatic analysis revealed that a number of the up-regulated genes in Oma1-deficient mice are associated with two metabolic pathways (peroxisome proliferator-activated receptors α and γ, and retinol pathway), which can be related to the obesity phenotype observed in these mutant mice (Supplementary Figure S4). Collectively, these results indicate that mutant mice deficient in OMA1 mitochondrial protease exhibit significant alterations in lipid metabolism, characterized by an increase in triglycerides and hepatic steatosis, as well as an improved glucose clearance likely derived from an increased sensitivity to insulin.
Energy balance and thermogenesis are altered in Oma1-deficient mice
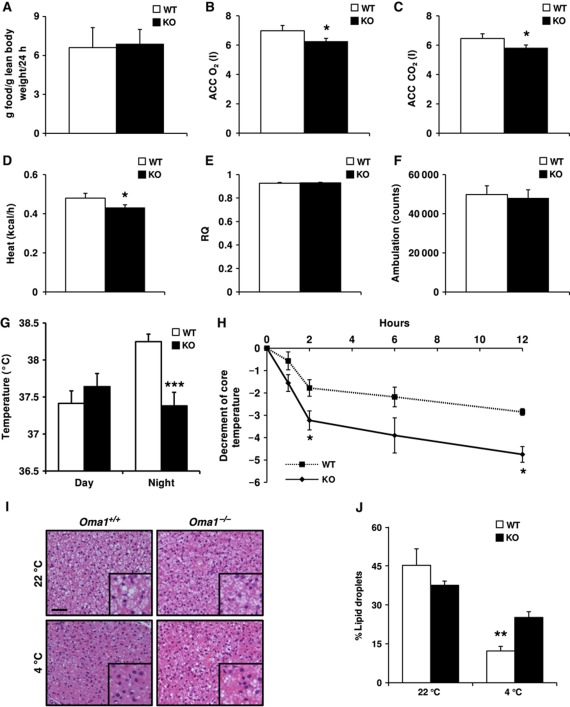
To determine the possible causes of the obesity and metabolic dysfunction observed in Oma1-deficient mice, we performed analyses of their food-intake and energy balance in mice maintained on a control chow diet. We did not find significant differences in food-intake of Oma1−/− mice relative to control littermates (Figure 3A). However, indirect calorimetry studies indicated that Oma1-deficient mice showed reduced oxygen consumption and CO2 production, which correlated with a significant reduction in heat production in these mice (Figure 3B–D). These alterations occurred in the absence of changes in respiratory quotient values (Figure 3E) and could not be explained by changes in ambulation (Figure 3F). Thus, we decided to study thermogenic activity of BAT under basal and cold-stress conditions. As expected, wild-type mice showed an increase in body temperature at night under basal conditions (22°C), which was not observed in Oma1-deficient mice (Figure 3G). To further evaluate their adaptive thermogenic response, Oma1+/+ and Oma1−/− mice were subjected to cold exposure at 4°C monitoring their temperature for 12 h. As can be seen in Figure 3H, Oma1−/− mice showed a significant decrease in body core temperature when compared with controls from 3 to 12 h. We analysed UCP1 protein levels after cold-stress but we did not find any differences, indicating that the observed failure in thermogenic response was not due to a decrease in UCP1-mediated uncoupling (data not shown). However, histological analysis of BAT showed that while control mice depleted lipid droplets under cold-stress, Oma1-deficient animals retained most of them (Figure 3I and J). Taken together, these results indicate that Oma1 ablation causes reduced energy expenditure under stress conditions, which may explain the susceptibility of these mice to obesity and their impaired adaptive thermogenic response.
Figure 3.
Decrease of energy expenditure and impaired thermogenesis in Oma1−/− mice. (A) Food-intake of 2-month-old Oma1+/+ and Oma1−/− mice kept on standard rodent chow (n=6). (B) Whole body oxygen consumption (ACC O2), (C) carbon dioxide production (ACC CO2), (D) heat production, (E) respiratory quotient (RQ) and (F) ambulatory movement of 2-month-old Oma1+/+ and Oma1−/− mice during dark period of 3 days kept on standard rodent chow (n=6–8). (G) Basal rectal temperature of Oma1+/+ and Oma1−/− mice measured during daytime (1000 hours) and night (2200 hours) for 3 consecutive days (n=5). (H) Body temperature curve of 2-month-old Oma1+/+ and Oma1−/− mice exposed to 4°C during 12 h (n=6). (I) Representation of BAT histology stained with H&E of Oma1-deficient mice and controls before and after cold exposure for 3 h. Scale bar: 50 μm. (J) The cumulative area occupied by all lipid droplets in each of the tissue sections was determined using image J and presented as a percentage of the total area analysed (n=4). *P<0.05; **P<0.01; ***P<0.001.
Next, we examined whether reduced respiration of Oma1-deficient mice could be explained by alterations in the expression of genes coding for mitochondrial proteins or for the nuclear co-activator PGC1α, an essential transcriptional regulator of mitochondria and oxidative metabolic programmes (Finck and Kelly, 2006; Uldry et al, 2006; Fernandez-Marcos and Auwerx, 2011). To this end, we obtained tissues from wild-type and Oma1-deficient mice chronically subjected to a high-fat diet (20 weeks) and analysed the expression of selected genes by real-time qPCR. BAT samples showed reduced expression of β-oxidation genes, Cpt1b and Vlcad in the Oma1-deficient group (Table I). Livers from Oma1-deficient mice showed a reduced expression of Ndufa9, Uqcrc2 and CoxIV (Table I). Furthermore, we detected increase in Pgc1a and decrease in Pgc1b, together with an increase in lipogenic genes Fasn and Scd1 in Oma1-deficient mice compared with controls (Table I). In addition, we also detected reduced expression of Sdha and Atp5a1 in skeletal muscle from Oma1-null mice (Supplementary Table S2). Collectively, these data indicate that Oma1 deficiency causes a reduced expression of nuclear genes encoding mitochondrial proteins, decrease in β-oxidation genes and increase in lipogenic genes, which may explain the low oxygen consumption and obesity phenotype of these animals. Notably, Oma1 deletion also led to a marked decrease in the expression of genes encoding proteins involved in mitochondrial fusion (Mfn2 or Opa1) or in mitochondrial fission (Drp1). In fact, Mfn2 was markedly down-regulated in BAT and WAT from Oma1-null mice, whereas Opa1 was down-regulated in BAT, WAT and skeletal muscle (Table I; Supplementary Table S2). Additionally, Drp1 was down-regulated in BAT, skeletal muscle and liver from Oma1-deficient mice (Table I; Supplementary Table S2).
Table 1. Expression of mitochondrial genes in BAT and liver of Oma1+/+ and Oma1−/− under high-fat diet.
| Brown adipose tissue |
Liver |
|||
|---|---|---|---|---|
| Oma1+/+ | Oma1−/− | Oma1+/+ | Oma1−/− | |
| Mitochondrial dynamics | ||||
| Mfn2 | 1±0.18 | 0.55±0.09* | 1±0.25 | 0.64±0.11 |
| Opa1 | 1±0.37 | 0.47±0.05 | 1±0.12 | 0.94±0.30 |
| Drp1 | 1±0.17 | 0.56±0.07* | 1±0.21 | 0.41±0.07* |
| OXPHOS genes | ||||
| Ndufa9 | 1±0.30 | 0.93±0.07 | 1±0.11 | 0.67±0.14* |
| SdhA | 1±0.42 | 0.64±0.10 | 1±0.14 | 1.18±0.33 |
| Uqcrc2 | 1±0.39 | 0.84±0.08 | 1±0.16 | 0.61±0.12* |
| CoxIV | 1±0.29 | 1.13±0.12 | 1±0.05 | 0.77±0.09* |
| Atp5a1 | 1±0.35 | 0.73±0.05 | 1±0.12 | 0.91±0.18 |
| Metabolic regulators | ||||
| Pgc1a | 1±0.11 | 0.82±0.24 | 1±0.01 | 1.62±0.19* |
| Pgc1b | 1±0.16 | 0.64±0.12 | 1±0.19 | 0.53±0.07* |
| β-Oxidation genes | ||||
| Mcad | 1±0.17 | 0.83±0.13 | 1±0.08 | 0.82±0.17 |
| Vlcad | 1±0.14 | 0.65±0.65* | 1±0.11 | 0.92±0.14 |
| Cpt1a | 1±0.15 | 1.07±0.21 | ||
| Cpt1b | 1±0.08 | 0.67±0.13* | ||
| Cpt2 | 1±0.11 | 0.82±0.24 | 1±0.10 | 0.84±0.11 |
| Lipogenic genes | ||||
| Fasn | 1±0.11 | 0.82±0.24 | 1±0.14 | 2.79±0.45** |
| Acc1 | 1±0.06 | 0.91±0.21 | 1±0.06 | 1.27±0.13 |
| Scd1 | 1±0.13 | 0.81±0.04 | 1±0.11 | 2.16±0.47* |
| Gene expression relative to β-actin. Data represent mean±s.e.m. of at least five mice per group. *P<0.05; **P<0.01. | ||||
Alteration of OPA1 processing in Oma1−/− mice
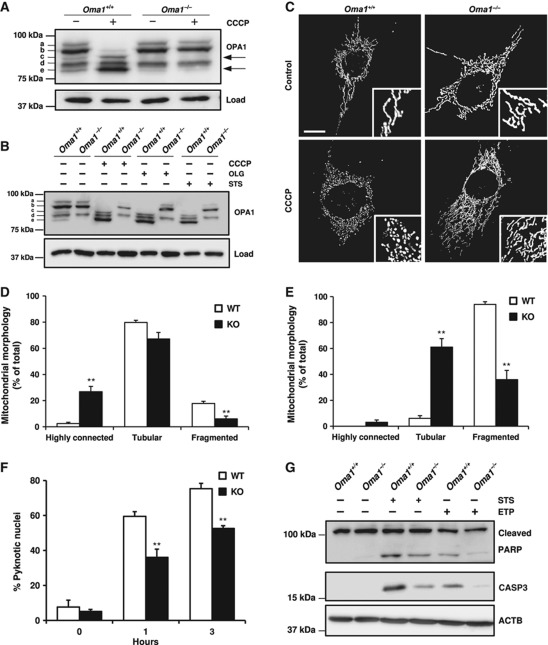
Recent in vitro studies have shown that OMA1 cooperates with m-AAA proteases in the proteolytic inactivation of OPA1 (Ehses et al, 2009; Head et al, 2009). To evaluate how mouse OMA1 induces this proteolytic inactivation, we first compared the levels of the different isoforms of OPA1 in murine embryonic fibroblasts (MEFs) from Oma1+/+ and Oma1−/− mice. In Oma1+/+ cells, we detected the five characteristic isoforms of OPA1 (labelled a–e) (Figure 4A). After treatment of these cells with the protonophore carbonylcyanide m-chlorophenylhydrazone (CCCP), an inducer of loss of mitochondrial membrane potential, we detected the conversion of bands b–e (Figure 4A). This proteolytic event involves the processing of large (L-OPA1) to short (S-OPA1) isoforms of OPA1 and protects cells from mitochondrial fusion under stress conditions. Interestingly, we could not detect the S-OPA1 isoform corresponding to band c in Oma1−/− MEFs, concomitant with a decrease in the production of band e (Figure 4A). Furthermore, we did not observe the cleavage of L-OPA1 (band b) to S-OPA1 isoforms (band e) in Oma1−/− cells treated with CCCP (Figure 4A). Similar results were obtained when cells were treated with staurosporine, a known apoptosis inducer, or with oligomycin, an inhibitor of ATP synthesis (Figure 4B). Analysis of tissue samples from Oma1-deficient mice also revealed alterations in OPA1 processing, characterized by the absence of isoform c in all cases (Supplementary Figure S5A and B). Interestingly, analysis of WAT samples from high-fat diet showed an increase in isoform e and a decrease in L-OPA1 isoforms in control mice, a characteristic pattern of stress conditions, whereas Oma1-deficient mice maintained all L-OPA1 isoforms intact (Supplementary Figure S5C). Accordingly, we conclude that OMA1 mediates the in vivo cleavage and inactivation of OPA1.
Figure 4.
Impaired OPA1 processing induces mitochondrial morphology alterations and apoptosis resistance in Oma1−/− mice. (A) Western blot analysis of OPA1 processing in Oma1+/+ and Oma1−/− MEFs after treatment with 20 μM of CCCP for 60 min. (B) Western blot analysis of OPA1 processing after treatment of Oma1+/+ and Oma1−/− MEFs with 20 μM CCCP, 20 μM oligomycine (OLG) and 1 μM staurosporine (STS) during 3 h. (C) Confocal images of Oma1+/+ and Oma1−/− MEFs in control cells and after CCCP treatment. Insets represent characteristic mitochondrial morphologies observed in each situation. Scale bar: 10 μm. (D) Mitochondrial morphology of Oma1+/+ and Oma1−/−. MEFs were visualized after transfection with a vector containing mito-DsRed, and (E) after treatment with 20 μM of CCCP during 3 h. More than 100 cells were scored. Bars represent mean values±s.e.m. and are shown as a percentage of the total, of three independent experiments (**P<0.01). (F) Percentage of apoptotic cells was determined by counting pyknotic nuclei after treatment of Oma1+/+ and Oma1−/− MEFs with 1 μM staurosporine during each time point. More than 150 cells were counted. Bars represent mean values±s.e.m. and are shown as a percentage of the total, of three independent experiments (**P<0.01). (G) Western blot analysis of PARP and activated caspase-3 as apoptosis markers in Oma1+/+ and Oma1−/− MEFs, after treatment of these cells with 1 μM staurosporine and 100 μM etoposide (ETP) during 3 and 16 h, respectively. Figure source data can be found in Supplementary data.
To further investigate the molecular consequences of Oma1 deletion on mitochondrial structure and function, we examined mitochondrial morphology in immortalized MEFs from Oma1+/+ and Oma1−/− mice. We observed that Oma1−/− MEFs show a lower percentage of mitochondrial fragmentation than control MEFs, as well as an increase in tubular connections (Figure 4C and D; Supplementary Figure S5D). We also examined the mitochondrial morphology of MEFs from mutant and control mice after treatment with CCCP. We observed that the CCCP-induced mitochondrial fragmentation is lower in Oma1−/− cells than in their corresponding controls (Figure 4E). Likewise, mutant cells predominantly maintain the tubular network morphology when compared with cells from wild-type mice (Figure 4E). Furthermore, after treatment with this inducer of membrane potential loss, we still observed a significant percentage of Oma1−/− cells that conserved the highly connected mitochondrial morphology (Figure 4E). Collectively, these results provide evidence that Oma1−/− cells are unable to perform the protease-mediated process of inactivation of OPA1 and are also refractory to the stress signals induced by CCCP, maintaining the mitochondrial tubular network morphology.
It has been previously reported that OPA1 is cleaved concomitantly with cytochrome c release during apoptosis (Cipolat et al, 2006; Baricault et al, 2007; Griparic et al, 2007). As can be seen in Figure 4B, proteolytic processing of OPA1 in Oma1-deficient cells was blocked after induction of apoptosis with staurosporine. To further evaluate the putative impact of Oma1 deficiency on apoptosis, Oma1−/− cells were treated with staurosporine and the number of apoptotic cells was determined. The percentage of pyknotic nuclei was significantly lower in Oma1-deficient cells than in controls (Figure 4F). Western blot analysis of PARP cleavage and activated caspase-3 levels confirmed the decreased sensitivity to apoptosis of Oma1-deficient cells after treatment with staurosporine and etoposide (Figure 4G). Taken together, these results indicate that Oma1 deficiency impairs cellular sensitivity to apoptosis.
Analysis of mitochondrial function in Oma1−/− cells
To determine whether the deficiency in OMA1 metalloproteinase has any effect on mitochondrial activity, we performed a series of functional experiments using Oma1−/− MEFs. First, we assessed the proliferation rate of Oma1-deficient MEFs but we did not observe significant differences with control cells (Supplementary Figure S6A). Similarly, we did not find significant differences in OXPHOS function in the absence of OMA1. In fact, coupled and CCCP-uncoupled respiration rates were comparable between Oma1+/+ and Oma1−/− cells, as well as oligomycin- and KCN-insensitive oxidation (Supplementary Figure S6B). Furthermore, the ratio between uncoupled and coupled respiration (Supplementary Figure S6C) and the polarography measurements of mitochondrial complexes did not show significant differences between Oma1+/+ and Oma1−/− cells (Supplementary Figure S6D). Moreover, ATP levels in mutant cells were very similar to those of control cells under both basal and oxidative stress conditions (Supplementary Figure S6E). Collectively, these results indicate that OMA1 does not play a major role in the maintenance of OXPHOS function. Analysis of mitochondrial inner membrane potential (ΔΨm) using TMRM fluorescence probe did not show differences between Oma1+/+ and Oma1−/− cells, in neither control nor CCCP-stimulated uncoupling conditions (Supplementary Figure S6F). However, we could detect a significant loss of mtDNA in Oma1-deficient cells (Supplementary Figure S6G). This decrease could not be explained by a decrease in mitochondrial mass as the activity of citrate synthase (Supplementary Figure S6H), a known marker of mitochondrial mass, and MitoTracker flow cytometry analysis (Supplementary Figure S6I) was comparable in cells from both genotypes. Thus, it is reasonable to speculate that the loss of OMA1 alters the copy number of mtDNA, likely as a consequence of defective OPA1 processing and the subsequent changes in the mitochondrial morphology.
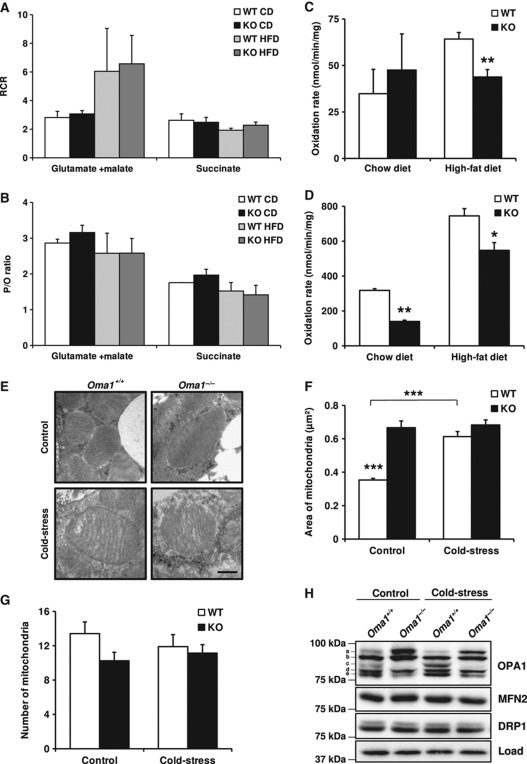
To study mitochondrial function in tissues, we analysed mitochondrial respiration rates in mitochondria isolated from livers of Oma1+/+ and Oma1−/− mice on control and high-fat diet. Thus, we measured the respiration rates in states 3 and 4 using two different combinations of substrates (glutamate+malate and succinate). Similar to the above cell analysis, we did not observe differences between the two genotypes in respiration control rate or ATP synthesis per O2 consumed—represented as P/O rate—under control and high-fat diets (Figure 5A and B; Supplementary Figure S7A). These results demonstrate that abnormalities observed in Oma1-deficient mice were not due to defects in the OXPHOS system. To analyse other possible pathways that could explain the phenotype observed in Oma1-deficient mice, we assessed fatty-acid utilization in the BAT and liver of Oma1−/− mice on both control and high-fat diets. As shown in Figure 5C and D, Oma1-deficient mice on a high-fat diet had a significant decrease in fatty-acid β-oxidation in BAT and liver compared with wild-type controls. Under a control diet, a significant decrease in fatty-acid utilization was found only in the BAT of Oma1−/− mice, indicating that the dysfunction in β-oxidation was due to primary defects in BAT and not a result of an inflammatory state caused by the obesity phenotype. However, and because the impaired β-oxidation was only observed in the livers of Oma1+/+ on a high-fat diet, we cannot exclude that inflammation may contribute to defective fatty-acid utilization in this tissue. Additionally, we measured citrate synthase activity in both liver and BAT from Oma1+/+ and Oma1−/− mice on control and high-fat diet, and did not find differences under these conditions between the two genotypes (Supplementary Figure S7B and C). These results demonstrate that differences in β-oxidation were due to specific mitochondrial defects and not attributable to a decrease in mitochondrial mass.
Figure 5.
Mitochondrial alterations in liver and BAT after obesity and cold-stress. Analysis of OXPHOS function in mitochondria isolated from liver of Oma1+/+ and Oma1−/− mice on control and high-fat diet. (A) Mitochondrial respiration control rate (RCR) and (B) P/O ratio, using glutamate+malate or succinate as substrates. Reduced fatty-acid utilization in Oma1-deficient mice. Mitochondrial fatty-acid oxidation was measured by the incubation of 14C-palmitate (100 μM) in liver (C) and BAT (D) extracts from mice under control diet and high-fat diet (n=4). (E) Electron microscopy (EM) images from BAT of Oma1+/+ and Oma1−/− mice on control and cold-stress. Scale bar: 0.4 μm. Mitochondrial morphology quantification in EM was measured with ImageJ, represented as (F) area of mitochondria and (G) number of mitochondria per field, using at least 12 different pictures of each condition. (H) Western blot analysis of OPA1, MFN2 and DRP1 in BAT samples extracted from mice after control and cold-stress treatment. Bars represent mean values±s.e.m. *P<0.05; **P<0.01; ***P<0.001. Figure source data can be found in Supplementary data.
Having identified a significant inability of Oma1-deficient mitochondria to oxidize fatty acids under control and high-fat diets in BAT, we next chose to explore the impact of Oma1 ablation on the morphology of mitochondria in this tissue by electron microscopy. Measurements of mitochondrial size showed a significant increase in mitochondrial area from Oma1−/− BAT compared with controls (Figure 5E and F). Moreover, we found that after cold-stress, Oma1+/+ BAT increased the size of mitochondria while Oma1−/− BAT maintained an identical mitochondrial size (Figure 5F). All of these changes were produced without significant differences in the mitochondrial number between control and Oma1-deficient mice (Figure 5G). Focusing on OPA1 processing after cold-stress, Oma1+/+ mice displayed a small but appreciable loss of isoform ‘a’ and a clear accumulation of isoform ‘c’, the isoform exclusively generated by OMA1 in response to cold-stress (Figure 5H). This pattern of OPA1 processing was absent in Oma1−/− BAT, indicating that cold-stress initiates OMA1-mediated processing of OPA1. Moreover, while OPA1 remodelling was detected in BAT, neither MFN2 nor DRP1 proteins showed a clear difference in either condition. Collectively, these results indicate that Oma1-null mice are unable to develop the appropriate mitochondrial dynamic response required for cold-stress-induced thermogenesis.
OMA1-OPA1 system is necessary for brown adipocytes function
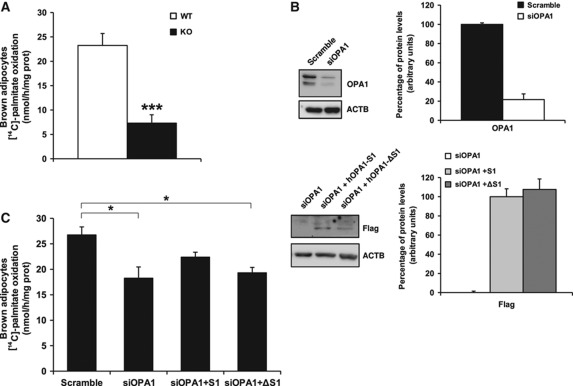
To explore the connection observed in BAT between OPA1 processing by OMA1 and β-oxidation function, we next analysed fatty-acid utilization in primary brown adipocytes from Oma1+/+ and Oma1−/− mice. Brown preadipocytes were isolated from newborn mice, immortalized and differentiated into mature brown adipocytes. We then analysed palmitate oxidation rates as an indicator of β-oxidation function in Oma1+/+ and Oma1−/− mature brown adipocytes. In agreement with our experiments in BAT (Figure 5D), Oma1-deficient adipocytes displayed significantly lower palmitate oxidation rates in comparison to wild-type control cells (Figure 6A). Next, to delineate the importance of OPA1 for brown adipocyte function, we ablated OPA1 protein levels in Oma1+/+ using siRNA and then analysed palmitate oxidation rates. This protocol involved first differentiating the primary brown preadipocytes for 2 days with insulin, T3, IBMX, dexamethasone and indomethacin, and then transfecting the cells with an siRNA targeting OPA1 mRNA or a scrambled control. The cells were maintained for a further 72 h with insulin and T3 to complete adipocyte differentiation and then analysed for palmitate oxidation rates. This approach reduced OPA1 protein levels by ∼75% in the final mature brown adipocytes (Figure 6B). Interestingly, knockdown of OPA1 decreased palmitate oxidation rates by ∼25% in Oma1+/+ brown adipocytes compared with cells transfected with the scrambled siRNA (Figure 6C). This indicates that OPA1 is required for the correct functioning of brown adipocyte β-oxidation. Further, to specifically demonstrate the relevance of OPA1 processing by OMA1 in brown adipocyte function, we restored OPA1 in these cells by reintroducing wild-type OPA1 or a mutant OPA1 isoform not cleavable by OMA1. Brown adipocytes were transfected with OPA1 siRNA to remove endogenous OPA1 protein as outlined previously, then 24 h later, we transfected a mammalian expression vector containing a wild-type full-length cDNA of OPA1 (OPA1-S1) or a mutant cDNA isoform of OPA1 without the S1 cleavage site that is utilized by OMA1 (OPA1-ΔS1) (Ishihara et al, 2006). Both constructs were insensitive to the siRNA targeting mouse Opa1 due to their human origin and lack of homology in the targeted region and contained a Flag epitope for detection by western blot (Figure 6B). Thus, reintroduction of full-length OPA1 in adipocytes partially restored the levels of palmitate oxidation (Figure 6C). However, mutant OPA1-ΔS1 did not demonstrate a similar ability to increase the levels of palmitate oxidation, displaying similar oxidation rates to those observed in OPA1-depleted cells (Figure 6C). These results indicate that β-oxidation in brown adipocytes requires the OMA1-OPA1 system, and an absence of OPA1 or its proteolytic regulator OMA1, impairs mitochondrial function.
Figure 6.
β-Oxidation in brown adipocytes depends on OMA1 and OPA1 function. (A) Analysis of palmitate oxidation rate in brown adipocytes obtained from Oma1+/+ and Oma1−/− mice (n=3). (B) OPA1, Flag-tag and actin western blot analyses from wild-type adipocytes after transfection with an siRNA scrambled control (Scramble) or Opa1 siRNA (siOPA1) (top panel, Opa1 western blot) or siOPA1 samples retransfected with hOPA1-S1 or hOPA1-ΔS1 (bottom panel, Flag western blot). To the right of each representative western blot, is the densitometry analysis from three independent experiments, normalized to actin levels. (C) Palmitate oxidation rate in wild-type brown adipocytes after 72 h of the above transfections. Bars represent mean values±s.e.m. of three independent experiments. *P<0.05; ***P<0.001. Figure source data can be found in Supplementary data.
In summary, our results indicate a pivotal role for the OMA1 protease in mitochondrial homeostasis by regulating the mitochondrial dynamics that is essential for lipid metabolism as well as to maintain body temperature and energy expenditure under cold-stress conditions.
Discussion
We describe herein the generation and phenotypic characterization of mutant mice deficient in OMA1, a mitochondrial metalloprotease proposed to be involved in protein quality control. Despite the wide tissue distribution of OMA1, the disruption of this gene does not cause the severe abnormalities observed in mice deficient in other mitochondrial proteases such as SPG7, AFG3L2 or PARL (Ferreirinha et al, 2004; Cipolat et al, 2006; Maltecca et al, 2008). However, further analysis of Oma1−/− mice revealed an unexpected contribution of this protease to metabolic homeostasis. In fact, Oma1-deficient mice exhibit a progressive increase in body weight when compared with control animals, and these differences are further enhanced when mice from both genotypes are maintained on a high-fat diet. Histological analysis showed a marked hypertrophy of adipocytes from Oma1−/− mice. These mutant animals also exhibited hepatic steatosis as well as a significant increase in plasma triglycerides and a decrease in circulating leptin and insulin levels. Moreover, glucose and insulin tolerance tests revealed that Oma1-null mice display improved glucose tolerance and increased insulin sensitivity. Furthermore, after high-fat diet induced obesity, Oma1-deficient mice lost glucose tolerance protection and developed insulin insensitivity, displaying higher levels of leptin than those found in obese controls. Finally, hepatic steatosis and triglyceride levels were also dramatically enhanced in mutant mice after high-fat diet.
The finding of this array of metabolic changes in Oma1−/− mice prompted us to explore their putative links to the absence of this mitochondrial metalloproteinase, which had not been previously associated with metabolic homeostasis. In this regard, there is growing evidence that a number of metabolic disorders are caused by alterations of mitochondrial function and dynamics (Detmer and Chan, 2007; Suen et al, 2008; Hyde et al, 2010; Westermann, 2010). There are many proteins implicated in the regulation of the appropriate balance between mitochondrial fission and fusion. In mammals, mitochondrial fission is largely controlled by the dynamin-related GTPase DRP1 (Chang and Blackstone, 2010), whereas mitochondrial fusion mainly depends on the activity of three large GTPases, the mitofusins (MFN1 and MFN2) and OPA1 (Hoppins and Nunnari, 2009; Zorzano et al, 2010). On the basis of these previous findings, we speculated that the absence of OMA1 could alter the levels of some of these proteins involved in modulating mitochondrial dynamics, shifting the balance towards fusion or fission events, and finally resulting in the metabolic alterations observed in Oma1-mutant mice. In fact, our proteomic analysis of cells and tissues from Oma1−/− mice revealed that OMA1 plays an in vivo non-redundant role in the proteolytic processing of OPA1, which leads to its functional inactivation under stress conditions, thus extending recent in vitro findings showing that OPA1 can be targeted by OMA1 (Ehses et al, 2009; Head et al, 2009). There are at least five OPA1 isoforms in mammalian cells, including two long forms (L-OPA1) and three short isoforms (S-OPA1) (Duvezin-Caubet et al, 2006; Ishihara et al, 2006; Griparic et al, 2007; Song et al, 2007). The precise molecular balance between these long and short isoforms is necessary for mitochondrial fusion to occur. However, under stress conditions such as loss of mitochondrial membrane potential, low levels of mitochondrial ATP or induction of apoptosis, L-OPA1 isoforms are cleaved to S-OPA1 isoforms and mitochondrial fusion is blocked (Song et al, 2007). Our finding that Oma1−/− MEFs lack one isoform (‘c’) and have a decrease in another isoform (‘e’) of the three S-OPA1 isoforms, clearly indicate that OMA1 is responsible for the in vivo proteolytic processing of L-OPA1 isoforms. Notably, even under stress conditions, Oma1−/− cells display the same pattern of OPA1 isoforms characterized by the lack of processing of L-OPA1, which finally results in the shift of mitochondrial dynamics towards fusion events. Tissue samples from Oma1-deficient mice also lack one S-OPA1 isoform in all cases (isoform ‘c’), although the pattern of OPA1 bands exhibits some differences with that of MEFs, which can be due to variations in compensatory or redundant proteolytic activities operating in the different cells and tissues from Oma1-null mice under normal conditions. The OPA1-processing deficiency was also clearly evident in BAT samples from mutant mice after cold-stress and in WAT samples after high-fat diet, as assessed by the maintenance of all L-OPA1 isoforms and the absence of any increase in S-OPA1 bands. Collectively, these results demonstrate that both cells and tissues from Oma1-null mice exhibit an in vivo impairment of OPA1 inactivation, which is especially significant under stress conditions. Consistent with this proteolytic processing deficiency, Oma1−/−-deficient cells show an increase of highly connected and less-fragmented mitochondria when compared with control cells. Likewise, after treatment of cells with CCCP, which induces loss of mitochondrial membrane potential, most mitochondria from Oma1-deficient cells maintain their archetypal tubular network, whereas those present in control cells show a fragmented morphology. We found the same protection against mitochondrial fission when Oma1-mutant cells were treated with a respiratory chain inhibitor such as oligomycin or with an apoptosis inducer such as staurosporine, demonstrating that even under different stress conditions, the absence of OMA1 imposes a pro-fusion phenotype in mitochondria from Oma1-mutant mice.
In addition to its mitochondrial pro-fusion activity, OPA1 plays an important role in the maintenance of cristae junctions of the organelle, protecting cells from apoptosis by preventing cytochrome c release and mitochondrial depolarization (Olichon et al, 2003; Cipolat et al, 2006; Frezza et al, 2006). Accordingly, we also examined whether the defects in L-OPA1 proteolytic processing observed in Oma1-deficient cells could have an impact on apoptosis in these cells. In fact, Oma1−/− cells display low sensitivity to apoptosis compared with control cells, thus demonstrating that the absence of OMA1 metalloprotease has a profound impact on both pro-fusion and anti-apoptotic activities of OPA1. The proposed implication of OMA1 in the OPA1-mediated control of cristae remodelling and cytochrome c release during apoptosis likely derives from its ability to cleave L-OPA1 isoforms. In fact, these transmembrane isoforms keep in check the cristae junctions through the formation of stabilizing oligomeric structures with soluble short isoforms of OPA1 (Cipolat et al, 2006; Frezza et al, 2006). Since Oma1-deficient cells maintain their L-OPA1 isoforms intact, even under stress conditions, and still have available some soluble S-OPA1 isoform, the tight structural organization of cristae junctions is completely preserved. This situation prevents the redistribution and release of cristae-stored cytochrome c and finally results in apoptosis impairment. Notably, we also observed a significant decrease of mtDNA levels in Oma1−/− cells, which was not a consequence of any putative mitochondrial mass decrease between normal and mutant cells. The recent finding that OPA1 is also linked to mtDNA stability (Landes et al, 2010) suggests that the OPA1-processing deficiency found in mitochondria from Oma1−/− cells could contribute to explain why these cells are prone to mtDNA loss.
Interestingly, despite all these mitochondrial abnormalities observed in Oma1−/− cells, including inhibition of mitochondrial fission and loss of mtDNA, mitochondria deficient in OMA1 metalloprotease do not display marked respiratory defects, indicating that this enzyme is dispensable for OXPHOS function. This somewhat paradoxical situation is not unprecedented, as mitochondria deficient in rhomboid PARL1 protease do not exhibit primary respiratory defects in a variety of cell types, although this enzyme is also involved in OPA1 processing (Cipolat et al, 2006). These observations reinforce the usefulness of in vivo models to define the specific functions of proteins, which form part of complex and redundant regulatory pathways like those involving proteolytic systems (Lopez-Otin and Bond, 2008; Lopez-Otin and Hunter, 2010). In fact, over recent years, several mitochondrial proteases from different catalytic classes, including the serine protease PARL1 and several ATP-dependent and -independent metalloproteases, have been implicated in the constitutive or regulated proteolytic processing of mammalian OPA1 (Cipolat et al, 2006; Ishihara et al, 2006; Griparic et al, 2007; Song et al, 2007). These studies were initially based on in vitro experiments involving cell cultures and pointed to a large degree of overlapping functions between candidate OPA1-processing proteases. However, the recent introduction of studies based on animal models of protease deficiency such as those presented herein, has finally demonstrated the non-redundant in vivo role of enzymes such as OMA1 in the proteolytic regulation of the mitochondrial function of the dynamin-related GTPase OPA1.
It is also remarkable that the absence of significant pathological alterations in adult mice deficient in OMA1 facilitated the identification of the obesity phenotype, which has led us to propose a new and unexpected role for this mitochondrial protease in metabolic control. How can we explain that the deficient OPA1 processing observed in Oma1-null mice leads to an obesity phenotype? Our data indicate that Oma1 deficiency causes reduced energy expenditure, which agrees with previous results, indicating that a reduction in energy expenditure causes obesity in animal models (Lowell et al, 1993; Feldmann et al, 2009). Accordingly, we suggest that the reduced energy expenditure in Oma1-deficient mice may account for their increased adipose mass. In addition, Oma1 deficiency causes a reduced expression of nuclear genes encoding for mitochondrial proteins and key metabolic transcription co-activators, and this may be instrumental in the reduced energy expenditure shown in Oma1-null mice. Moreover, deregulation in lipid metabolism due to defects in β-oxidation genes observed in Oma1-deficient mice contributes to the obesity and thermoregulation defects. In this respect, we can speculate that altered OPA1 processing induces changes in mitochondrial dynamics, modifying expression levels of dynamic proteins to compensate or counteract this deficiency. Thus, under physiological conditions and in the absence of any relevant stressor, the lack of OMA1 induces changes in mitochondrial dynamics without seriously affecting any physiological function. In this situation, Oma1-deficient mice display a modest increase in body weight, improvement in glucose metabolism, hepatic steatosis, decrement in energy expenditure and low body temperature. However, under stress conditions such as a high-fat diet, the lack of OMA1 induces obesity, loss of glucose metabolism improvement and decrease in β-oxidation that induce the marked increase in the expression of genes of the lipogenic pathway and manifests as the significant hepatic steatosis observed in Oma1-deficient mice. Likewise, cold-stress induces impaired thermogenic response diminishing core body temperature, due to defects in β-oxidation and dysfunction in mitochondrial dynamics in response to cold-stress. Interestingly, a recent in vitro study using 3T3-L1 adipocytes has described a new function for OPA1 as a dual-specificity A-kinase anchoring protein, which interacts with perilipin and PKA forming a supramolecular complex involved in the lipolysis of lipid droplets (Pidoux et al, 2011). We did not observe any change in perilipin phosphorylation in Oma1-deficient BAT in comparison to normal tissue (data not shown), although this does suggests that perilipin functions normally in Oma1−/− BAT and likely does not contribute to the numerous defects we have described in these mutant mice, it may function in other tissues, for example white adipose, a possibility that will require further detailed examination in the future. Finally, and in relation to the putative implications of these findings for human metabolic control, several genome-wide linkage studies have found evidence of a quantitative trait locus for human obesity phenotypes in the region harbouring OMA1 (Saar et al, 2003; Choquette et al, 2008). Nevertheless, further epidemiological studies will also be necessary to clarify the putative relevance of OMA1 mutations or polymorphisms in abnormal weight control and glucose metabolism in humans.
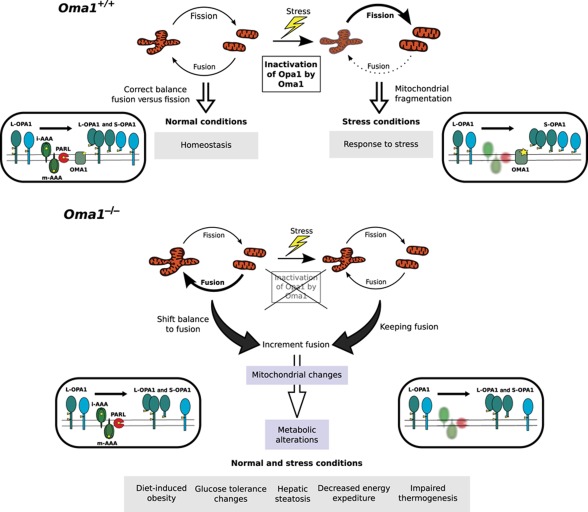
In summary, the generation and characterization of Oma1-deficient mice has contributed to clarifying the in vivo functional role of this metalloproteinase in mitochondrial quality control and metabolic homeostasis (Figure 7). The first steps in these processes are performed by mitochondrial peptidases, which play direct roles in the degradation of misfolded and damaged proteins or participate in the proteolytic processing of proteins that form part of the mitochondrial dynamics machinery. OMA1 belongs to this second category of mitochondrial proteases due to its ability to target OPA1. Accordingly, Oma1-deficient cells have marked alterations in mitochondrial dynamics due to the incomplete processing of L-OPA1. This perturbation of the fusion–fission equilibrium in Oma1−/− cells has clear implications for metabolic homeostasis and causes a series of energy metabolic derangements, which finally result in abnormal weight control, hepatic steatosis and defective thermogenesis. The absence of OMA1 is especially significant under stress conditions, indicating that an intact OMA1-OPA1 system is essential for developing the appropriate adaptive response to different metabolic stressors such as high-fat diet or cold-shock. However, we cannot discard that, despite all of these metabolic derangements observed, other potential substrates/interacting proteins for OMA1 may be involved in this process. The marked transcriptional changes observed in genes of lipid and glucose metabolic pathways and the substantial alterations in circulating blood parameters found in Oma1-mutant mice, are likely part of an exacerbated or anomalous adaptive response, which finally fails in the absence of OMA1 and leads to the phenotypes observed in these mice including obesity and defective thermogenesis. Hopefully, the identification of OMA1 as a proteolytic enzyme involved in the regulation of energy metabolism may open new ways for a better understanding of human obesity, a major medical problem that is reaching epidemic dimensions worldwide.
Figure 7.
Schematic model summarizing the functional relevance of OMA1 in mitochondrial dynamics and metabolic homeostasis.
Materials and methods
Generation of Oma1−/− mice
We first constructed the targeting vector in pKO scrambler V916 (Lexicon Genetics). A 4.3-kb SalI–NotI fragment from 5′-flanking region, exon 1 and part of intron 1 was used as the 5′-homologous region, whereas a 3.2-kb XhoI–ClaI fragment containing part of intron 2 and exon 3 was used as the 3′-region of homology. The 2.4-kb neo cassette was used as a positive marker and replaced a 2.9-kb fragment containing exon 2 of the Oma1 gene. The thymidine kinase marker was used as negative selection (Supplementary Figure S1A). The targeting vector was linearized by digestion with NotI, electroporated into G4 embryonic stem cells, and selected for homologous recombination with G418 and ganciclovir. Positive clones were screened by Southern blot after PvuII digestion of genomic DNA and probed with radiolabelled 3′-external probe. A 7.5-kb fragment was detected from wild-type allele and a 4.3-kb fragment from mutant allele. The targeted ES cells clones were expanded and subsequently injected into blastocysts to generate chimeras. Chimeric males were mated with C57BL/6 female mice and the offspring heterozygous for Oma1 were used to generate homozygous null mice. In all cases, mice genotypes were determined by PCR analysis of tail DNA.
Animal care
All animal procedures were approved in accordance with the guidelines of the Committee for Animal Experimentation of the Universidad de Oviedo. For diet-induced obesity, 4-week-old Oma1-deficient mice and their wild-type littermates (C57BL/6/129Sv) were kept in microisolation cages on a 12-h day–night cycle and fed a high-fat diet containing 42% fat (Harlan TD 88137). Mice were weighted once a week for all the experiments. After 24 weeks, mice were sacrificed by cervical dislocation.
Histological analysis
Tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and stored in 70% ethanol. Fixed tissues were embedded in paraffin by standard procedures. Blocks were sectioned (5 μm) and stained with haematoxylin and eosin (H&E). For adipose tissue evaluation, portions of gonadal and subscapular fat pads and pieces of skin were processed as described above. The number of adipocytes and their mean diameter were determined in 3 μm tissue sections by computer-assisted image analysis. For each sample, different sections were analysed and 100 adipocytes were measured. The area of cells was calculated considering the maximum and minimum diameter of each adipocyte. For triglyceride detection, liver samples were fixed with 4% formaldehyde and left overnight at 4°C. After that, samples were treated with 15% sucrose in PBS for 4 h, and then with 30% sucrose solution overnight, embedded in Tissue-Tek OCT compound (Sakura Finetechnical Co. Ltd.) and stored at −70°C. Finally, samples were sectioned at 5 μm thickness with cryostat (CM3050 S, Leica) and cryosections were stained with Oil Red O. For BAT droplets quantification, pictures from BAT sections stained with H&E were analysed with Image J software.
Blood and plasma parameters
Animals fed on regular and high-fat diet were fasted overnight and used for measurements of blood and plasma parameters. Blood glucose was measured with Accu-Chek glucometer (Roche Diagnostics) using blood from the tail vein. For all the other measured parameters, blood was extracted directly from heart after anaesthetizing mice with halothane. Plasma was obtained as previously described. Briefly, blood was immediately centrifuged after collection at 3000 g and 4°C, and the supernatant was collected and stored at −80°C until analysis. For plasma insulin, leptin and free fatty-acid measurements, we used Millipore ELISA Kits. All protocols were performed according to the manufacturer's instructions. Levels of hepatic transaminases, cholesterol and triglycerides were determined in the Servicio de Bioquímica, Hospital Universitario Central de Asturias, Oviedo, Spain.
Glucose and insulin tolerance test
Prior to studies, mice were fasted overnight. For IPGTT, mice received an intraperitoneal injection of glucose (2 mg/g body weight). In IPITT studies, mice received an intraperitoneal injection of 1 U of insulin per kg of body weight. Blood glucose levels were determined as describe above. Areas under the curve during IPGTT and IPITT were then calculated by the linear trapezoidal method.
Indirect calorimetry
Energy expenditure and ambulation were assessed using Micro-Oxymax multiple sensor (Columbus Instruments). Oma1+/+ and Oma1−/− mice kept on standard rodent chow were housed individually in Oxymax chambers and kept on a 12:12-h light–dark cycle. Constant airflow (0.5 l/min) was drawn through each chamber. After 1 day of acclimation, O2 consumption, CO2 production, RQ, heat and ambulation parameters were recorded during 72 h, using software provided by Columbus Instruments.
RNA preparation
Collected tissue was immediately homogenized in TRI reagent (Sigma-Aldrich, St Louis, MO, USA) and processed in the same day through alcohol precipitation according to the manufacturer's instructions. RNA pellets were then washed in cold 80% ethanol and stored at −80°C until further use. Following resuspension of RNA in nuclease-free water (Ambion, Austin, TX, USA), the samples were quantified and evaluated for purity (260/280 nm ratio) using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and 100 mg of each sample was further purified using RNeasy spin columns according to the manufacturer's instructions (Qiagen, Valencia, CA, USA).
Transcriptional profiling and gene expression analysis
The full transcriptome analysis was conducted using Affymetrix Mouse Gene 1.0 ST Array according to the protocols recommended by the manufacturer (Affymetrix, Santa Clara, CA, USA). Total RNA was isolated as described above from mice tissues maintained under the same high-fat diet and light/dark cycle conditions. The RNA integrity was assessed using Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). Labelling and hybridizations were performed according to the protocols from Affymetrix. Washing and scanning were performed using Fluidics Station 400 and GeneChip Scanner (Affymetrix). After scanning, raw data were processed with RMAExpress (http://RMAExpress.bmbolstad.com), using default settings. DAVID, Ingenuity and OpenOffice software were used for data analysis. The DAVID web portal was used to calculate statistical enrichment of KEGG pathways and Gene Ontology biological processes for each group (Dennis et al, 2003; Huang da et al, 2009). The network analyses were generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems; http://www.ingenuity.com).
Real-time quantitative PCR
cDNA was synthesized using 1–5 μg of total RNA, 0.14 mM random hexamer primer, 0.2 mM of each deoxynucleoside triphosphate and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Quantitative reverse transcription–PCR (qRT–PCR) was carried out in triplicate for each sample using 20 ng of cDNA, TaqMan® Universal PCR master mix (Applied Biosystems, San Francisco, CA, USA), and 1 μl of the specific TaqMan custom gene expression assay for the gene of interest (Applied Biosystems). To quantitate gene expression, PCR was performed at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 60°C for 30 s and 72°C for 30 s using an ABI Prism 7300 sequence detector system. As an internal control, gene expression was normalized to the mouse β-actin gene using the Mouse β-actin Endogenous Control (VIC®/MGB Probe, Primer Limited) TaqMan gene expression assay (Applied Biosystems). Relative expression of the analysed genes was calculated according to the manufacturer's instructions. Briefly, the analysed gene expression was normalized to β-actin in wild-type or Oma1−/− derived samples, using the following formula: the mean value of 2ΔCTgene (Gene of interest)−ΔCTgene (β-actin) for at least six different wild-type animals were considered 100% for each analysed gene and the same values for Oma1−/− mice tissues were referred to those values as previously described.
Western blotting
Mice tissues were immediately frozen in liquid nitrogen after extraction, then homogenized in 50 mM Tris buffer, pH 7.4, containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA and complete protease inhibitor cocktail (Roche Applied Science). Once homogenized, tissue extracts were centrifuged at 13 000 g and 4°C for 5 min, and supernatants were collected. The protein concentration of the supernatant was evaluated by bicinchoninic acid technique (BCA protein assay kit; Pierce Biotechnology Inc.). A protein sample (15 μg) was loaded on SDS–polyacrylamide gels. After electrophoresis, gels were electrotransferred onto PVDF filters, and the filters were blocked with 5% non-fat dried milk in TBS-T (TBS with 0.05% Tween-20) and incubated with primary antibodies following the commercial instructions. After three washes with TBS-T, filters were incubated with the corresponding secondary antibody in 1.5% milk in TBS-T, and developed with Immobilon Western Chemiluminescent HRP substrate (Millipore) in a LAS-3000 Imaging System (Fujifilm). The antibodies against OPA1 and DRP1 were from BD Bioscience, and the antibody against MFN2 was from Abcam. All the other antibodies used in this work were from Cell Signaling.
Cell culture and microscopy analysis
Primary MEFs were extracted from E13.5 embryos. Briefly, embryos were sterilized with ethanol, washed with PBS and triturated with razor blades. Samples were then incubated in Dulbecco's modified Eagle's medium (DMEM) (Gibco) overnight at 37°C and 5% CO2. The next day, cultured cells were trypsinized and washed. Finally, MEFs were incubated at 37°C and 5% CO2 and used for the corresponding experiments. Adult murine fibroblasts (AFs) were extracted from 12-week-old mice ears. Ears were sterilized with ethanol, washed with PBS and triturated with razor blades. Samples were then incubated with 600 μl of 4 mg/ml collagenase D (Roche Applied Science) and 4 mg/ml dispase II (Roche Applied Science) in DMEM (Gibco) for 45 min at 37°C and 5% CO2. After filtering and washing, 6 ml of DMEM with 10% fetal bovine serum (Invitrogen), and 1% antimycotic-antibiotic (Invitrogen) were added, and the mixture was incubated at 37°C and 5% CO2. Once extracted, cells were cultured in DMEM containing 10% fetal bovine serum at 37°C and 5% CO2. Brown preadipocytes were isolated from newborn mice by collagenase digestion and were differentiated to mature adipocytes as previously described (Fasshauer et al, 2000). Immortalized MEFs, AFs and preadipocytes were generated by SV40 transformation. For fluorescence microscope analyses, mitochondrial morphology was examined after transfection of vector containing mito-DsRed, and images were obtained using a laser-scanning confocal microscope (TCS-SP2-AOBS; Leica) using HCX PL APO Ibd BL × 63 NA 1.4 objective lens. Images were acquired with LCS Suite version 2.61 (Leica), and then modified with ImageJ, using the maxima projection of 0.2 μm stacks images after convolve, median filter and despeckle modification. Brightness/contrast was adjusted using ImageJ.
Transmission electron microscopy
BAT samples were dissected from control and cold-stress-induced mice and fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 1 h at 4°C. After three washes in 0.1 M Sorensen's phosphate buffer, samples were postfixed with 1% osmium tetroxide for 1 h and rinsed three times in Sorensen's buffer. Tissues were dehydrated in graded ethanol and infiltrated in 50% propylene oxide/50% resin for 1 h, then placed in resin for 1 h, and embedded in a resin-based mould for polymerization overnight in a 37°C oven. Ultra-thin sections (85 nm) were taken from each sample and analysed on a Jeol (JEM-1011) transmission electron microscope at 80 kV. Images were modified using ImageJ and Gimp 2.6 software.
Proliferation assays
Growth capacity was determined in six-well test plates in which 6 × 104 cells were plated per well in 2.5 ml of DMEM medium and incubated at 37°C for 72 h. Cells were daily counted by using a Neubauer chamber. Population doubling time was calculated using the algorithm provided by http://www.doubling-time.com.
Mitochondrial ATP content
Cellular ATP levels were determined using the ATP determination kit (Molecular probes, Invitrogen) according to the instructions of the manufacturer. Briefly, MEFs were collected by trypsinization and lysed using PLB (Promega). The lysates were centrifuged at 12 000 g and 4°C for 5 min, and supernatants were collected. The protein concentration of the supernatant was evaluated by bicinchoninic acid technique (BCA protein assay kit, Pierce Biotechnology, Rockford, IL, USA). We used 3 μg of lysated material for ATP determination with a D-luciferin/firefly luciferase reaction mix, and luminescence was measured in a Luminometer TD20/20 and compared with a freshly prepared ATP standard curve.
Mitochondrial DNA copy number quantification
We quantified mtDNA by real-time PCR using ABI PRISM 7300 Sequence Detector System (Applied Biosystems) and TaqMan Universal PCR master mix (Applied Biosystems). Total DNA was used as a template and amplified with specific oligodeoxynucleotides for mt-Co2 and Sdha. We calculated the mtDNA copy number per cell using Sdha amplification as a reference for nuclear genome (Moreno-Loshuertos et al, 2006).
Citrate synthase activity
We determined mitochondrial citrate synthase activity in whole MEFs lysates and tissues homogenates. Briefly, MEFs were collected by trypsinization, washed twice with PBS and resuspended in buffer A (0.25 M sucrose and 20 mM MOPS-KOH, pH 7.4). Then, we added digitonin to a final concentration of 0.1 mg/ml, and cells were kept on ice for 5 min and centrifuged at 5000 g and 4°C for 5 min. The pellets were resuspended in buffer B (0.25 M sucrose, 1 mM EDTA and 20 mM MOPS-KOH, pH 7.4), kept on ice 5 min and centrifuged again at 10 000 g. The final mitochondrial fraction pellets were resuspended in 10 mM of potassium phosphate buffer (pH 7.4) and samples were freezed-thawed three times in liquid nitrogen before use. For tissue analysis, homogenates were prepared as described in palmitate oxidation protocol. Citrate synthase activity was determined spectrophotometrically at 30°C as the reduction rate of 100 μM DTNB (5,5′-dithio-bis-2-nitrobenzoic acid) at 412 nm (ε=13.8/mM/cm) after the addition of 400 μM acetyl-Coenzyme A and 500 μM oxalacetate (Sigma-Aldrich).
Oxygen consumption measurements
Endogenous respiration measurements in intact cells or polarography in digitonin-permeabilized cells were performed using a Clark type oxygen electrode (Hansatech) as previously described (Hofhaus et al, 1996; Acin-Perez et al, 2003). Briefly, for the O2 consumption determinations in intact cells to measure the maximum respiration capacity, exponentially growing cells were collected by trypsinization and centrifugation, and resuspended at 6.5 × 106 cells/ml in 0.75 ml of DMEM containing glucose (4.5 g/l), supplemented with 10% FBS. The cell suspension was transferred to the electrode's 1.5 ml water-jacketed chamber containing a small magnetic bar, thermostated at 37°C. Recording of oxygen consumption was carried out for 150 s. Then, 65 nmol 2,4-dinitrophenol (DNP; Sigma) were added to uncouple the respiration and oxygen consumption was monitored for an additional 150 s to determine the maximum O2 consumption. For polarographic measurements of the individual respiratory complexes, cells were collected by trypsinization and centrifugation, and resuspended at 2–5 × 106 cells/ml in Respiration Buffer (250 mM HEPES, pH 7.1; 250 mM sucrose, 10 mM MgCl2, 1 mM ADP and 2 mM potassium phosphate). Cells were permeabilized using 10–20 μg digitonin per 106 cells and the oxygen consumption rates were determined using specific substrates and inhibitors for each of the respiratory complexes: glutamate+malate and rotenone for complex I; succinate+glycerol-3-phosphate and antimycin A for complex III; TMPD and KCN for complex IV.
Palmitate oxidation measurement
Oxidation of [1-14C]-palmitic acid by tissue homogenates was performed as previously described (Hirschey et al, 2010). Briefly, liver and BAT samples were extracted and homogenized in sucrose/Tris/EDTA buffer, incubated for 120 min in the reaction mixture (pH 8.0) containing [1-14C]-palmitic acid, and measured for acid-soluble metabolites. Palmitate oxidation rates in primary brown adipocytes were quantified as described (Antinozzi et al, 1998) and corrected for total cellular protein content.
Statistical analysis
All experimental data are reported as mean and the error bars represent the standard error of the mean (s.e.m.). Differences between mean values were analysed by two-tailed Student's t-test. A value of P<0.05 was considered significant. Statistical significant differences are shown with asterisks.
Accession numbers
The Affymetrics GeneChip data set is publicly available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE27525.
Supplementary Material
Acknowledgments
We thank G Mariño, AR Folgueras, A Astudillo and JM Freije for helpful comments and T Langer for OPA1 expression plasmids. We also thank FV Álvarez, JC Fernández and E Lapuente-Brun for help in blood analysis and cell culture, and M Fernández, D Puente, A Moyano, R Feijoo and MS Pitiot for excellent technical assistance. This work was supported by grants from Ministerio de Ciencia e Innovación-Spain and the European Union (FP7-Microenvimet). The Instituto Universitario de Oncología was supported by Obra Social Cajastur-Asturias and Acción Transversal del Cáncer-RTICC, Spain. CL-O is an Investigator of the Botin Foundation.
Author contributions: PMQ and AJR performed the experiments and analysed the data; FR participated in the generation of knockout mice; DS and AZ designed and carried out energy expenditure and ambulation experiments in mice; EFV and JAE carried out and analysed oxygen consumption measurements; JRP collaborated in blood analysis; MSFG analysed histological samples; JAV carried out electronic microscopy analysis; PMQ, AJR and CLO conceived, designed and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acin-Perez R, Bayona-Bafaluy MP, Bueno M, Machicado C, Fernandez-Silva P, Perez-Martos A, Montoya J, Lopez-Perez MJ, Sancho J, Enriquez JA (2003) An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum Mol Genet 12: 329–339 [DOI] [PubMed] [Google Scholar]
- Antinozzi PA, Segall L, Prentki M, McGarry JD, Newgard CB (1998) Molecular or pharmacologic perturbation of the link between glucose and lipid metabolism is without effect on glucose-stimulated insulin secretion. A re-evaluation of the long-chain acyl-CoA hypothesis. J Biol Chem 273: 16146–16154 [DOI] [PubMed] [Google Scholar]
- Baricault L, Segui B, Guegand L, Olichon A, Valette A, Larminat F, Lenaers G (2007) OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313: 3800–3808 [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93: 973–983 [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C (2010) Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann NY Acad Sci 1201: 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC (2009) Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 18: R169–R176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquette AC, Lemieux S, Tremblay A, Chagnon YC, Bouchard C, Vohl MC, Perusse L (2008) Evidence of a quantitative trait locus for energy and macronutrient intakes on chromosome 3q27.3: the Quebec Family Study. Am J Clin Nutr 88: 1142–1148 [DOI] [PubMed] [Google Scholar]
- Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS (2005) Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 46: 2347–2355 [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, Derks C, Dejaegere T, Pellegrini L, D’Hooge R, Scorrano L, De Strooper B (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126: 163–175 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Detmer SA, Chan DC (2007) Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8: 870–879 [DOI] [PubMed] [Google Scholar]
- Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, Veneziano L, Sacco T, Boda E, Brussino A, Bonn F, Castellotti B, Baratta S, Mariotti C, Gellera C, Fracasso V et al. (2010) Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat Genet 42: 313–321 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, Neupert W, Reichert AS (2006) Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem 281: 37972–37979 [DOI] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Ueki K, Kriauciunas KM, Benito M, White MF, Kahn CR (2000) Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J Biol Chem 275: 25494–25501 [DOI] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J (2009) UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Auwerx J (2011) Regulation of PGC-1{alpha}, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93: 884S–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreirinha F, Quattrini A, Pirozzi M, Valsecchi V, Dina G, Broccoli V, Auricchio A, Piemonte F, Tozzi G, Gaeta L, Casari G, Ballabio A, Rugarli EI (2004) Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J Clin Invest 113: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Kelly DP (2006) PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189 [DOI] [PubMed] [Google Scholar]
- Fulda S, Galluzzi L, Kroemer G (2010) Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G (2004) The pathophysiology of mitochondrial cell death. Science 305: 626–629 [DOI] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, van der Bliek AM (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 187: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV Jr, Alt FW, Kahn CR, Verdin E (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G, Shakeley RM, Attardi G (1996) Use of polarography to detect respiration defects in cell cultures. Methods Enzymol 264: 476–483 [DOI] [PubMed] [Google Scholar]
- Hoppins S, Nunnari J (2009) The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta 1793: 20–26 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hyde BB, Twig G, Shirihai OS (2010) Organellar vs cellular control of mitochondrial dynamics. Semin Cell Dev Biol 21: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K (2006) Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25: 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser M, Kambacheld M, Kisters-Woike B, Langer T (2003) Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J Biol Chem 278: 46414–46423 [DOI] [PubMed] [Google Scholar]
- Koppen M, Langer T (2007) Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 42: 221–242 [DOI] [PubMed] [Google Scholar]
- Landes T, Leroy I, Bertholet A, Diot A, Khosrobakhsh F, Daloyau M, Davezac N, Miquel MC, Courilleau D, Guillou E, Olichon A, Lenaers G, Arnaune-Pelloquin L, Emorine LJ, Belenguer P (2010) OPA1 (dys)functions. Semin Cell Dev Biol 21: 593–598 [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Bond JS (2008) Proteases: multifunctional enzymes in life and disease. J Biol Chem 283: 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Hunter T (2010) The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer 10: 278–292 [DOI] [PubMed] [Google Scholar]
- Lowell BB, S-Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS (1993) Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366: 740–742 [DOI] [PubMed] [Google Scholar]
- Maltecca F, Aghaie A, Schroeder DG, Cassina L, Taylor BA, Phillips SJ, Malaguti M, Previtali S, Guenet JL, Quattrini A, Cox GA, Casari G (2008) The mitochondrial protease AFG3L2 is essential for axonal development. J Neurosci 28: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Rizzuto R (2010) Signaling pathways in mitochondrial dysfunction and aging. Mech Ageing Dev 131: 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA (2006) Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet 38: 1261–1268 [DOI] [PubMed] [Google Scholar]
- Navarro CL, Cadinanos J, De Sandre-Giovannoli A, Bernard R, Courrier S, Boccaccio I, Boyer A, Kleijer WJ, Wagner A, Giuliano F, Beemer FA, Freije JM, Cau P, Hennekam RC, Lopez-Otin C, Badens C, Levy N (2005) Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum Mol Genet 14: 1503–1513 [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G (2003) Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278: 7743–7746 [DOI] [PubMed] [Google Scholar]
- Orrenius S, Gogvadze V, Zhivotovsky B (2007) Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol 47: 143–183 [DOI] [PubMed] [Google Scholar]
- Pidoux G, Witczak O, Jarnaess E, Myrvold L, Urlaub H, Stokka AJ, Kuntziger T, Tasken K (2011) Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J 30: 4371–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K, Geller F, Ruschendorf F, Reis A, Friedel S, Schauble N, Nurnberg P, Siegfried W, Goldschmidt HP, Schafer H, Ziegler A, Remschmidt H, Hinney A, Hebebrand J (2003) Genome scan for childhood and adolescent obesity in German families. Pediatrics 111: 321–327 [DOI] [PubMed] [Google Scholar]
- Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y (2002) The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7: 653–662 [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ (2008) Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Langer T (2008) Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J 27: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM (2006) Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 3: 333–341 [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C (2005) Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437: 564–568 [DOI] [PubMed] [Google Scholar]
- Varela I, Pereira S, Ugalde AP, Navarro CL, Suarez MF, Cau P, Cadinanos J, Osorio FG, Foray N, Cobo J, de Carlos F, Levy N, Freije JM, Lopez-Otin C (2008) Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med 14: 767–772 [DOI] [PubMed] [Google Scholar]
- Voos W (2009) Mitochondrial protein homeostasis: the cooperative roles of chaperones and proteases. Res Microbiol 160: 718–725 [DOI] [PubMed] [Google Scholar]
- Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39: 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W, Procaccio V (2010) Mitochondrial energetics and therapeutics. Annu Rev Pathol 5: 297–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B (2010) Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884 [DOI] [PubMed] [Google Scholar]
- Zorzano A, Liesa M, Sebastian D, Segales J, Palacin M (2010) Mitochondrial fusion proteins: dual regulators of morphology and metabolism. Semin Cell Dev Biol 21: 566–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.