Abstract
EMBO J 31 9, 2076–2089 March 13 2012
EMBO J 31 9, 2090–2102 March 13 2012
It is well known that somatic and germ cells use different cohesin complexes to mediate sister chromatid cohesion, but why different isoforms of cohesin also co-exist within somatic vertebrate cells has remained a mystery. Two papers in this issue of The EMBO Journal have begun to address this question by analysing mouse cells lacking SA1, an isoform of a specific cohesin subunit.
When one cell divides into two, many things have to go right for the two daughter cells to receive identical copies of their mother cell’s genome. It has long been recognized that sister chromatid cohesion, the physical connection established during DNA replication between newly synthesized sister DNA molecules, is one of these essential prerequisites for proper chromosome segregation. It is this cohesion that enables the bi-orientation of chromosomes on the mitotic or meiotic spindle, and thus makes their symmetrical segregation possible. Cohesion is mediated by cohesin, a multi-subunit protein complex, which is thought to connect sister DNA molecules by embracing them as a ring (Figure 1; reviewed in Peters et al, 2008). It is well established that cohesin complexes differ between somatic and germ cells, where they are needed for the proper separation of sister chromatids and of homologous chromosomes, respectively. What has been largely ignored, however, is that even within somatic vertebrate cells there are different forms of cohesin, containing mutually exclusive variable subunits: either SA1 or the closely related SA2 protein (also known as STAG1 and STAG2, respectively), and either Pds5A or the related Pds5B subunit (Peters et al, 2008). Why is that? Two papers from the Losada lab (Remeseiro et al, 2012a, 2012b) have begun to address this question by generating mouse cells lacking the SA1 gene, revealing unexpected insights into the functions of SA1 subunit-containing cohesin complexes (cohesin-SA1).
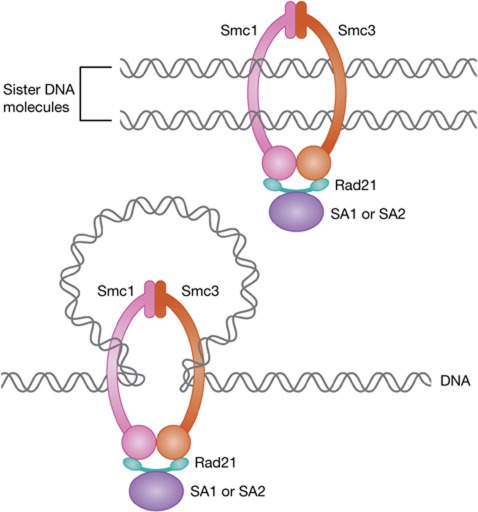
Figure 1.
Schematic drawing illustrating how the SA1 and SA2 proteins interact in a mutually exclusive manner with three core subunits of cohesin (Smc1, Smc3, Rad21) that form a ring-like structure. It has been proposed that these complexes mediate cohesion by trapping the two sister DNA molecules inside the cohesin ring (above), and that cohesin rings might affect chromatin structure by forming or stabilizing intra-chromatid loops (below). Cohesin is thought to influence gene regulation at least in part by mediating chromatin looping.
Although cohesin is best known for its role in sister chromatid cohesion, it is clearly also needed for homologous recombination-mediated DNA repair and for gene regulation. Much of what we know about these functions comes from studies in yeast and fruit flies, organisms with only a single SA1/SA2-related mitotic subunit (Scc3 in budding yeast), and only a single Pds5 subunit. It is therefore plausible that, like many other genes during vertebrate evolution, SA1/SA2 and Pds5A/Pds5B have arisen by gene duplication to constitute paralogs, with functional differences between them assumed to be subtle. Consistently, absence of either Pds5A or Pds5B causes only mild, if any, defects in sister chromatid cohesion, and mice lacking either protein can develop to term, although they die shortly after birth owing to multiple organ defects (Zhang et al, 2007, 2009). First indications that the situation may be different for the Scc3-related subunits came from Canudas and Smith (2009), who reported that RNAi depletion of SA1 and SA2 from HeLa cells caused defects in telomere and centromere cohesion, respectively. The generation of mice lacking either one or both alleles of the SA1 gene has now allowed a more systematic and thorough analysis of SA1 function (Remeseiro et al, 2012a, 2012b).
One of the most striking results obtained in these studies is that most mice lacking SA1 die around day 12 of embryonic development, clearly showing that the function of SA1 cannot be fulfilled by SA2, despite the fact that SA2 is substantially more abundant in somatic cells than SA1 (Holzmann et al, 2010). What could this SA1-specific function be? Losada and colleagues report observations, which imply that SA1 does not have just one, but possibly several important functions in different processes. First, the authors confirm the previous observation that SA1 is required for cohesion specifically at telomeres, while likely collaborating with SA2 in chromosome arms or centromeric regions. Furthermore, telomeres have an unusual morphology in mitotic chromosomes lacking SA1 (Remeseiro et al, 2012a), reminiscent of a fragile-site phenotype previously reported in telomeres with DNA replication defects (Sfeir et al, 2009), and SA1 is indeed required for efficient telomere duplication. Depletion of sororin, a protein that is required for cohesin’s ability to mediate sister chromatid cohesion, also causes a fragile-site phenotype at telomeres. These findings imply that SA1’s role in telomere cohesion is important for efficient telomere replication, perhaps, as the authors speculate, because telomere cohesion may help to stabilize or re-start stalled replication forks, or because cohesion-dependent homologous recombination might be involved in repair of DNA double strand breaks created by collapsed replication forks. Interestingly, cells lacking SA1 frequently show chromosome bridges in anaphase, often fail to divide, and either die or become bi-nucleated. The exact origin of chromosome bridges is difficult to determine, but previous studies have found such bridges often associated with fragile sites on chromosomes; treatment with low doses of DNA replication inhibitors was shown to increase the frequency of such bridges (Chan et al, 2009), and similar observations were indeed made by Remeseiro et al (2012a) in mouse embryonic fibroblasts. It is therefore plausible that the telomere cohesion defect observed in SA1-lacking cells leads to incomplete telomere replication, which in turn results in the formation of anaphase chromosome bridges and subsequent cytokinesis defects. Losada and colleagues further speculate that these chromosome segregation defects could underlie the increased frequency of spontaneous development of various tumours in mice containing just one instead of two SA1 alleles (Remeseiro et al, 2012a). This is an attractive interpretation since tetraploidy and aneuploidy are thought to contribute to the rate with which tumour cells can evolve; however, Losada and colleagues report SA1 deficiency to cause defects also in other cohesin functions, which may therefore as well contribute to tumour formation.
To further understand why SA1 cannot be fulfilled by SA2, Losada and colleagues also analysed the distribution of these proteins in the non-repetitive parts of the mouse genome by chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq). The results of these experiments, published in the second of the two papers (Remeseiro et al, 2012b), raise the interesting possibility that cohesin-SA1 associates more frequently with gene promoters than cohesin-SA2. However, the fact that different antibodies have to be used for any ChIP-based comparison of the distribution of two proteins makes it difficult to know to what degree observed differences might be due to different antibody efficiency. Obviously, such limitations do not exist if the distribution of one and the same protein is analysed under different conditions, and in such an experimental setting, Remeseiro et al indeed make some striking observations. When SA1 is absent, SA2 does not detectably change in abundance, but its distribution in the genome does, in that more than half of all SA2-binding sites in SA1-deficient cells differ from those bound in wild-type cells. Most SA2-binding sites in SA1-deficient cells are in intergenic regions, and CTCF, a zinc finger protein often co-localizing with cohesin and implicated in its gene regulation function (Peters et al, 2008), appears to be absent at many of these sites. It presently remains a mystery why cohesin-SA2 changes its distribution so dramatically in the absence of SA1, but the observation that gene promoters are more frequently occupied by cohesin in the presence of SA1 than in its absence raises the possibility that cohesin-SA1 may have a specific role in gene regulation. This possibility is particularly interesting in light of a recent study that found hardly any change in gene expression upon re-expression of SA2 in SA2-deficient human glioblastoma cells (Solomon et al, 2011), despite the fact that cohesin is thought to regulate numerous genes. With this in mind, Remeseiro et al analysed gene expression in mouse cells and indeed found 549 genes to be mis-regulated in the absence of SA1, in striking contrast to the above-mentioned comparison of human SA2-deficient or proficient cells that found only 19 genes to change in expression levels (Solomon et al, 2011). Obviously direct comparisons will be essential to analyse further the specific roles of SA1 and SA2 in gene regulation, but the current evidence raises the interesting possibility that SA1 may have a particularly important role in gene regulation, whereas cohesin-SA2 is dedicated to creating arm and centromeric cohesive structures for chromosome segregation.
That is not to say that cohesin-SA1 cannot mediate sister chromatid cohesion. It almost certainly can, as it is essential for cohesion at telomeres (Canudas and Smith, 2009; Remeseiro et al, 2012a). Likewise, it would be wrong to assume that we now fully understand why SA1 and SA2 co-exist in somatic vertebrate cells, and what their precise functions is. There are many things we do not understand. For example, if SA2 has little or no role in gene regulation, as the Solomon et al (2011) study implies, why does SA2 nevertheless interact directly with CTCF (Xiao et al, 2011), its gene regulation collaborator? How do cohesin-SA1 and cohesin-SA2 complexes further differ in their genomic distributions and their functions depending on whether they contain either Pds5A or Pds5B, constituting really not just two but four distinct cohesin complexes? The work by Losada and colleagues represents an important step towards understanding these questions, but there is still a long and presumably exciting way to go to understand how different cohesin complexes control the mammalian genome.
Footnotes
The author declares that he has no conflict of interest.
References
- Canudas S, Smith S (2009) Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Holzmann J, Fuchs J, Pichler P, Peters JM, Mechtler K (2010) Lesson from the stoichiometry determination of the cohesin complex; a short protease mediated elution increases the recovery from crosslinked antibody-conjugated beads. J Proteome Res 10: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A (2012a) Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A (2012b) A unique role of cohesin-SA1 in gene regulation and development. EMBO J 31: 2090–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G (2011) Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 31: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Kulkarni S, Deardorff MA, Uzielli ML, Dorsett D, Beebe DC, Jay PY, Heuckeroth RO, Krantz I, Milbrandt J (2009) Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One 4: e5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J (2007) Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134: 3191–3201 [DOI] [PubMed] [Google Scholar]