Abstract
Alternative splicing leads to the expression of multiple isoforms of the subunits (IFNAR1 and IFNAR2) of the type I IFN receptor. Here we describe two transcripts representing extracellular forms of ovine IFNAR1 and show that soluble extracellular forms of both IFNAR2 and IFNAR1, prepared in recombinant form in Escherichia coli, have antiviral (AV) activity in the absence of IFN. Exposure of Madin-Darby bovine kidney cells to the extracellular domain (R2E) of IFNAR2 at concentrations as low as 10 nM afforded complete protection against vesicular stomatitis virus and led to the rapid activation of the transcription factors ISGF3 and GAF. Although R2E can bind IFN (Kd ≈70 nM), activity was observed irrespective of whether or not ligand was present. R2E was inactive on mouse L929 cells but active on L929 cells expressing a membraneanchored, ovine/human chimeric IFNAR2 with an ovine extracellular domain. The data suggest that AV activity is conferred by the ability of soluble R2E to associate with the transfected IFNAR2 subunit rather than resident murine IFNAR1. Soluble extracellular forms of IFNAR1 have lower AV activity than R2E on Madin-Darby bovine kidney cells but are less species-specific and protect wild-type L929 cells as efficiently as the transfected cell line, presumably by interacting with one of the murine receptor subunits.
Keywords: alternative splicing, cross-linking, cytokine, IFN-τ, sheep
Soluble forms of cytokine membrane receptors (1–5) usually are produced either by proteolytic cleavage from a membrane form or by alternative splicing of mRNA precursor. Because these soluble forms often can bind ligand, they initially were proposed to be antagonists, blocking or, at the very least, reducing cytokine potency by competing with their membrane-anchored homologs for common ligands. As a result, recombinant, extracellular portions of some receptors are being used to therapeutically reduce inflammatory, autoimmune, and other pathological events mediated by cytokines (6–9). Another potential function for soluble receptor forms is to protect ligand from degradation or excretion (10). A soluble receptor subunit also may convert a ligand-resistant target into a sensitive one by substituting for an absent endogenous binding subunit (11, 12). In this paper, we report another functional possibility, namely that the soluble extracellular domains of receptor subunits, in this instance those of type I IFN receptors (IFNARs), can mimic the activity of a cytokine even when the cytokine itself is absent.
The type I IFN includes at least four subtypes: IFN-α, -β, -ω, and -τ. They share a common receptor, for which they compete in binding experiments (13–15). The receptor consists of two subunits, IFNAR1 and IFNAR2 (16) (Fig. 1). IFNAR1 has been cloned from human (17), bovine (18, 19), ovine (19, 20), murine (21), and chicken cells (22). A soluble form of human (hu) IFNAR2 was first purified as an IFN-α binding protein from urine (IFNAR2a; ref. 23 and references therein). Its identification led to the cloning of its own cDNA and those of two membrane-associated forms, huIFNAR2b and huIFNAR2c (23, 24). Although all three forms bind type I IFN, only the long form (IFNAR2c) participates in signal transduction (25–29). The functions of huIFNAR2a (the extracellular soluble form) and huIFNAR2b, which possesses only a short intracellular domain, remain unclear. The latter, when coexpressed with the long form acts in a dominant negative manner (30). Because huIFNAR2 generally is regarded as the major IFN-binding component of the receptor complex (16, 17), the soluble form of IFNAR2 has the potential to compete with the membrane-anchored form for ligand, provided that its affinity is sufficiently high (31).
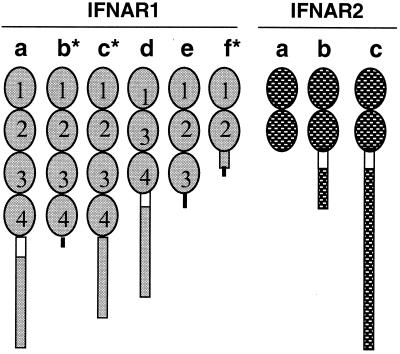
Figure 1.
Different forms of IFNAR1 and IFNAR2. The cDNA of the membrane-anchored full-length IFNAR1 and IFNAR2 with a single transmembrane domain (empty box) have been cloned from human, mouse, sheep, cattle, and chicken. The various cDNA representing short forms of IFNAR1 (human, sheep) and IFNAR2 (human, mouse) also have been identified either by hybridization screening of cDNA libraries or by RT-PCR in several species. The three IFNAR2 forms have been termed IFNAR2a, IFNAR2b, and IFNAR2c, which reflects the order of their cloning. We suggest that the IFNAR1 variants be named in the same manner. The extracellular domains of full-length IFNAR1 and IFNAR2 are organized, respectively, into four and two Ig-like subdomains, each about 100 aa long (SD 100; ovals in the diagram) (16, 32, 33). Both polypeptides belong to the class II cytokine receptor superfamily. The short variants are either membrane-anchored with shorter cytoplasmic domains or soluble proteins with either two, three, or four SD100 subdomains. Those identified by RT-PCR amplification without full characterization of the entire ORF are marked with *. Novel amino acid sequences arising as a result of frame shifts are shown as black lines. The fact that IFNAR1 and IFNAR2 each have a single gene in human (22, 42) and that the missing regions of these variants encompass whole exons indicate that the mRNA of the variants arise from alternative splicing of the same precursor RNA.
A typical feature of cytokine receptors is that they have either two or four Ig-like subdomains of about 100 aa (SD100) comprising their extracellular portion (32, 33). In the case of IFNAR2a above, both SD100 subdomains have been retained. Alternatively spliced variants for huIFNAR1 also have been identified, including one (huIFNAR1d, Fig. 1) that lacks the second of the four SD100 subdomains (34). Two other inferred forms possess all four SD100 domains but lack the transmembrane and cytoplasmic domains (IFNAR1b and IFNAR1c). Like huIFNAR2a, they are presumed to be soluble (35) (Fig. 1). The functions of these soluble receptor forms are unknown.
Methods
Reagents.
Glutathione S-transferase (GST)-fusion-protein expression vector pGEX-2T was purchased from Amersham Pharmacia. Vector pET15b, which provides no fusion tag, and the host strain BL21(DE3)pLysS were bought from Novagen. Taq DNA polymerase was obtained from GIBCO/BRL. The pGEM-T easy cloning kit and all of the other enzymes were products of Promega. Oligonucleotides and their particular uses are listed in Table 1. Anti-STAT-1 (SC-346) and anti-STAT-2 (SC-476) were obtained from Santa Cruz Biotechnology.
Table 1.
Oligonucleotides used for RT-PCR, preparation of recombinant proteins, and EMSA
| Names | Sequences |
|---|---|
| Group 1 | |
| 606 | 5′-AGTTAAAGCAGAACTGCGT-3′ |
| 741 | 5′-CAGAGGGTATCTTCTGGAC-3′ |
| swk2s | 5′-GTAGAGTTGGTTGCTATAGT-3′ |
| mo4a | 5′-TTAGGATC CTCTTGTAAGACTCCCTAAT-3′ |
| Group 2 | |
| ror2s | 5′-AGACGCGGATCC TCGTATGTTGTGCCTGATCT-3′ |
| ror2a | 5′-CCGCTAGAATTCTTA TGTAGCAGGTTCTGA TGACT-3′ |
| ro2is | 5′-ATTCGAGGATCC AAACGGATTGGTTATATATG-3′ |
| ro2ia | 5′-GTCTCAGAATTC TTAATTAAAATTTTTCAGAT-3′ |
| ror1s | 5′-TAACGCGGATCC GCAAATCTGAAGTCTGAAAA-3′ |
| ror1a | 5′-GTTCTAGAATTCTCA CCAGGTTTTGGAAGTGTT-3′ |
| ror1a2 | 5′-GTTCTAGAATTCTCA TGGGGG ACCAATCTG-3′ |
| ror1va | 5′-GTTCTAGAATTCTCA GAGAAATAGTTTCACTT-3′ |
| Group 3 | |
| GASs | 5′-GATTTTCCCAGAAAAGGAAC-3′ |
| ISREs | 5′-TTTACAAACAGCAGGAAATAGAAACTTAAGAGAAATACA-3′ |
Group 1 was used for RT-PCR detection of ovIFNAR1f and ovIFNAR1e; group 2 was used for preparing recombinant proteins; and group 3 (sense only shown) for EMSA.
Cloning of Ovine (ov)IFNAR1e and ovIFNAR1f.
The procedure for cloning ovIFNAR1e by screening a uterine endometrial cDNA library has been described (19). ovIFNAR1f was identified in endometrium by reverse transcriptase (RT)-PCR. Two rounds of PCR amplification with nested primers were performed. In the first round, primers 606 and 741 (Table 1) were used under the following PCR conditions (94°C for 1 min; 55°C for 1 min; 72°C for 1 min; 30 cycles). In the second round of PCR, two different primers, swk2s and mo4a (Table 1), were used (94°C for 52 sec; 55°C for 31 sec; 25 cycles). The amplified product was cloned by using the pGEM-T easy kit from Promega and then sequenced.
Recombinant Proteins.
The cDNA sequences representing different regions of ovIFNAR1, ovIFNAR2, and ovIFNAR1e were amplified by PCR and cloned into either the pGEX2T or pET15b vectors and transformed into BL21 cells. Expression was induced by 1 mM isopropylthio-β-d-galactoside at 3°C for 4 h during midlog phase growth. Cells were harvested by centrifugation, resuspended in buffer A (50 mM Tris, pH 8.0/1 mM EDTA/100 mM NaCl/1 mM PMSF), and disrupted by passing through a French press. Inclusion bodies were collected by centrifugation, washed three times with buffer B (2 M urea in buffer A), and solubilized in either buffer C (8 M urea in buffer A) or buffer D (6 M guanidine hydrochloride in buffer A). The concentrations of solubilized protein solutions were adjusted to 0.1 mg/ml and refolded by dialysis in either buffer A plus 10% glycerol or PBS containing 1 mM reduced glutathione and 5 mM oxidized glutathione. Proteins that remained soluble were concentrated to ≈1 mg/ml.
Antiviral (AV) Assays.
Soluble proteins were added to the first-row wells of 96-well plates and then diluted in series (1:3) in DMEM containing 10% FBS. Madin-Darby bovine kidney (MDBK) cells (ATCC CCL22) or L929 cells (parental or transfected) were seeded (5 × 105 cells) into each well. After 16 h, they had formed monolayers. The medium was removed, and vesicular stomatitis virus in serum-free DMEM was added to each well (1 plaque-forming unit/cell). After 1 h at 37°C, the medium was removed and replaced with medium containing 10% FBS. After 24–30 h, surviving cells were stained by Gentian violet (36).
Electrophoretic Mobility-Shift Assays (EMSA).
Close to confluent MDBK cells on serum-free medium were exposed to either R2E (10 μM) or bovine IFN-α1 (500 units/ml, ≈10 nM) for 15 min. Control cells were untreated. The cells were washed twice with cold PBS, and either whole cell (36, 37) or crude nuclear extracts (37) were prepared for use in EMSA analysis. The interferon-stimulated regulatory elements probe was derived from the 9–27 gene (36, 37). The GAS (gamma-activated sequence) probe was based on the promoter of the Fc-γ receptor gene (36, 38). Probes were labeled with 32P. Either whole-cell extracts (2 μl; ≈5 μg of protein) or nuclear extracts (6 μl; 3 μg protein) were incubated with 1 μg of poly(dIdC) for 10 min, and labeled probe (30,000 cpm) was added. To identify transcription factors that bound the probes, some reactions were spiked with either anti-STAT-1 or anti-STAT-2 Igs (each 2 μg) or with rabbit Igs (2 μg) as a control. After 20 min, the mixture was analyzed on a 4% 0.5× Tris-borate-EDTA polyacrylamide gel.
32P Labeling of IFN-τ.
To label ovIFN-τ4, a protein kinase A phosphorylation site was introduced into the amino terminus of the protein (39, 40) by using the pGEX-2TK vector. The soluble GST-fusion protein was purified on a glutathione affinity column, and the GST removed by thrombin cleavage. The phosphorylation reaction mixture (in 50 μl) consisted of 5 μl of kinase buffer (200 mM Tris⋅HCl, pH 7.5/1 M NaCl/120 mM MgCl2), 20 μl γ-[32P]-ATP (6,000 Ci/mmol, 10 μCi/μl), 1 μg phosphorylatable IFN-τ, 45 μg bovine heart protein kinase A (Sigma). After 30 min at 37°C, the reaction was stopped by adding 350 μl of inhibitory buffer (10 mM sodium phosphate/10 mM sodium pyrophosphate/10 mM EDTA, pH 7.0) containing 1 mg/ml BSA. Unincorporated γ-[32P]-ATP was removed by ultrafiltration. The IFN, whose AV activity was unchanged, had a specific activity of 75 μCi/μg.
Affinity Cross-Linking of R2E and [32P]-IFN-τ4.
GST-R2E fusion protein or free R2E were added to [32P]-IFN-τ4 (41) and the binding reaction was allowed to proceed at 4 C for 1 h. The bivalent cross-linker bis(sulfosuccinimidyl) suberate (BS3, Pierce) then was added to a final concentration of 5 mM. After standing in ice for 30 min, the BS3-ligand-receptor complexes was analyzed by SDS/PAGE and detected on x-ray film.
Stable Transfection of L929 Cells with the ov/hu (oh) Chimeric IFNAR2.
Mouse L929 cells were stably transfected with a functional IFNAR2c that possessed an ovIFNAR2 extracellular and transmembrane domain and a human intracellular domain (41). Control of expression was through the human elongation factor 1 α promoter. The cell line used exhibited 14,000 ± 460 ovIFN-τ binding sites per cell (42).
Gel Filtration Analysis of Refolded R2E Protein.
Refolded recombinant proteins were applied to a (1.5 × 100 cm) Sephadex G-200 column (Amersham Pharmacia) in PBS buffer at various dilutions of chaotropic agents. The apparent Mr of the protein peaks were estimated by comparison with proteins of known molecular weight.
Results
Cloning of ovIFNAR1e and ovIFNAR1f.
Full-length cDNAs for ovIFNAR1 previously have been characterized (19, 20). Another cDNA, cloned from an ovine endometrial cDNA library (19), lacked the region encoding the fourth SD100 subdomain, the entire transmembrane domain, and the cytoplasmic domain of IFNAR1 (Fig. 1) and had three novel amino acids (LFL) to the end, was named ovIFNAR1e. When aligned with the huIFNAR1 genomic sequence (43), the missing region of the cDNA extended from the beginning of intron 7 to the end of intron 10. ovIFNAR1e transcripts were difficult to detect by standard RT-PCR, but detection became possible when using nested primers in conjunction with reduced extension times. Interestingly, ovIFNAR1e was more abundant in kidney and liver than in endometrium but absent in pituitary (data not shown).
RT-PCR amplification from ovine endometrial RNA revealed another short transcript (ovIFNAR1f), which, unlike that for IFNAR1e, was absent in liver and kidney. This product lacked the region from the beginning of intron 6 to the end of intron 10 and represented only the first and second SD100 subdomains and the first 31 amino acid residues of the third plus six new amino acids to the end (ISLISH) (Fig. 1).
Production of Recombinant Proteins.
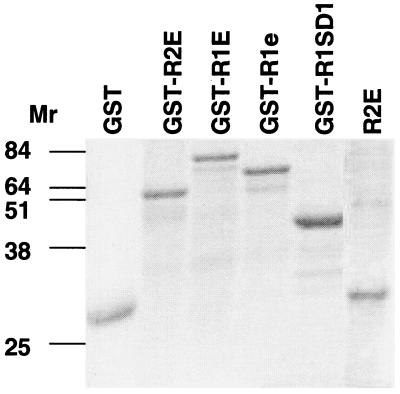
The various soluble, extracellular forms were expressed in E. coli, either as free proteins or as fusion products coupled to GST (Fig. 2). The ones studied here were the extracellular domains of ovIFNAR2 (GST-R2E and R2E) and IFNAR1a (GST-R1aE), ovIFNAR1e (GST-R1e), the first SD100 domain of ovIFNAR1 (GST-R1SD1), and, as a control, the cytoplasmic domain of ovIFNAR2 (GST-R2C). Only the latter proved to be soluble. It was purified directly by affinity chromatography on glutathione-Sepharose. The remainder were solublized in either 8 M urea or 6 M guanidinium hydrochloride and then refolded. When required, the GST portions of the fusion proteins were cleaved by thrombin. Overall yields were generally low (1–5 mg/liter culture) because as much as 90% of the protein precipitated during the refolding step.
Figure 2.
SDS/PAGE of recombinant proteins solubilized from washed inclusion bodies. Analysis was performed in 10% PAGE gels, which were stained with Coomassie blue. GST lane is the 26,000 Mr carboxyl terminus of GST. GST-R1SD1 is the GST-fusion protein of the first SD1 domain of IFNAR1. GST-R1e lacks the fourth SD100 domain of the extracellular domain of IFNAR2. (Fig. 1). Unconjugated R2E was prepared in the pET vector.
Cross-Linking of IFN-τ to R2E.
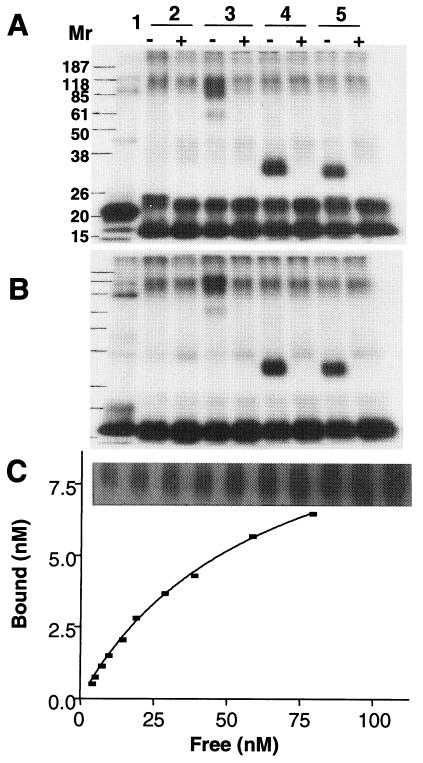
[P32]-OvIFN-τ4 was tested for its ability to bind R2E by cross-linking (Fig. 3 A and B). Both GST-R2E and free R2E formed complexes with ovIFN-τ4. Based on the band intensities, free R2E bound ligand more efficiently than GST-R2E. The binding data (Fig. 3C) for R2E fitted a one-site binding model. The calculated Kd for the association of R2E and ovIFN-τ4 was 70 ± 5 nM (n = 3), which is higher than that for ovIFN-τ4 binding to membrane-anchored ovIFNAR2 (in the absence of IFNAR1) in mouse fibroblasts (2.5 nM; data not shown). The electrophoretic mobilities of the ligand-receptor complexes were higher than anticipated (Fig. 3 A and B). For example, the theoretical Mr of the GST-R2E-[32P]-ovIFN-τ4 complex is 72,000, whereas the value determined from the gel was about half that, presumably because cross-linked proteins were unable to unfold completely in SDS and mercaptoethanol.
Figure 3.
Binding of IFN to refolded R2E. The ligand-receptor complex was cross-linked with BS3 and analyzed by SDS/PAGE under either nonreducing (A) or reducing (B) condition. Radioactive bands were visualized by autoradiography. Sample 1: [32P]-IFN-τ4 only. Sample 2: [32P]-IFN-τ4 plus BS3. Sample 3: GST-R2E plus [32P]-IFN-τ4 and BS3. Sample 4: R2E derived from thrombin cleavage of GST-R2E plus [32P]-IFN-τ4 and BS3. Sample 5: R2E produced in the pET15B vector plus [32P]-IFN-τ4 and BS3. Lanes labeled + or − indicate whether or not an excess amount of unlabeled IFN-τ4 (100×) was included in the cross-linking reaction. Molecular weight markers are on the left. (C) Determination of the Kd for binding of R2E to ovIFN-τ4. Cross-linked ligand-receptor complexes at different concentrations of [32P]-IFN-τ4 were resolved in SDS/PAGE, identified by autoradiography (panel above the graph) and counted by liquid scintillation. The amount of 32P in the band relative to the amount of 32P loaded then was calculated. The Kd value was determined by fitting the plot of specifically bound versus free ligand concentrations to the equation bound = Bmax [L]/(Kd + [L]), where [L] is the concentration of free ligand and Bmax the maximum binding sites. Calculations were performed by the graphpad prism version 3.02 for Windows, GraphPad Software, San Diego, http://www.graphpad.com. The experiment was performed three times.
AV Activities of Soluble Receptor Domains.
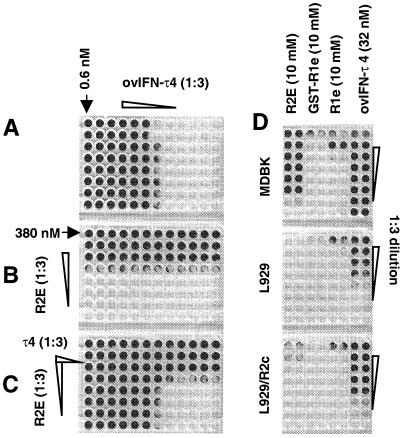
Recombinant R2E was anticipated to antagonize the AV effect of type I IFN on MDBK cells. However, rather than reducing the protection resulting from treatment with ovIFN-τ4, R2E provided a marked increase in AV activity when the two proteins were combined together (Fig. 4C). When R2E was used alone, half-maximal protection of the MDBK cells was observed 14 nM (Fig. 4B). ovIFN-τ4 alone provided protection at 0.8 pM (Fig. 4A), about the same as published values (44). Importantly, the combined effects of R2E and ovIFN-τ4 were additive and not synergistic (Fig. 4C). Bacterial lysate, GST, GST-R2C (the intracellular domain of the receptor), and R2E, which had not been through the refolding steps, each failed to provide AV protection (data not shown), indicating that the effects of ovR2E were specific and a feature of the refolded protein. Free R2E had higher AV activity than the GST-R2E fusion protein from which it was derived (Table 1). The increase in AV activity observed after thrombin cleavage was not caused by the presence of thrombin because the enzyme alone provided no protection (data not shown). R2E, produced by using the pET15b(+) vector, had an AV activity of 12 nM, closely matching the activity of R2E produced by cleavage from the GST-fusion protein (Table 2).
Figure 4.
The extracellular domain of the ovIFNAR2 subunit (R2E) has intrinsic AV activity. (A) ovIFN-τ4 was diluted from left to right in a 1:3 series. (B) Thrombin-cleaved GST-R2E was diluted from top to bottom in a 1:3 series. (C) The treatments were combined (ovIFN-τ4, left to right; R2E, top to bottom). Protected cell monolayers take up the Gentian violet stain. (D) A comparison of the effects of R2E, GST-R2e, R1e, and ovIFN-τ4 on MDBK cells (Top), mouse L929 cells (Middle), and L929 cells stably transfected with a chimeric receptor (R2C) containing the extracellular domain of ovIFNAR2 (19) and the intracellular domain of huIFNAR2c (24) (Bottom).
Table 2.
The AV activities of soluble IFNAR proteins
| Proteins | AV activities, nM |
|---|---|
| GST-R2E | 126 |
| R2E* | 14 |
| R2E† | 12 |
| GST-R1e | 3,300 |
| R1e* | 1,100 |
| GST-R1E | 1,000 |
| GST-R1SD1 | 3,000 |
Insoluble proteins from inclusion bodies were solubilized with 8 M urea and refolded in 50 mM Tris⋅HCl, pH 8.0 and 100 mM NaCl. Data are averages from three experiments, each carried out in duplicate and performed within 2 weeks of refolding the proteins (which were stored at 4°C). The GST-R2E corresponds to preparation 1 in Table 3. All assays (based on a series of 1∶3 dilutions) gave identical results. Hence, there are no error terms.
Produced from GST-fusion protein.
Produced with pET15b vector without GST.
To check whether the AV activity of R2E was unique to this subunit of the type I IFN receptor, GST-fusion proteins of ovIFNAR1 extracellular domains were prepared. R1aE, R1e, and R1SD1 contain four, three, and one SD100 subdomain, respectively (Fig. 1). Each protein had low, but measurable, AV activity (Table 2). Values were about 2 orders of magnitude lower than that of R2E. The protection offered by free R1e was higher than that of its GST-fusion protein (≈3-fold). The activities observed seemed not to be related to the number of SD100 subdomains present in the proteins tested.
Renaturation Over Time.
Freshly prepared GST-R2E often had very low AV activity, whichever renaturation conditions were used (Table 3). This activity usually increased on sterile storage of the preparation at 4°C, although the final activity achieved varied considerably. At least one preparation (preparation 2) with very low AV activity was able to form complexes with ovIFN-τ4 in the cross-linking assay, suggesting that different domains of the protein might fold independently.
Table 3.
AV activity of R2E increases upon storage
| Preparation and form | Date refolded | Refolding conditions | Ligand binding | Initial AV activity, nM | Final AV activity, nM | Increased activity, fold |
|---|---|---|---|---|---|---|
| 1, GST-RE | 4/23/98 | 1 | N.P. | 126 | 26 | 5 |
| 2, R2E | 1/27/99 | 2 | + | Undetectable | 340 | — |
| 3, R2E | 2/24/99 | 1 | N.P. | Undetectable | 13 | — |
| 4, GST-R2E | 3/18/99 | 2 | + | 960 | 54 | 18 |
| 5, GST-R2E | 3/18/99 | 1 | N.P. | Undetectable | 90 | — |
| 6, GST-R2E | 5/14/99 | 2 | + | 85 | 3 | 28 |
| 7, GST-R2E | 5/14/99 | 2 | + | 1400 | 9 | 155 |
| 8, GST-R2E | 5/01/00 | 2 | − | Undetectable | Undetectable | — |
Samples 6 and 7 were stored frozen until February 27, 2000 and at 4°C until determination of final AV activity. All others were maintained aseptically at 4°C until assayed. Proteins were prepared as either GST-fusion proteins or unconjugated in the pET15b vector. Initial activity was determined within 7 days from refolding. Final AV activity and ligand-binding activity were determined on June 20, 2000. N.P., assay was not performed because preparations were in Tris buffer. Refolding conditions: 1, 50 mM Tris⋅HCl (pH 8.0), 100 mM NaCl; 2, PBS, 10% (vol/vol) glycerol, 1 mM reduced glutathione 5 mM oxidized glutathione.
Activation of Signal Transducers and Activators of Transcription (STATs).
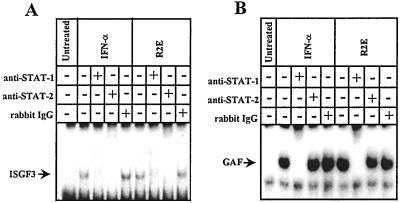
The AV activity ascribed to R2E above did not distinguish effects mediated directly through activation of the Janus kinase (JAK)-STAT signal transduction pathway from ones that might have occurred through secondary induction of IFN. In the case of the former, there should be rapid activation of STAT proteins (36, 45, 46). Type I IFNs typically induce the formation of both ISGF3 (IFN-stimulated gene factor-3), consisting of STAT1, STAT2, and p48 (ISGF3γ), and STAT1a homodimer (GAF or IFN-γ activated factor) (47). As shown in Fig. 5, treatment of MDBK cells with 10 μM R2E for 15 min induced the formation of ISGF3 and GAF, suggesting an immediate and direct effect on the signal induction cascade.
Figure 5.
Activation of the Janus kinase-STAT signal transduction pathway by R2E. MDBK cells were either left untreated or exposed to R2E (10 μM) or to 500 units/ml of bovine IFN-α1 for 15 min before EMSA analysis with the ISRE (A) and GAS (B) probes. Whole-cell extracts were prepared and incubated with radioactive probes before EMSA. Lanes indicate extracts from untreated cells, cells treated with bovine IFN-α1, and cells treated with R2E. The positions of the bands containing ISGF3 and GAF, which can be dissociated with excess probe (not shown) and supershifted by anti-STAT antisera, are arrowed. Additional controls, in which cells were exposed to bacterial lysate (250 ng/ml), revealed no evidence for STAT activation and are not shown. Experiments in which cells were exposed to R2E for 10 min also provided a positive STAT activation. Experiments with nuclear extracts gave identical results.
Mediation of R2E AV Activity Requires Membrane-Anchored IFNAR2.
A murine L929 cell clone was stably transfected to express a chimeric ohIFNAR2 receptor, which possessed the extracellular and transmembrane domains of ovIFNAR2 and the cytoplasmic domain of huIFNAR2c (cloned from Daudi cells) (42). For reasons not understood, the complete ovIFNAR2 subunit is inactive when transfected into mouse cells. It binds IFN but is incapable of activating the Janus kinase/STAT signal transduction pathway in the presence ovine IFN (data not shown), presumably because its intracellular domain cannot interact with the mouse cell signal transduction apparatus. However, the presence of the chimeric receptor (ohIFNAR2c) with the huIFNAR2c cytoplasmic domain provided a cell line that was responsive to both ovine and bovine IFN (42) and R2E. Whereas R2E provided no protection to parental L929 cells, it protected the cells expressing the transfected receptor when supplied at a concentration of 1.1 μM (Fig. 4D). Even though this activity was about 80 times lower than that observed on MDBK cells, there was also a similar (≈100-fold) drop in ovIFN-τ4 potency on the same transfected mouse cell line when compared with the MDBK cell line. These experiments suggest that the AV activity of R2E is mediated by the membrane-anchored ohIFNAR2 receptor subunit and not through the endogenous mouse receptor complex. Interestingly, R1e had low, but measurable, activity on L929 cells. This activity on mouse fibroblasts was only about three times lower than that observed on MDBK cells, and it was not enhanced by the presence of the ohIFNAR2c chimeric receptor subunit.
Relative Activity of Monomeric and Oligomeric Forms of R2E.
When gel filtration chromatography of refolded R2E was conducted in presence of a low concentration of guanidine hydrochloride (1 mM), the GST-R2E eluted as a monomer with an apparent Mr of 52,000 (data not shown). However, if the concentration of chaotropic reagent was reduced to 1 μM, the protein eluted as a broad peak beginning at the void volume of the column. It clearly consisted of soluble but mainly aggregated forms. However, the AV activities of the monomeric and aggregated forms were quite similar (data not shown).
Discussion
Soluble receptor proteins prepared in E. coli (31, 48, 49) and yeast (50, 51), and representing either portions of or the entire extracellular domains of huIFNAR1 (47, 49), huIFNAR2 (31, 50), and murine IFNAR2 (51), have been studied primarily as potential antagonists of type I IFN action. Nguyen et al. (48) reported that the GST-fusion protein of huIFNAR1 extracellular domain inhibited the AV and antiproliferative activities of huIFN-α1 and competed with the cell surface receptor for binding the IFN. Arduini et al. (50) deglycosylated huIFNAR2 extracellular domain, obtained from the yeast Pichia pastoris and demonstrated that it formed a binary complex with huIFN-β1a. Piehler and Schreiber (31) used an E. coli preparation of huIFNAR2 extracellular domain that formed a 1:1 complex with IFN (huIFN-α2) with a Kd of 10 nM. It inhibited the AV activity of IFN-α effectively over a wide range of concentrations. The murine R2E prepared by Hardy et al. (51) was an antagonist of a number of IFN activities on murine cells, but boosted IFN growth inhibition of thymocytes null for the IFNAR2 gene. They suggested that soluble murine R2E, although normally an antagonist of IFN action, might, under certain circumstances, interact with membrane-anchored IFNAR1in presence of IFN to activate the signal transduction machinery and behave as an agonist. None of these investigators reported any intrinsic AV activity of their soluble receptor preparations. Possibly such activity was simply overlooked or precluded by the design of the experiments.
ovR2E was not an effective antagonist of type I IFN activity. It failed to inhibit the AV activity of ovIFN-τ4 (Fig. 4) even though it was capable of binding the protein (Fig. 3). A likely explanation is that the Kd of the soluble receptor for IFN-τ4 (70 nM) is sufficient for it to be detected in a binding assay but too high for it to compete effectively for ligand with the IFNAR1/IFNAR2 membrane-anchored receptor of MDBK cells, which binds IFN-τ with a Kd of ≈0.3 nM (unpublished data). There is also the possibility that the AV activity of R2E might be caused by a structural element in the recombinant protein that is distinct from the one responsible for binding IFN. Nevertheless, it is puzzling why we have been able to observe AV activity with ovR2E, whereas others, working on components from different species, have not. It is possibly the unique structure of the ovine receptors or a particular conformation reached during refolding that led to the observed properties.
The AV activity of R2E was almost certainly mediated through a membrane-anchored receptor because the Janus kinase-STAT signal transduction pathway was rapidly activated (Fig. 5). This conclusion is reinforced by the fact that R2E only had activity on murine L929 cells when they expressed the ohIFNAR2c chimeric receptor. Presumably, R2E was able to interact with the transfected IFNAR2 receptor but not with murine IFNAR1 or IFNAR2. Lewerenz et al. (52) have previously proposed that dimerization can occur between two IFNAR subunits in the presence of IFN-β, and Pattyn et al. (53) concluded that ligand-induced dimerization of IFNAR2 subunits can be sufficient to up-regulate several IFN-effector genes with classical interferon-stimulated regulatory elements and provide limited AV protection in the absence of IFNAR1 signaling. Finally, Kotenko et al. (54) dimerized IFNAR2 intracellular domains by means of IFN-γR1/IFNAR2 chimeras and obtained full STAT activation without the involvement of the IFNAR1 intracellular domain.
Although the soluble IFNAR1 variants had lower AV activity than R2E, activity on MDBK cells was still observed in the μM range (Table 2). The AV mechanism for R2E and R1e also might be different given the following three observations (1). The AV activity of R2E on MDBK cells was at least 80-fold higher than that of R1e (2). R2E was more species-specific than R1e. For example, activity of R2E on MDBK cells was at least 700-fold higher than on L929 cells, whereas activity of R1e on MDBK cells was only 3-fold higher than on L929 cells (3). The presence of the anchored ohIFNAR2c receptor subunit in L929 cells strongly enhanced the AV activity of R2E but had no effect on that of R1e (Fig. 4D). Conceivably R1e was able to interact with resident murine IFNAR1 or IFNAR2 subunits to induce an AV response. The data suggest that extracellular soluble forms of the receptors might, depending on their local concentration, provide a constitutive threshold of IFN signaling in absence of the ligand itself.
Acknowledgments
We thank Dr. Limin Liu, who originally noted the AV effect, Dr. Alan Ealy for preparing phosphorylatable ovIFN-τ4, and Drs. S. Kleman and N. Mathialagan who contributed to the original cloning of ovIFNAR1e. We are grateful to Dr. Gilles Uze for providing a critical review of the manuscript. The research was supported by National Institutes of Health Grant HD21896.
Abbreviations
- AV
antiviral
- EMSA
electrophoretic mobility-shift assay
- GST
glutathione S-transferase
- IFNAR
interferon receptor
- MDBK
Madin–Darby bovine kidney
- hu
human
- ov
ovine
- oh
ovine/human
- RT
reverse transcriptase
- BS3
bis(sulfosuccinimidyl) suberate
- STAT
signal transducer and activator of transcription.
Footnotes
References
- 1.Rose-John S, Heinrich P C. Biochem J. 1994;300:281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heaney M L, Golde D W. Blood. 1996;87:847–857. [PubMed] [Google Scholar]
- 3.Fernandez-Botran R, Chilton P M, Ma Y. Adv Immunol. 1996;63:269–336. doi: 10.1016/s0065-2776(08)60858-5. [DOI] [PubMed] [Google Scholar]
- 4.Kelso A. Immunol Cell Biol. 1998;76:300–317. doi: 10.1046/j.1440-1711.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Botran R. Crit Rev Clin Lab Sci. 1999;36:165–224. doi: 10.1080/10408369991239196. [DOI] [PubMed] [Google Scholar]
- 6.Ozmen L, Gribaudo G, Fountoulakis M, Gentz R, Landolfo S, Garotta G. J Immunol. 1993;150:2698–2705. [PubMed] [Google Scholar]
- 7.Mullarkey M F, Leiferman K M, Peters M S, Caro I, Roux E R, Hanna R K, Rubin A S, Jacobs C A. J Immunol. 1994;152:2033–2041. [PubMed] [Google Scholar]
- 8.Ozmen L, Roman D, Fountoulakis M, Schmid G, Ryffel B, Garotta G. Eur J Immunol. 1995;25:6–12. doi: 10.1002/eji.1830250103. [DOI] [PubMed] [Google Scholar]
- 9.Weinblatt M E, Kremer J M, Bankhurst A D, Bulpitt K J, Fleischmann R M, Fox R I, Jackson C G, Lange M, Burge D J. N Engl J Med. 1999;340:253–259. doi: 10.1056/NEJM199901283400401. [DOI] [PubMed] [Google Scholar]
- 10.Baumann G, Shaw M A, Buchanan T A. Metabolism. 1989;38:330–333. doi: 10.1016/0026-0495(89)90119-4. [DOI] [PubMed] [Google Scholar]
- 11.Silacci P, Dayer J M, Desgeorges A, Peter R, Manueddu C, Guerne P A. J Biol Chem. 1998;273:13625–13629. doi: 10.1074/jbc.273.22.13625. [DOI] [PubMed] [Google Scholar]
- 12.Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, Matsuda T, Hirano T, Kishimoto T. Cell. 1989;58:573–581. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]
- 13.Flores I, Mariano T M, Pestka S. J Biol Chem. 1991;266:19875–19877. [PubMed] [Google Scholar]
- 14.Li J, Roberts R M. J Biol Chem. 1994;269:13544–13550. [PubMed] [Google Scholar]
- 15.Niu P D, Lefevre F, Mege D, La Bonnardiere C. Eur J Biochem. 1995;230:200–206. doi: 10.1111/j.1432-1033.1995.tb20551.x. [DOI] [PubMed] [Google Scholar]
- 16.Moyemen K E, Lewerenz M, Reboul J, Luftalla G, Uze G. J Interferon Cytokine Res. 1999;19:1069–1098. doi: 10.1089/107999099313019. [DOI] [PubMed] [Google Scholar]
- 17.Uze G, Lutfalla G, Gresser I. Cell. 1990;60:225–234. doi: 10.1016/0092-8674(90)90738-z. [DOI] [PubMed] [Google Scholar]
- 18.Lim J K, Langer J A. Biochim Biophys Acta. 1993;1173:314–319. doi: 10.1016/0167-4781(93)90129-2. [DOI] [PubMed] [Google Scholar]
- 19.Han C S, Mathialagan N, Klemann S W, Roberts R M. Endocrinology. 1997;138:4757–4767. doi: 10.1210/endo.138.11.5530. [DOI] [PubMed] [Google Scholar]
- 20.Kaluz S, Fisher P A, Kaluzova M, Sheldrick E L, Flint A P. J Mol Endocrinol. 1996;17:207–215. doi: 10.1677/jme.0.0170207. [DOI] [PubMed] [Google Scholar]
- 21.Uze G, Lutfalla G, Bandu M T, Proudhon D, Mogensen K E. Proc Natl Acad Sci USA. 1992;89:4774–4778. doi: 10.1073/pnas.89.10.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reboul J, Gardiner K, Monneron D, Uze G, Lutfalla G. Genome Res. 1999;9:242–250. [PMC free article] [PubMed] [Google Scholar]
- 23.Novick D, Cohen B, Rubinstein M. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 24.Domanski P, Witte M, Kellum M, Rubinstein M, Hackett R, Pitha P, Colamonici O R. J Biol Chem. 1995;270:21606–21611. doi: 10.1074/jbc.270.37.21606. [DOI] [PubMed] [Google Scholar]
- 25.Owczarek C M, Hwang S Y, Holland K A, Gulluyan L M, Tavaria M, Weaver B, Reich N C, Kola I, Hertzog P J. J Biol Chem. 1997;272:23865–23870. doi: 10.1074/jbc.272.38.23865. [DOI] [PubMed] [Google Scholar]
- 26.Lutfalla G, Holland S J, Cinato E, Monneron D, Reboul J, Rogers N C, Smith J M, Stark G R, Gardiner K, Mogensen K E, et al. EMBO J. 1995;14:5100–5108. doi: 10.1002/j.1460-2075.1995.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen B, Novick D, Barak S, Rubinstein M. Mol Cell Biol. 1995;15:4208–4214. doi: 10.1128/mcb.15.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cutrone E, Langer J A. FEBS Lett. 1997;404:197–202. doi: 10.1016/s0014-5793(97)00129-4. [DOI] [PubMed] [Google Scholar]
- 29.Kim S H, Cohen B, Novick D, Rubinstein M. Gene. 1997;196:279–286. doi: 10.1016/s0378-1119(97)00240-0. [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer L M, Basu L, Pfeffer S R, Yang C H, Murti A, Russell-Harde D, Croze E. J Biol Chem. 1997;272:11002–11005. doi: 10.1074/jbc.272.17.11002. [DOI] [PubMed] [Google Scholar]
- 31.Piehler J, Schreiber G. J Mol Biol. 1999;289:57–67. doi: 10.1006/jmbi.1999.2726. [DOI] [PubMed] [Google Scholar]
- 32.Bazan J F. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoreau E, Petridou B, Kelly P A, Djiane J, Mornon J P. FEBS Lett. 1991;282:23–31. doi: 10.1016/0014-5793(91)80437-8. [DOI] [PubMed] [Google Scholar]
- 34.Cook J R, Cleary C M, Mariano T M, Izotova L, Pestka S. J Biol Chem. 1996;271:13448–13453. doi: 10.1074/jbc.271.23.13448. [DOI] [PubMed] [Google Scholar]
- 35.Abramovich C, Ratovitski E, Lundgren E, Revel M. FEBS Lett. 1994;338:295–300. doi: 10.1016/0014-5793(94)80287-4. [DOI] [PubMed] [Google Scholar]
- 36.Alexenko A P, Leaman D W, Li J, Roberts R M. J Interferon Cytokine Res. 1997;17:769–779. doi: 10.1089/jir.1997.17.769. [DOI] [PubMed] [Google Scholar]
- 37.Binelli M, Subramaniam P, Diaz T, Johnson G A, Hansen T R, Badinga L, Thatcher W W. Biol Reprod. 2001;64:654–665. doi: 10.1095/biolreprod64.2.654. [DOI] [PubMed] [Google Scholar]
- 38.Wilson K C, Finbloom D S. Proc Natl Acad Sci USA. 1992;89:11964–11968. doi: 10.1073/pnas.89.24.11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B L, Langer J A, Schwartz B, Pestka S. Proc Natl Acad Sci USA. 1989;86:558–562. doi: 10.1073/pnas.86.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X X, Li B L, Langer J A, Van Riper G, Pestka S. Anal Biochem. 1989;178:342–347. doi: 10.1016/0003-2697(89)90650-7. [DOI] [PubMed] [Google Scholar]
- 41.Ealy A D, Green J A, Alexenko A P, Keisler D H, Roberts R M. Biol Reprod. 1998;58:566–573. doi: 10.1095/biolreprod58.2.566. [DOI] [PubMed] [Google Scholar]
- 42.Han C S. Ph.D. Dissertation. Columbia: University of Missouri; 2000. [Google Scholar]
- 43.Lutfalla G, Gardiner K, Proudhon D, Vielh E, Uze G. J Biol Chem. 1992;267:2802–2809. [PubMed] [Google Scholar]
- 44.Alexenko A P, Ealy A D, Roberts R M. J Interferon Cytokine Res. 1999;19:1335–1341. doi: 10.1089/107999099312795. [DOI] [PubMed] [Google Scholar]
- 45.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 46.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 47.Decker T, Lew D J, Darnell J E. Mol Cell Biol. 1991;11:5147–5153. doi: 10.1128/mcb.11.10.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen N Y, Sackett D, Hirata R D C, Levy D E, Enterline J C, Bekisz J B, Hirata M H. J Interferon Cytokine Res. 1996;16:835–844. doi: 10.1089/jir.1996.16.835. [DOI] [PubMed] [Google Scholar]
- 49.Yoon S, Hirata R D C, Nguyen N Y, Curi R, Russo M, Hirata M H. Braz J Med Biol Res. 2000;33:771–778. doi: 10.1590/s0100-879x2000000700007. [DOI] [PubMed] [Google Scholar]
- 50.Arduini R M, Strauch K L, Runkel L A, Carlson M M, Hronowski X, Foley S F, Young C N, Cheng W, Hochman P S, Baker D P. Protein Sci. 1999;8:1867–1877. doi: 10.1110/ps.8.9.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardy M P, Owczarek C M, Trajanovska S, Liu X, Kola I, Hertzog P J. Blood. 2001;97:473–482. doi: 10.1182/blood.v97.2.473. [DOI] [PubMed] [Google Scholar]
- 52.Lewerenz M, Mogensen K E, Uze G. J Mol Biol. 1998;282:585–599. doi: 10.1006/jmbi.1998.2026. [DOI] [PubMed] [Google Scholar]
- 53.Pattyn E, Van Ostade X, Schauvliege L, Verhee A, Kalai M, Vandekerckhove J, Tavernier J. J Biol Chem. 1999;274:34838–34845. doi: 10.1074/jbc.274.49.34838. [DOI] [PubMed] [Google Scholar]
- 54.Kotenko S V, Izotova L S, Mirochnitchenko O V, Lee O C, Pestka S. Proc Natl Acad Sci USA. 1999;96:5007–5012. doi: 10.1073/pnas.96.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]