Abstract
Autophagy defends the mammalian cytosol against bacterial infection.1-3 Efficient pathogen engulfment is mediated by cargo-selecting autophagy adaptors that rely on unidentified pattern-recognition or danger receptors to label invading pathogens as autophagy cargo, typically by poly-ubiquitin coating.4-9 Here we show that galectin-8, a cytosolic lectin, is a danger receptor that restricts Salmonella proliferation. Galectin-8 monitors endo-lysosomal integrity and detects bacterial invasion by binding host glycans exposed on damaged Salmonella-containing vacuoles. By recruiting NDP52 galectin-8 activates anti-bacterial autophagy. Galectin-8-dependent recruitment of NDP52 to Salmonella-containing vesicles is transient and followed by ubiquitin-dependent NDP52 recruitment. Since galectin-8 also detects sterile damage to endosomes or lysosomes, as well as invasion by Listeria or Shigella, we suggest galectin-8 serves as a versatile receptor for vesicle-damaging pathogens. Our results illustrate how cells deploy the danger receptor galectin-8 to combat infection by monitoring endo-lysososomal integrity based on the specific lack of complex carbohydrates in the cytosol.
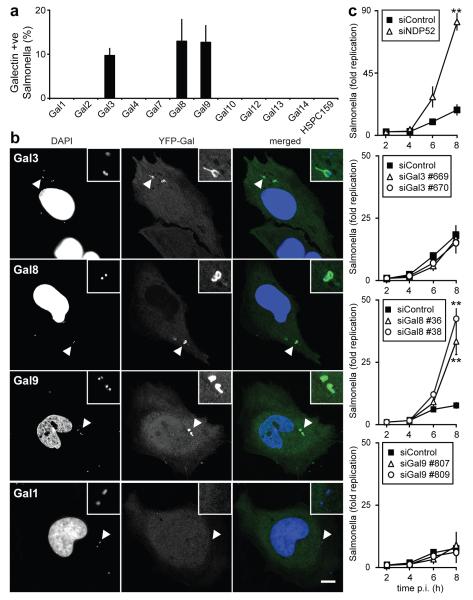
Galectins are beta-galactoside-binding lectins that accumulate in the cytosol before being secreted via a leader peptide-independent pathway.10,11 The best-characterized functions of galectins are performed extracellularly, where they bind glycans to modulate cellular behaviour. However, the occurrence of galectins in the cytosol, which under physiological conditions is devoid of complex carbohydrates, makes them prime candidates for a role as danger and/or pattern recognition receptors. Galectin-3 accumulates on damaged bacteria-containing vesicles, although the functional consequences of its recruitment remain unknown.12,13 We screened a panel of human galectins for their ability to detect invasion by S.Typhimurium. At 1h post infection (p.i.) galectin-3, 8, and 9 accumulated on about 10% of intracellular S.Typhimurium (Fig.1a,b,S1a), of which 90% were associated with LAMP1 (Fig.S1b). Recruitment of galectins peaked between 1 and 2h p.i. (Fig.S1c). Since galectin-3, 8, and 9 were recruited to SCVs, we used siRNAs to test whether their depletion causes hyperproliferation of S.Typhimurium. Cells lacking galectin-8 or NDP52, but not galectin-3 and/or 9, failed to suppress proliferation of S.Typhimurium (Fig.1c,S2a-c,S3a). Microscopic analysis confirmed that the greater bacterial burden of cells lacking galectin-8 was caused by enhanced proliferation rather than differential uptake of bacteria (Fig.S3b). Hyperproliferating bacteria in cells lacking galectin-8 appeared mainly in a LAMP1-negative compartment (Fig.S3c), consistent with colonization of the cytosol. We conclude that galectin-8 is an anti-bacterial restriction factor.
Figure 1. Galectin-8 responds to infection by S.Typhimurium and restricts bacterial proliferation.
A-B) Analysis of HeLa cells stably expressing YFP fused to the indicated galectins and infected with S.Typhimurium for 1h. A) Percentage of bacteria coated by the indicated galectins. YFP-positive bacteria were counted by microscopy. Mean and s.d. of triplicate HeLa cultures, n>100 bacteria per coverslip. B) Confocal micrographs. Arrowheads, bacteria shown in insets.
C) Kinetics of fold replication for S.Typhimurium in HeLa cells transfected with the indicated siRNAs. Bacteria were counted on the basis of their ability to form colonies on agar plates. Mean and s.d. of triplicate HeLa cultures and duplicate colony counts. siRNAs are further characterized in Supplementary Figure 2a-c.
** P<0.01, Student’s t-test. Scale bar 10 μm.
Since autophagy provides anti-bacterial protection to cells, the decoration of SCVs with galectins might be an autophagy-inducing signal, analogous to ubiquitin-coating.14,15 We therefore tested binding of galectins to autophagy receptors that restrict the proliferation of S.Typhimurium, i.e. NDP52, p62, and Optineurin.7-9 We found in a LUMIER assay that galectin-8 and NDP52 interacted specifically (Fig.2a,S4a). Binding was confirmed by precipitating endogenous NDP52 with Flag-tagged galectin-8 (Fig.2b).
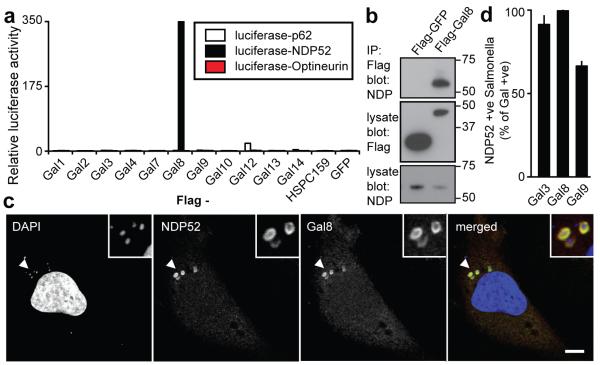
Figure 2. Galectin-8 binds NDP52.
A) LUMIER binding assay: Normalized ratio between luciferase activity bound to beads and present in lysates. Lysates of 293ET cells expressing NDP52, p62, or Optineurin each fused to luciferase, and the indicated Flag-tagged galectins were incubated with anti-Flag beads. Flag-tagged proteins are further characterized in Supplementary Figure S4a.
B) Lysates of 293ET cells, expressing Flag-tagged proteins as indicated, were immunoprecipitated with anti-Flag beads. Lysates and precipitates (IP) were blotted for the presence of Flag-tagged proteins and endogenous NDP52.
C) Confocal images of HeLa cells infected with S.Typhimurium for 1h and stained with antisera against NDP52 and galectin-8. Arrowheads, bacteria shown in insets.
D) Colocalization of NDP52 with galectin-positive bacteria in HeLa cells stably expressing YFP fused to the indicated galectins, infected with S.Typhimurium and stained with NDP52 antiserum 1h after infection. Mean and s.d. of duplicate HeLa cultures, n>100 bacteria per coverslip, representative of 2 independent experiments. Scale bar 10 μm.
SCVs double-labelled by endogenous galectin-8 and NDP52 were prominent in Salmonella-infected HeLa cells (Fig.2c). In cells expressing YFP-tagged galectins the majority of galectin-positive SCVs had accumulated NDP52 (Fig.2d,S5a). Furthermore, at 1h p.i. NDP52 and galectin-8 co-localized tightly in a pattern distinct from p62 or ubiquitin ‘microdomains’16 (Fig.S5b,c).
To further characterize the interaction between galectin-8 and NDP52 we determined their respective binding sites. Galectin-8 contains two carbohydrate-recognition domains (CRD) (Fig.S6a). NDP52 bound galectin-8Δ1-228, equivalent to the second CRD, but not Galectin-81-228 (Fig.S6b). NDP52 harbours a SKICH domain, a coiled coils-forming region, and an ubiquitin-binding zinc finger (Fig.S6a). Galectin-8 bound NDP521-393 but not NDP521-370 (Fig.S6c). The NDP52 fragment spanning residues 370 to 393 is therefore essential for binding galectin-8. This fragment, as well as NDP52372-380, purified as GST-fusion proteins, bound galectin-8 (Fig.S6d). A point mutation within NDP52372-380 (L374A) abrogated binding to galectin-8, without compromising binding to ubiquitin when introduced into full-length NDP52 (Fig.S6d,e) Binding of galectin-8 to NDP52 is direct since the purified proteins interacted (Fig.S6f).
To determine whether one monomer of the NDP52-galectin-8 heteromeric complex recruits the other partner, the accumulation of galectins on SCVs in cells depleted of NDP52 or TBK1 was analysed (Fig.3a, Fig.S2). Galectin-3, 8, and 9 re-distributed normally to SCVs in siRNA treated cells. In contrast, in cells depleted of galectin-8 NDP52 did not localize to SCVs at 1h p.i, a phenotype that was complemented upon expression of siRNA-resistant galectin-8 (Fig.3b,S7). Cells lacking galectin-3 or galectin-9 had no defect in recruiting NDP52 to SCVs. The recruitment of NDP52 to SCVs is therefore specifically mediated by galectin-8, while NDP52 and TBK1 are dispensable for the accumulation of galectins on SCVs.
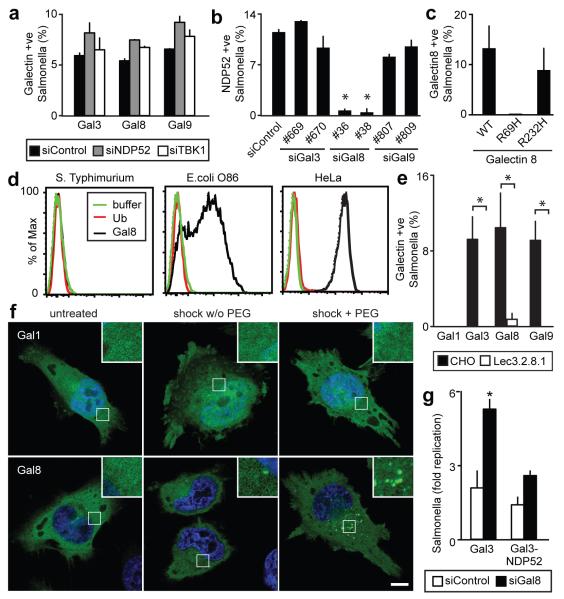
Figure 3. Galectin-8 is a danger receptor that senses cytosolic host glycans and recruits NDP52 to restrict Salmonella proliferation.
A) Percentage of S.Typhimurium coated by the indicated galectins. HeLa cells stably expressing YFP-tagged galectins were treated with the indicated siRNAs. YFP-positive bacteria were counted by microscopy at 1h post infection. siRNAs are further characterized in Supplementary Figure 2.
B) Analysis of HeLa cells treated with the indicated siRNAs and stained with NDP52 antiserum. NDP52-positive bacteria were counted by microscopy 1h after infection with S.Typhimurium. siRNAs are further characterized in Supplementary Figure 2.
C) Percentage of bacteria coated by the indicated galectin-8 alleles. HeLa cells stably expressing the indicated galectin-8 alleles fused to YFP were infected with S.Typhimurium. YFP-positive S.Typhimurium were counted by microscopy at 75min p.i.
D) Binding of galectin-8 to bacteria and HeLa cells. The indicated bacteria and HeLa cells were incubated with His-GST-ubiquitin, His-GST-galectin-8 or buffer as indicated, followed by murine anti-His antibody and PE-labelled anti-mouse serum.
E) Percentage of S.Typhimurium coated by the indicated galectins. Wild type CHO cells and mutant Lec3.2.8.1 cells stably expressing YFP-tagged galectins were infected with S.Typimurium. YFP-positive bacteria were counted by microscopy at 1h post infection.
F) Confocal images of HeLa cells expressing the indicated YFP-tagged galectins. Cells were left untreated or were exposed to hypertonic conditions, with or without PEG as indicated, followed by hypotonic shock.
G) Fold replication of S.Typhimurium in HeLa cells expressing the indicated galectin-3 variants and transfected with the indicated siRNAs. At 2 and 6h after infection, cells were lysed and bacteria counted on the basis of their ability to form colonies on agar plates. Galectin-3 proteins are further characterized in Supplementary Figure 4b. Mean and s.d. of duplicate coverslips (A-C,E) or triplicate HeLa cultures and duplicate colony counts (G). >100 bacteria counted per coverslip. Data are representative of at least two repeats. * P<0.05, Student’s t-test. Scale bar 10 μm
Rupture of SCVs exposes the cytosol to host glycans and microbial carbohydrates, either or both of which may cause galectin-8 accumulation at SCVs. The requirement for carbohydrate binding by galectin-8 was tested using point mutations in either CRD.17 In contrast to galectin-8R232H, galectin-8R69H did not accumulate at SCVs, proving that the N-terminal CRD is required for carbohydrate-dependent recruitment of galectin-8 to SCVs (Fig.3c). To test whether the carbohydrates detected by galectin-8 are of microbial origin, binding of recombinant galectin-8 to bacteria in vitro was analyzed. Galectin-8 did not bind to S.Typhimurium but stained blood-group B positive bacteria (E.coli O86)17 (Fig.3d), suggesting that galectin-8, when accumulating on SCVs, recognizes host glycans. The occurrence of galectin-8 ligands in host cells was confirmed by staining HeLa cells with recombinant galectin-8 (Fig.3d). Direct evidence that host glycans recruit galectins to SCVs was obtained from experiments with CHO-Lec3.2.8.1 cells18, which lack mature glycans and in which recruitment of galectins to SCVs was severely impaired (Fig.3e). The detection of host glycans on damaged vesicles by galectin-8 suggests it is not a receptor specific for S.Typhimurium. We therefore tested whether sterile damage to vesicles is detected by galectins. Osmotic damage of endosomes induced dense puncta formed by galectin-3, 8, and 9 but not by galectin-1 (Fig.3f,S8). Damage to lysosomes by GPN treatment resulted in the initial loss of lysotracker staining, followed by the appearance of galectin-3, 8, and 9 speckles (Fig.S9a). In contrast to damaged SCVs and endosomes, burst lysosomes were also detected by galectin-1, suggesting compartment-specific differences in the distribution of galectin ligands. GPN failed to induce speckles of galectin-8R69H (Fig.S9b), thereby indicating that binding of glycans to the N-terminal CRD of galectin-8 is required to detect lysosomal damage. The capacity of galectin-3, 8, and 9 to detect vesicle damage by binding exposed host glycans suggests their ability to sense the invasion of cells by a wide range of vesicle-damaging pathogens. Indeed, galectin-3, 8, and 9 also accumulated around Gram-positive L.monocytogenes and Gram-negative S.flexneri (Fig.S10), proving that these galectins detect the invasion of cells by phylogenetically distant bacteria. We conclude that galectin-3, 8, and 9 are danger receptors that sense the exposure of host glycans on ruptured membranes and thereby monitor the integrity of the endosomal / lysosomal compartment.
To test whether the recruitment of NDP52 to SCVs is essential for the anti-bacterial function of galectin-8, we depleted cells of galectin-8 and targeted NDP52 artificially to SCVs by fusing it to galectin-3 (Fig.3g, Fig.S4b). S.Typhimurium hyperproliferated in galectin-8-depleted cells, while expression of galectin-3 fused to NDP52 restored the cells’ restrictive capacity. Artificial targeting of NDP52 to SCVs therefore compensates for the lack of galectin-8, which strongly suggests that the recruitment of NDP52 via galectin-8 is essential to efficiently antagonize bacterial invasion. NDP52 restricts the proliferation of S.Typhimurium by targeting bacteria for autophagy.8 Since galectin-8 recruits NDP52 to SCVs, we investigated whether galectin-8 is required upstream of NDP52 for the induction of anti-bacterial autophagy. First, we confirmed that at 1h p.i. S.Typhimurium that had been sensed by galectin-8, and therefore had acquired an NDP52 coat, were taken up into LC3-positive autophagosomes (Fig.4a). Such an outcome was predicted from the pairwise co-localization of NDP52 with galectin-8- and LC3-positive bacteria (Fig.2d, S11). We then tested whether depletion of galectin-8 impairs autophagy of S.Typhimurium. In the absence of galectin-8 fewer bacteria were targeted by LC3 (Fig.4b) and of the remaining LC3-positive bacteria fewer had accumulated NDP52 (Fig.S11). In contrast, galectin-8 recruitment to SCVs did not require autophagy as it occurred undisturbed in ATG5−/− fibroblasts (Fig.S12). We conclude that the danger receptor galectin-8, by recruiting NDP52, directs autophagy towards invading bacteria.
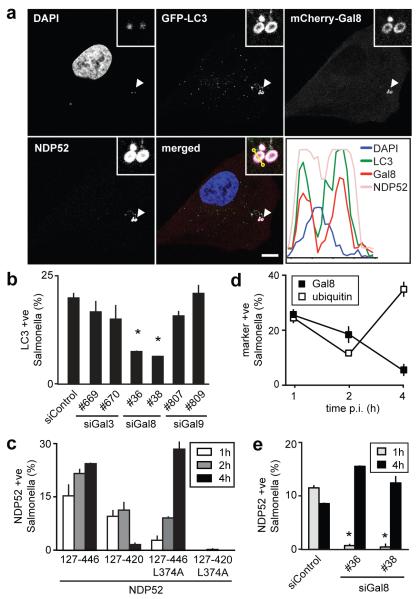
Figure 4. The anti-bacterial effect of galectin-8 is mediated by autophagy.
A) Confocal micrograph of HeLa cells expressing GFP-LC3 and mCherry-galectin-8, stained for NDP52 1h after infection with S.Typhimurium. The lower right panel contains a fluorescence line scan along the yellow line in the merge inset. Arrowheads, bacteria shown in insets.
B) Percentage of GFP-LC3 positive S.Typhimurium at 1h p.i. in HeLa cells treated with the indicated siRNAs.
C) Percentage of bacteria positive for NDP52. HeLa cells expressing the indicated NDP52 variants fused to YFP were infected with S.Typhimurium.
D) Percentage of bacteria positive for the indicated markers. HeLa cells, either wild type or expressing YFP-galectin-8 as indicated, were infected with S.Typhimurium. Ubiquitin was detected by antibody staining.
E) HeLa cells treated with the indicated siRNAs were infected with S.Typhimurium and stained for NDP52 at the indicated time points.
Fluorescent bacteria were counted by microscopy at the indicated time points. Graphs, representing at least two independent repeats, show mean and s.d. of duplicate coverslips for which >200 bacteria were counted. * P<0.05, Student’s t-test, scale bar 10 μm.
The recruitment of NDP52 to invading bacteria is mediated by two signals, the newly-discovered carbohydrate-dependent galectin-8 pathway and the previously known ubiquitin-dependent pathway.8 Their differential contribution to the recruitment of NDP52 to S.Typhimurium was investigated by analyzing NDP52 mutants selectively disabled to bind galectin-8 and / or ubiquitin. For accurate scoring, NDP52ΔSKICH was used as this truncated allele is distributed diffusely throughout the cytosol. NDP52127-446 associated with SCVs at all time points investigated (Fig.4c). Deleting the C-terminal ubiquitin-binding zinc finger (NDP52127-420) impaired the recruitment of NDP52 to S.Typhimurium at late but not early time points. In contrast, NDP52127-446L374A, which lacks affinity for galectin-8 but not ubiquitin (Fig.S6e), did not co-localize with bacteria at 1h p.i. but accumulated progressively over time (Fig.4c). NDP52 is therefore recruited to SCVs in two phases – an early transient surge driven by galectin-8 and a later wave depending on ubiquitin. The kinetics of galectin-8 and ubiquitin recruitment to SCVs support this model as SCVs are marked by Galectin-8 only at early time points, while ubiquitin marks persist (Fig.4d). Direct proof for early galectin-8-dependent and late galectin-8-independent recruitment of NDP52 to S.Typhimurium was obtained from cells depleted of galectin-8, in which NDP52 and bacteria co-localized at 4h but not at 1h p.i. (Fig.4e). NDP52127-420L374A, deficient in binding to ubiquitin and galectin-8, did not translocate to SCVs at any time point tested (Fig.4c). Taken together, NDP52 relocates to SCVs in response to two signals, which are active against bacteria at different stages of invasion. The early response to invading bacteria requires the galectin-8-dependent pathway, while the zinc finger-dependent pathway dominates at later time points.
The discovery of the galectin-8/NDP52 pathway helps to understand why most intracellular bacteria avoid the cytosol and prefer vesicular compartments. The cytosol appears protected by synergistic layers of antibacterial defence that activate autophagy at distinct steps of the invasion process. An early line of defence comprises the accumulation of diacylglycerol on bacteria-containing vesicles, which subsequently become the target of autophagy.19 Bacteria escaping the DAG pathway and exposing host glycans on their damaged vacuoles are targeted by galectin-8 and NDP52, as described in this work. A third layer of defence coats invading bacteria with poly-ubiquitin.6-9,20 Neither the enzymatic machinery for nor the substrate of ubiquitylation have been identified, although LRSAM1, a RING-finger E3 ubiquitin ligase, contributes to autophagy of S.Typhimurium.21 Peptidoglycan and septin-cages surrounding cytosolic bacteria also contribute to autophagy.22-25 Defects in this intricate network of autophagy-inducing defence pathways are likely to cause susceptibility to infection and promote inflammation, for example in Crohn’s disease.26-29 Galectin-8 is positioned strategically at the cellular entry point for a variety of pathogens and is therefore expected to have shaped pathogen evolution.
Supplementary Material
Acknowledgments
We thank J. Kendrick-Jones (MRC Laboratory of Molecular Biology), A. Geerlof (European Molecular Biology Laboratory Heidelberg), N. Mizushima (Tokyo University), and P. Stanley (Albert Einstein College of Medicine) for kindly sharing reagents.
APPENDIX
Methods Summary
Galectins were cloned as YFP fusions and transduced into HeLa cells. HeLa cells were infected with S.Typhimurium 12023. For confocal microscopy cells were fixed in paraformaldehyde. Bacterial growth was assessed by gentamycin protection assay. Knockdowns were accomplished with Stealth™ siRNAs. LUMIER assays were performed as described.30 For flow-cytometric analysis samples were incubated with lysates of E. coli expressing His:GST fusion proteins, followed by anti-His antibody and goat-anti-mouse serum. Statistical testing was performed using two-tailed Student’s t-test.
Full Methods
Antibodies
Antibodies were from QIAGEN (Penta-His), Developmental Studies Hybridoma Bank (Lamp1), BD Transduction Laboratories (p62), Santa Cruz (Gal8-H80, TBK1-C100), R&D Systems (galectin-8), Transduction Laboratories (NDP52, for western blots), Enzo Life Science (ubiquitin FK2), Sigma (ATG5, FLAG M2), Dabco (HRP-conjugated reagents), Jackson ImmunoResearch Laboratories (goat-anti-mouse-PE), and Invitrogen (Alexa-conjugated anti-mouse and anti-rabbit antisera). The antiserum against NDP52 used for immunofluorescence was a kind gift from John Kendrick-Jones.
Plasmids
M5P or closely related plasmids were used to produce recombinant MLV for the expression of proteins in mammalian cells.32 pETM plasmids were gifts from A. Geerlof. Open reading frames encoding human galectins, NDP52, p62, Optineurin, ubiquitin, ATG5, and LC3C were amplified by PCR or have been described.8,31 Mutations were generated by PCR and verified by sequencing.
Bacteria
S.Typhimurium (strain 12023), provided by David Holden, was grown overnight in LB and sub-cultured (1:33) in fresh LB for 3.5 hours prior to infection. HeLa cells in 24 well plates were infected with 20 μl of such cultures for 15 minutes at 37 degrees. Following two washes with warm PBS and an incubation with 100 μg/ml gentamycin for two hours cells were cultured in 20 μg/ml gentamycin. To enumerate intracellular bacteria, cells from triplicate wells were lysed in 1 ml cold PBS containing 0.1 % Triton X100. Serial dilutions were plated in duplicate on TYE agar.
S. flexneri M90T, provided by Chris Tang, was grown overnight in Tryptic Soy Broth (TSB) and sub-cultured (1:100) in fresh TSB for 2 hours prior to infection. Bacteria were resuspended in warm IMDM and 100 μl were added to HeLa cells in 24 well plates. Samples were centrifuged for 10 minutes at 2000 rpm. Following an incubation at 37 °C for 30 minutes, cells were washed with warm PBS and cultured in 100 μg/ml gentamycin for 2 hours and 20 μg/ml thereafter.
L. monocytogenes strain EGD (BUG 600), provided by Pascale Cossart, was grown overnight in Brain Heart Infusion (BHI) at 30 °C with shaking. 500 μl of diluted cultures (1:333) were added to HeLa cells in 24 well plates, which were centrifuged at 2000 rpm for 10 minutes. Cells were incubated for 1 hour at 37 °C, washed with warm PBS and cultured in media supplemented with 100 μg/ml gentamycin for the next hour and 20 μg/ml gentamycin thereafter.
Cell culture
Cells were grown in IMDM supplemented with 10% FCS at 37°C in 5% CO2. HeLa cells were obtained from the European Collection of Cell Cultures, CHO and Lec3.2.8.118 from P. Stanley, ATG5−/− MEFs33 from N. Mizushima.
RNAi
5×104 cells per well were seeded in 24 well plates. The following day, cells were transfected with 40 pmol of siRNA (Invitrogen) using Lipofectamine 2000 (Invitrogen) in Optimem medium (Invitrogen). Optimem was replaced with complete IMDM medium after four hours and experiments were performed after three days. siRNAs targeted the following sequences:
siNDP52 5′UUCAGUUGAAGCAGCUCUGUCUCCC8
siGAL8#36 5′CCCACGCCUGAAUAUUAAAGCAUUU
siGAL8#38 5′GGACAAAUUCCAGGUGGCUGUAAAU
siGAL3#669 5′AAGCCCAAUGCAAACAGAAUUGCUU
siGAL3#670 5′GAGAACAACAGGAGAGUCAUUGUUU
siGAL9#807 5′GGCUUCAGUGGAAAUGACAUUGCCU
siGAL9#809 5′UGUGCAACACGAGGCAGAACGGAGG
siTBK1 5′GACAGAAGUUGUGAUCACA(TT)34
To render galectin-8 resistant to siGal8#38 silent mutations (underlined) GGATAAGTTTCAAGTCGCAGTTAAT were introduced by PCR and confirmed by sequencing.
Immunoprecipitation and Western blot
Post-nuclear supernatants from 2 x 106 HeLa cells expressing Flag-tagged proteins were obtained following lysis (150 mM NaCl, 0.1% Triton-X100, 20 mM Tris-HCl (pH 7.4), 5 mM EDTA and proteinase inhibitors). Protein complexes were immunoprecipitated for 2 hours with FLAG agarose prior to washing. Samples were eluted with FLAG peptide and separated on 4-12% denaturing Bis-Tris gels (Invitrogen). Visualization following immunoblotting was performed using ECL detection reagents (Amersham Bioscience).
LUMIER assays.30,35
Binding assays with pairs of putative interactors, one fused to luciferase and the other fused to GST or Flag, were performed in lumier lysis buffer (150 mM NaCl, 0.1% Triton-X100, 20 mM Tris-HCl (pH 7.4), 5% glycerol, 5 mM EDTA and proteinase inhibitors). GST-fusion proteins were immobilized on beads before incubation with the luciferase tagged binding partner for two hours. For Flag-based assays, both proteins were expressed in 293ET cells and immobilized using Flag-agarose. After washing in lysis buffer, proteins were eluted with glutathione or Flag peptide in Renilla lysis buffer (Promega). Relative luciferase activity represents the ratio of activity eluted from beads and present in lysates.
FACS
To examine the binding of galectin-8, bacteria in stationary phase or HeLa cells were washed in PBSF (PBS, 2% FCS) and incubated for 30min at 4°C with cleared lysates of E. coli expressing His:GST fusion proteins, followed by incubations with anti-His antibody and PE-conjugated goat-anti-mouse serum. Bacteria were fixed in 4% paraformaldehyde before analysis.
Sterile damage to vesicles
Endosomes were lysed by exposing cells for 10 minutes to hypertonic medium (0.5M sucrose in PBS, with or without 10% PEG100), followed by two PBS washes and an incubation in 60% PBS for 3 minutes.36 Cells were returned to complete medium for 20 minutes, before being fixed in paraformaldehyde. For live imaging of lysosomal damage, cells were labelled for 1h with 100nM LysoTracker Red (Invitrogen), washed with PBS, incubated in Leibovitz L15 medium and, after acquisition of the first image, exposed to 333μM glycyl-L-phenylalanine 2-naphthylamide (GPN)37.
Microscopy
HeLa cells were grown on glass cover slips prior to infection. Following infections, cells were washed twice with warm PBS and fixed in 4% paraformaldehyde in PBS for 30 minutes. Cells were washed twice in PBS and then quenched with PBS pH 7.4 containing 1M glycine and 0.1% Triton X100 for 30 minutes prior to blocking for 30 minutes in PBTB (PBS, 0.1% Triton X100, 2% BSA). Cover slips were incubated with primary, followed by secondary antibodies for one hour in PBTB before being mounting in medium containing DAPI (Vector Laboratories). At least 100 events per slide were scored in quantitative assays. Confocal images were taken with a 63×1.4 objective on either a Zeiss 710 or a Zeiss 780 microscope. Live imaging was performed on a Nikon Eclipse Ti equipped with an Andor Revolution XD system and a Yokogawa CSU-X1 spinning disk unit.
References for Methods
- 31.Bloor S, et al. Signal processing by its coil zipper domain activates IKK gamma. Proc Natl Acad Sci USA. 2008;105:1279–1284. doi: 10.1073/pnas.0706552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randow F, Sale JE. Retroviral transduction of DT40. Subcell Biochem. 2006;40:383–386. doi: 10.1007/978-1-4020-4896-8_30. [DOI] [PubMed] [Google Scholar]
- 33.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald KA, et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 35.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 36.Shaughnessy LM, Lipp P, Lee K-D, Swanson JA. Localization of protein kinase C epsilon to macrophage vacuoles perforated by Listeria monocytogenes cytolysin. Cell Microbiol. 2007;9:1695–1704. doi: 10.1111/j.1462-5822.2007.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg TO, Strømhaug PE, Berg T, Seglen PO. Separation of lysosomes and autophagosomes by means of glycyl-phenylalanine-naphthylamide, a lysosome-disrupting cathepsin-C substrate. Eur J Biochem. 1994;221:595–602. doi: 10.1111/j.1432-1033.1994.tb18771.x. [DOI] [PubMed] [Google Scholar]
References
- 1.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 5.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7 doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin A, Jiang X, Birmingham C, So N, Brumell J. Recognition of Bacteria in the Cytosol of Mammalian Cells by the Ubiquitin System. Current Biology. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 8.Thurston TLM, Ryzhakov G, Bloor S, Von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 9.Wild P, et al. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science. 2011 doi: 10.1126/science.1205405. doi:10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houzelstein D, et al. Phylogenetic analysis of the vertebrate galectin family. Mol Biol Evol. 2004;21:1177–1187. doi: 10.1093/molbev/msh082. [DOI] [PubMed] [Google Scholar]
- 11.Rabinovich GA, Toscano MA. Turning "sweet" on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 2009;9:338–352. doi: 10.1038/nri2536. [DOI] [PubMed] [Google Scholar]
- 12.Paz I, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01415.x. doi:10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 13.Dupont N, et al. Shigella Phagocytic Vacuolar Membrane Remnants Participate in the Cellular Response to Pathogen Invasion and Are Regulated by Autophagy. Cell Host and Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Randow F. How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy. 2011;7 doi: 10.4161/auto.7.3.14539. [DOI] [PubMed] [Google Scholar]
- 15.Shahnazari S, Brumell JH. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Current Opinion in Microbiology. 2011;14:68–75. doi: 10.1016/j.mib.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:22–26. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.PATNAIK S, STANLEY P. Lectin‐Resistant CHO Glycosylation Mutants. Methods in Enzymology. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 19.Shahnazari S, et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host and Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins CA, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng ACY, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci USA. 2010:1–8. doi: 10.1073/pnas.1000093107. doi:10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostowy S, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host and Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 25.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 26.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 27.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 30.Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 2007;26:3180–3190. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.