Abstract
During meiotic cell division, proper chromosome synapsis and accurate repair of DNA double strand breaks (DSBs) are required to maintain genomic integrity, loss of which leads to apoptosis or meiotic defects. The mechanisms underlying meiotic chromosome synapsis, DSB repair and apoptosis are not fully understood. Here, we report that the chromodomain-containing protein MRG-1 is an important factor for genomic integrity in meiosis in Caenorhabditis elegans. Loss of mrg-1 function resulted in a significant increase in germ cell apoptosis that was partially inhibited by mutations affecting DNA damage checkpoint genes. Consistently, mrg-1 mutant germ lines exhibited SPO-11-generated DSBs and elevated exogenous DNA damage-induced chromosome fragmentation at diakinesis. In addition, the excessive apoptosis in mrg-1 mutants was partially suppressed by loss of the synapsis checkpoint gene pch-2, and a significant number of meiotic nuclei accumulated at the leptotene/zygotene stages with an elevated level of H3K9me2 on the chromatin, which was similarly observed in mutants deficient in the synaptonemal complex, suggesting that the proper progression of chromosome synapsis is likely impaired in the absence of mrg-1. Altogether, these findings suggest that MRG-1 is critical for genomic integrity by promoting meiotic DSB repair and synapsis progression in meiosis.
Keywords: apoptosis, DSB repair, synapsis, meiosis, genomic integrity, C. elegans
Introduction
Genomic integrity is a prerequisite for the survival, development and propagation of eukaryotic organisms. The genome is, however, frequently attacked by either endogenous or exogenous DNA-damaging agents that generate a wide variety of DNA lesions. To maintain the integrity of their genomes, eukaryotic organisms have developed multiple evolutionarily conserved strategies to repair the lesions caused by DNA-damaging insults 1. When DNA lesions are too severe to repair, cells are destined to undergo apoptosis so as to avoid their deleterious propagation. Defective DNA repair or abnormal apoptosis leads to genomic instability, which is a hallmark of cancer 2. On the other hand, the maintenance of genomic integrity is required for faithful transmission of genetic information from parents to progeny through meiosis, a specialized cell division program in which DNA replicates once followed by two successive cell divisions. During the prophase of meiosis I, chromosomes undergo a series of programmed events, including sister chromatid cohesion, pairing of homologous chromosomes, synapsis, and homologous recombination 3. Defects in any of these steps may result in aberrant chromosome segregation, leading to sterility, X chromosome non-disjunction, aneuploidy, or embryonic lethality 3, 4.
The regulatory mechanisms underlying the maintenance of genomic integrity in the process of meiosis are evolutionarily conserved. In the nematode C. elegans, genetic and cell biological studies have revealed key components required for distinct chromosomal events in the prophase of meiosis I 3, 4, 5. Following DNA replication, chromosome condensation begins and homologous chromosomes pair and align longitudinally. The meiosis-specific protein REC-8 and some COH (cohesion protein homologs) paralogs are essential for sister chromatid cohesion, while the zinc-finger proteins HIM-8, ZIM-1, -2, and -3 promote chromosome pairing and synapsis 3, 5, 6. The proteinaceous synaptonemal complex (SC), which consists of the lateral elements HTP-1, -2, -3, and HIM-3 and the central elements SYP-1, -2, -3 and -4, forms a zipper-like structure to hold homologous chromosomes together along their entire lengths. Loss-of-function mutations (lf) in SC components lead to synaptic failure, which manifests as the presence of up to 12 univalents in diakinesis oocytes, compared with 6 bivalents representing six well-paired homologous chromosomes in wild-type oocytes 3, 5, 7, 8. Concomitant with the morphological reorganization of chromosomes, DNA molecules also undergo programmed double strand breaks (DSBs) and their repair results in the formation of crossovers between homologous chromosomes. DSBs are normally induced by an evolutionarily conserved topoisomerase-like endonuclease, SPO-11, loss of which leads to failure in crossover. Consequently spo-11 mutants show 12 univalents in oocytes in C. elegans 9. In contrast, loss of rad-51, which encodes a RecA-related protein required for DSB repair, causes severe defects in chromosome morphology 10. Likewise, mutants for other genes required for DSB repair display phenotypes of altered chromosome morphology, aneuploidy, sensitivity to DNA damage, and developmental retardation or even embryonic lethality, all of which are indicative of chromosomal instability 4. In addition, defective DSB repair in C. elegans germ cells manifests as the high-incidence-of-males (Him) phenotype owing to X chromosome non-disjunction. Notably, while meiotic DSBs are required for SC formation and thus synapsis in yeast, plants and mammals, DSB formation and synapsis are largely independent of one another in C. elegans 9, 11, 12. This makes C. elegans an excellent model organism for identifying novel factors and further dissecting their functions in either synapsis or meiotic DSB repair, or both.
In the C. elegans hermaphrodite germ line, either synaptic failure or defective DSB repair leads to an elevation in apoptosis, indicating that apoptosis is an essential cellular event for maintenance of genomic integrity 13, 14. Nevertheless, synaptic failure and defective DSB repair appear to trigger the cell death program by distinct signaling pathways. DNA lesions resulting from either defective meiotic DSB repair or exogenous genotoxic stress are sensed by checkpoint genes hus-1, mrt-2/rad1 and/or clk-2/rad5, which transduce death signals to the effector protein CEP-1, the C. elegans homolog of mammalian p53 14. As a transcription factor, CEP-1 upregulates the expression of the cell death initiator EGL-1, a BH3-only protein homologous to the mammalian p53 targets PUMA and Noxa, to induce apoptosis through an evolutionarily conserved cell death machinery 15, 16, 17. In comparison, synaptic failure originating from defects in the pairing center or mutations in the SC triggers apoptosis through the synapsis checkpoint PCH-2, a C. elegans homolog of the yeast AAA-adenosine triphosphatase (AAA-ATPase) Pch2 13, though it remains unknown how PCH-2 mediates death signals to the death initiator. The requirement of distinct death signaling pathways by synaptic failure or defective DSB repair further provides a basis for analyzing the function of genes involved in genomic integrity.
Here, we report the identification of MRG-1 as an important factor for genomic integrity during the meiotic process in C. elegans. Worm MRG-1 is a homolog of the mammalian mortality factor-related protein MRG15, which is conserved in yeast, plants, and Drosophila 18, 19. In mammals, MRG15 is a chromodomain-containing protein that potentially regulates both transcriptional silencing and activation by associating with both histone deacetylases and histone acetyltransferase (HAT) complexes 20. Deletion of Mrg15 in mouse causes embryonic lethality, improper organogenesis and proliferation, and defective DNA damage repair 21. Nevertheless, it remains elusive how MRG15 is involved in these processes at the molecular level. In C. elegans, mrg-1 was found to promote embryo survival, and maternal MRG-1 is required for proliferation and immortality of the primordial germ cells (PGC) 18. MRG-1 is essential for maintaining repression of transgenes in the maternal germ line by associating with chromatin in autosomes but not the X chromosome 18. Remarkably, repression of several X-linked genes is dependent on autosome-associated MRG-1 in conjunction with the “maternal-effect sterile” protein MES-4, a methyltransferase that modifies H3K36 22. These findings suggest that MRG-1 acts at the chromatin level to regulate germline potential and immortality. In the present study, we found that loss of mrg-1 function leads to excessive apoptosis in the C. elegans germ line, which requires both DNA damage checkpoint genes, such as clk-2/rad5, and the chromosome synapsis checkpoint gene pch-2. Consistent with this, we found that meiosis is defective in mrg-1 mutants, probably due to defects in DSB repair and synapsis. These results suggest that MRG-1 plays an important role in maintenance of genomic integrity during meiosis.
Results
Loss of mrg-1 function causes increased germ cell apoptosis
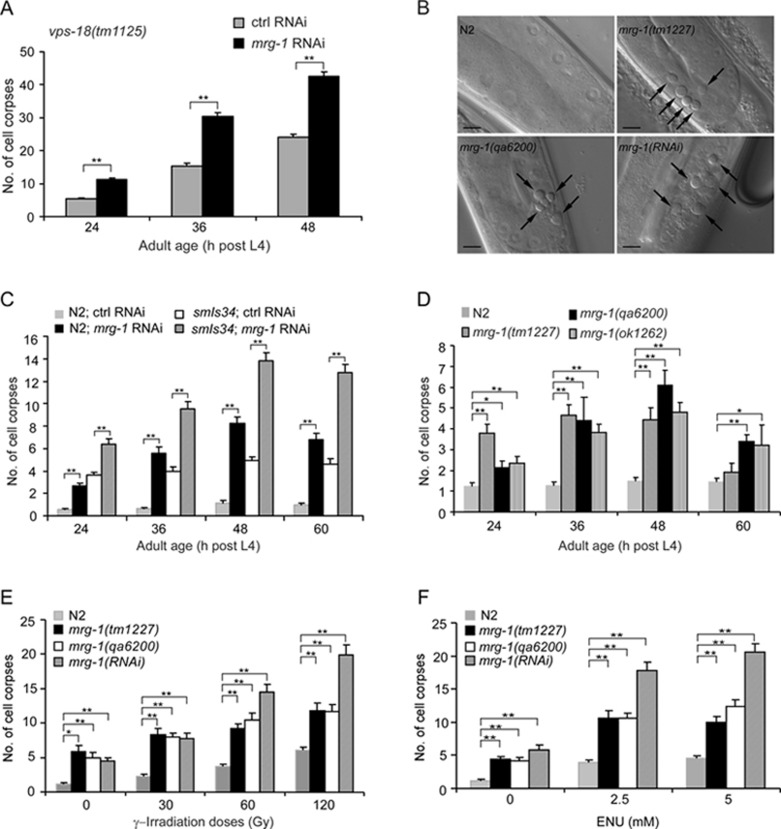
To identify novel factors involved in germ cell apoptosis in C. elegans, we performed a genome-wide RNA interference (RNAi) screen for genes that when inactivated can reduce or enhance the number of germ cell corpses in vps-18(tm1125) mutants, which accumulate germ cell corpses as a result of defective corpse degradation 23. We found that RNAi inactivation of mrg-1, a germline mortality-related gene 18, 19, enhanced the number of germ cell corpses in vps-18(tm1125) mutants in an age-dependent manner (Figure 1A). In wild-type worms or worms carrying an integrated array (smIs34) expressing the phagocytic receptor CED-1 fused with green fluorescence protein (CED-1::GFP) 24, mrg-1 RNAi also led to an obvious increase in germ cell corpses as viewed by either formation of disc-like structures under differential interference (DIC) optics or formation of CED-1::GFP rings at all adult ages examined (Figure 1B and 1C). Similarly, mrg-1 deletion mutants, ok1262, qa6200, and tm1227 18, showed a significantly higher level of apoptosis than wild-type worms at most adult ages examined (Figure 1B and 1D). Notably, the number of germ cell corpses in mrg-1(RNAi) worms appeared higher than in mrg-1 deletion mutants (Figure 1C and 1D, and see Figure 1E and 1F), probably because mrg-1 RNAi had a less severe effect on the proliferation of germ cells than mrg-1 deletion such that mrg-1(RNAi) worms possessed more germ cells competent for apoptosis than the deletion mutants. For example, mrg-1(RNAi) worms contained an average of 142.3 ± 8.1 (SEM, standard error of the mean; n = 14) mitotic cells per gonad arm, compared with 197.8 ± 7.1 (n = 14) mitotic cells per gonad arm in wild type, whereas mrg-1(qa6200) hermaphrodites only had an average of 101.3 ± 2.8 (n = 13) mitotic cells per gonad arm. These results indicate that loss of mrg-1 function induced apoptosis in the C. elegans germ line.
Figure 1.
Loss of mrg-1 function induces germ cell apoptosis. (A) mrg-1 RNAi enhances the number of germ cell corpses in vps-18(tm1125) mutants. (B) Representative DIC images of germ cell corpses in wild-type (N2), mrg-1(qa6200), mrg-1(tm1227) and mrg-1(RNAi) worms. Arrows indicate cell corpses. Bars, 10 μm. (C) Time course analyses of germ cell corpses in wild-type and smIs34 worms treated with mrg-1 RNAi. In wild type, cell corpses were viewed by formation of disc-like structures under DIC optics. In smIs34 worms, cell corpses were viewed by formation of CED-1::GFP rings. (D) mrg-1 deletion mutants show increased germ cell apoptosis. (E, F) Quantification of germ cell apoptosis induced by γ-irradiation (E) and ENU (F) in mrg-1 mutants. Cell corpses were scored and analyzed as described in Materials and Methods. Error bars represent standard error of the mean (SEM). Comparisons were performed using unpaired t-test. *P < 0.05, **P < 0.001.
C. elegans MRG-1 is homologous to yeast Eaf3/Alp13, Drosophila dMrg15, and mammalian MRG15, all of which are implicated in repair of DNA DSBs caused by exogenous DNA-damaging agents 19, 25, 26, 27, 28. During meiosis of C. elegans hermaphrodite, failure to repair programmed DSBs or artificially induced breaks leads to apoptotic cell death. To test whether loss of mrg-1 function confers sensitivity to DNA damage, we exposed mrg-1(lf) worms to DNA-damaging agents and examined germ cell apoptosis. Both γ-irradiation, which mainly induces DSBs, and N-ethyl-N-nitrosourea (ENU), which causes a wide spectrum of DNA lesions, induced a significant increase in germ cell corpses in mrg-1(lf) worms in a dose- or concentration-dependent manner compared with wild-type worms (Figure 1E and 1F). This suggests that loss of mrg-1 function likely causes defective DNA repair in germ cells, which in turn results in excessive apoptosis following exposure to DNA-damaging insults.
The increase in germ cell apoptosis in the absence of mrg-1 partially requires DNA damage checkpoint signaling
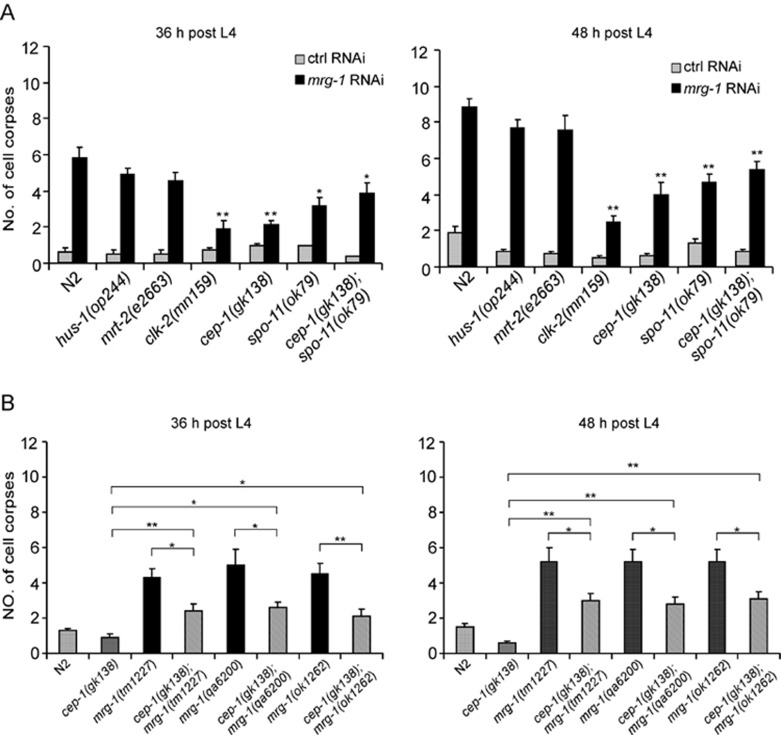
To understand the mechanisms underlying the increased apoptosis in mrg-1 mutants, we investigated whether the DNA damage signaling pathway plays a role. In C. elegans, germ cell apoptosis resulting from irreparable DNA lesions is dependent on DNA damage checkpoint genes that function in two parallel groups to mediate death signals to the downstream effector CEP-1/p53, with hus-1, mrt-2/rad1 in one group and clk-2/rad5 in the other. Mutations in these genes suppress germ cell apoptosis in response to DNA damage 15, 29, 30, 31, 32. Since mrg-1(RNAi) worms showed excessive germ cell apoptosis similarly to other mrg-1 mutants (Figure 1), we performed mrg-1 RNAi in mutants of the DNA damage checkpoint genes hus-1, mrt-2, and clk-2. Interestingly, the numbers of germ cell corpses in mrg-1 RNAi-treated hus-1(op244) and mrt-2(e2663) mutants were not obviously decreased compared with that in mrg-1 RNAi-treated wild type at the adult ages examined, whereas mrg-1 RNAi-induced cell corpses were substantially reduced, though not totally blocked, in clk-2(mn159) germ lines (Figure 2A), suggesting that the DNA damage checkpoint gene clk-2/rad5 is partially required for germ cell apoptosis in the absence of mrg-1. Similarly, germ cell corpses were significantly reduced but not completely eliminated in mrg-1 RNAi-treated cep-1(gk138) deletion mutants and double mutants of cep-1(gk138) with individual mrg-1 alleles (Figure 2A and 2B). Based on these observations, we reasoned that the increase in germ cell apoptosis in mrg-1(lf) worms were partially due to endogenous DNA lesions. In agreement with this, we found that the number of germ cell corpses in mrg-1 RNAi-treated spo-11(ok79) mutants, in which no DSBs were generated (see below) was only reduced to a similar level to that in cep-1(gk138);mrg-1(RNAi) worms (Figure 2A). Furthermore, in double mutants of cep-1(gk138) with spo-11(ok79), mrg-1 RNAi still caused a significant increase in germ cell apoptosis compared with control RNAi treatment (Figure 2A). Altogether, these data suggest that germ cell apoptosis in the absence of mrg-1 is partially attributable to defective DSB repair that signals through the DNA damage checkpoint gene clk-2/rad5 to CEP-1/p53.
Figure 2.
Mutations affecting DNA damage checkpoint genes partially suppress germ cell apoptosis induced by mrg-1 RNAi. (A) Germ cell corpses in mrg-1 RNAi-treated mutants including hus-1(op244), mrt-2(e2663), clk-2(mn159), cep-1(gk138), spo-11(ok79) and cep-1(gk138);spo-11(ok79) were quantified and analyzed at the adult ages of 36 h (left) and 48 h (right) post L4 stage, respectively. Comparisons were made between N2 and other strains treated with mrg-1 RNAi using unpaired t-test. *P < 0.05, **P < 0.001. (B) Quantification of germ cell corpses in N2, cep-1(gk138) and double mutants of cep-1(gk138) with mrg-1(tm1227), mrg-1(qa6200) and mrg-1(ok1262) at 36 and 48 h post L4 stage. *P < 0.05, **P < 0.001.
Loss of mrg-1 function affects DSB repair
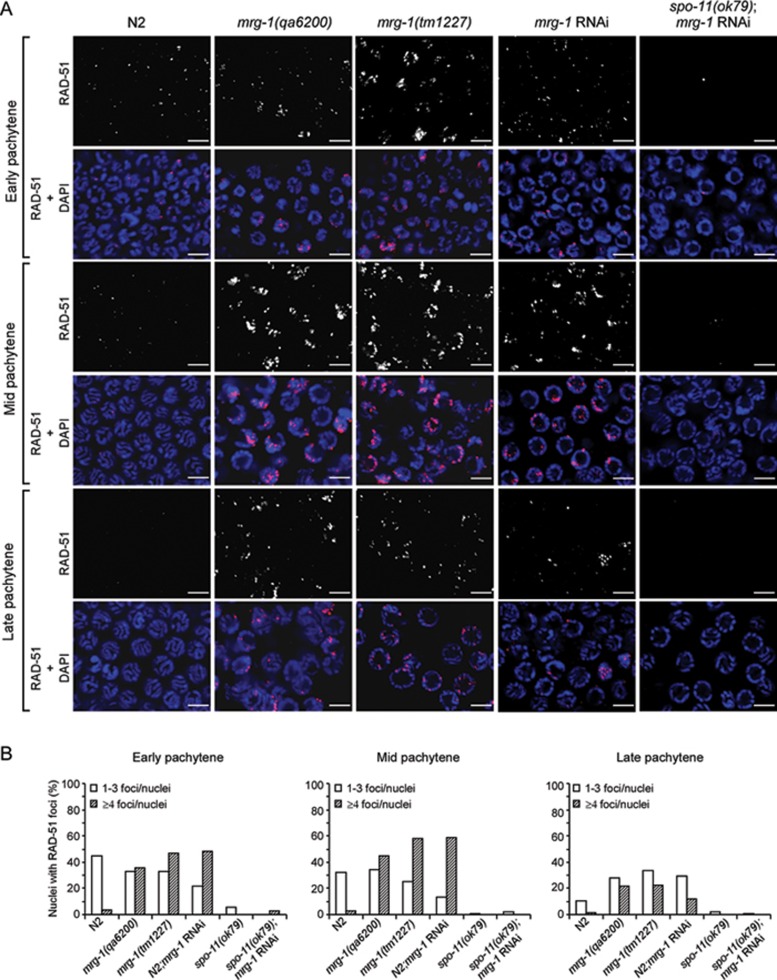
As the increased germ cell apoptosis in mrg-1 mutants is partially dependent on the DNA damage signaling pathway, we next investigated whether germline nuclei of mrg-1 mutants indeed contained excessive unrepaired DSBs. To this end, we analyzed RAD-51 foci in consecutive germline regions as described previously 33. RAD-51 is a RecA recombinase required for strand invasion/exchange during HR-mediated DSB repair by binding to single-stranded DNA with a 3′ overhang and forming a nucleoprotein filament 34. In wild-type germ lines, RAD-51 foci mostly appeared at the early pachytene stage, which gradually reduced in number during the mid pachytene stage and could barely be detected at the late pachytene stage, indicating that DSBs are mostly repaired before meiotic progression to the late pachytene stage (Figure 3A and 3B). In mrg-1(lf) and mrg-1(RNAi) germ lines, however, RAD-51 foci greatly increased in the early pachytene stage, which peaked in the mid pachytene stage and persisted until the late pachytene stage (Figure 3A and 3B). In contrast, RAD-51 foci were barely induced in mrg-1 RNAi-treated spo-11(ok79) null mutants, indicating that the accumulation of DSBs in mrg-1 mutants is dependent on SPO-11 (Figure 3A and 3B). Thus loss of mrg-1 function leads to accumulation of DSBs that likely result from a failure of timely processing.
Figure 3.
mrg-1 mutants accumulate RAD-51 foci in the germ line. (A) Representative images of RAD-51 foci and merged images of RAD-51 foci and DAPI staining are shown for early, mid and late pachytene nuclei of N2, mrg-1(qa6200), mrg-1(tm1227), mrg-1(RNAi), and spo-11(ok79);mrg-1(RNAi) worms. Bars, 5 μm. (B) Quantification of RAD-51 foci in early, mid and late pachytene stages of germ lines in worms as indicated. For each region, at least 20 nuclei per gonad arm were analyzed for a total of ≥ 3 gonads.
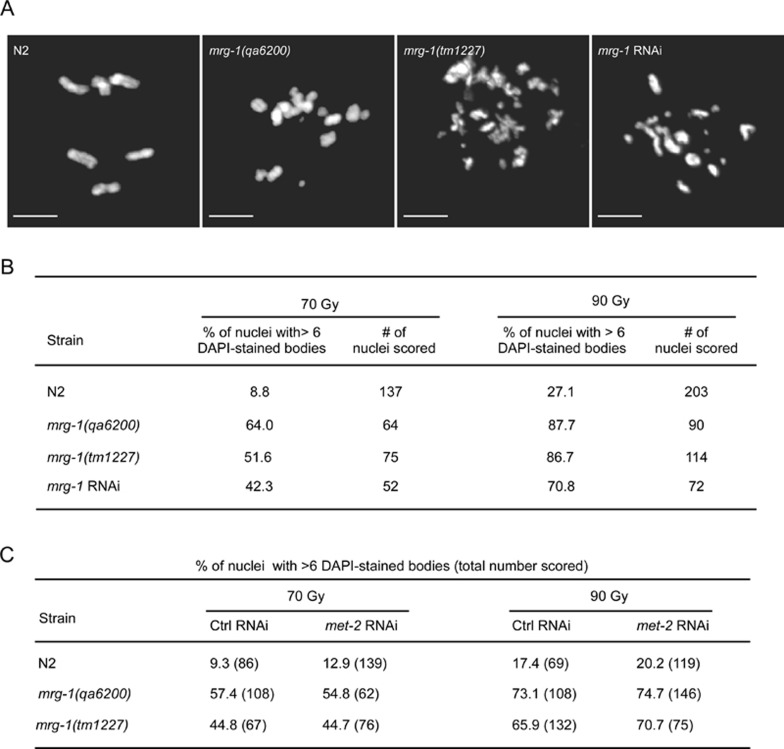
To further determine the role of MRG-1 in DSB repair in C. elegans, we treated mrg-1(lf) worms at L4 stage with γ-irradiation and examined the appearance of fragmented chromosomes in oocytes using a procedure described previously 35. In wild-type worms, γ-irradiation of 70 and 90 Gy induced chromosome fragmentation in 8.8% and 27.1% oocytes, respectively (Figure 4A and 4B). In mrg-1(lf) and mrg-1(RNAi) worms, the proportion of oocytes with fragmented chromosomes following γ-irradiation were greatly increased, ranging from 42.3%-64.0% and 70.8%-87.7% with irradiation of 70 and 90 Gy, respectively (Figure 4A and 4B). In addition, an irradiation of mrg-1(qa6200) L1 worms with a lower dose of γ-ray (50 Gy) led to only about 40% worms growing to sterile adults, compared with around 93% of N2 L1 worms growing to fertile adults with the same treatment (data not shown). Altogether, these findings suggest that mrg-1 plays an important role in DSB repair to maintain genomic integrity.
Figure 4.
γ-irradiation induced chromosome fragmentation in N2 and mrg-1(lf) worms. (A) Representative images of chromosomes in oocytes of N2 and mrg-1(lf) worms treated with γ-irradiation of 70 Gy. Bars, 5 μm. (B) Quantification of chromosome fragmentation in oocyte nuclei with γ-irradiation of 70 and 90 Gy, respectively. (C) Quantification of chromosome fragmentation in oocyte nuclei of met-2 RNAi-treated worms as indicated after exposure to γ-irradiation of 70 and 90 Gy, respectively. The percentages of nuclei with > 6 DAPI-stained bodies are shown and the total numbers of nuclei scored are shown in parentheses.
mrg-1 mutants exhibit meiotic defects
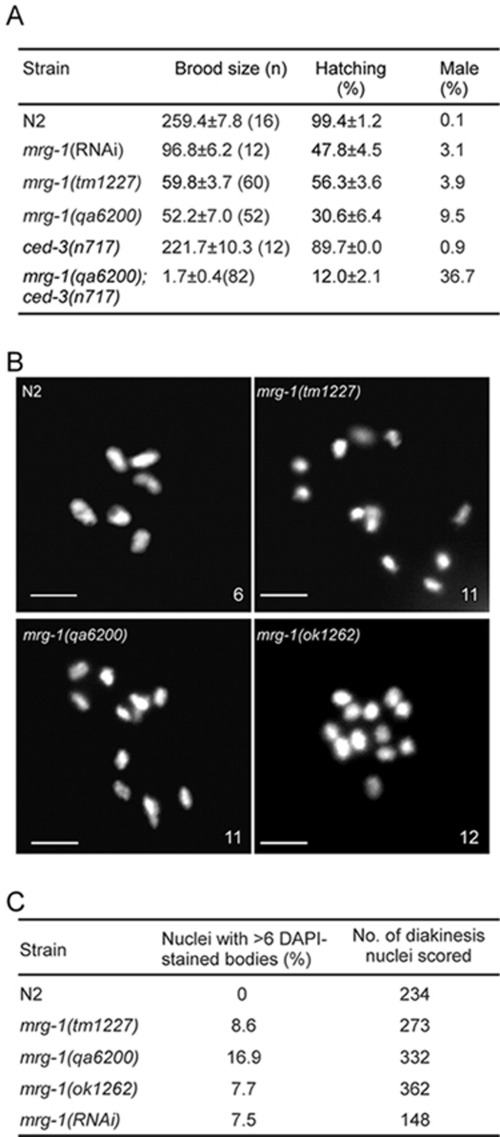
Our finding that mrg-1 is important for DSB repair in meiosis prompted us to further examine whether mrg-1 mutants exhibited meiotic phenotypes such as high incidence of males (Him), which reflects the frequency of X chromosome non-disjunction, and increased number of 4′,6-diamidino-phenylindole (DAPI)-stained bodies at diakinesis, which reflects defective crossover formation and/or DNA repair 4. It was shown previously that mrg-1 is required for proliferation and immortality of PGC and the brood sizes of mrg-1(lf) and mrg-1(RNAi) worms were greatly reduced, with a substantial proportion of embryos inviable 18 (Figure 5A). Among the surviving progeny, however, around 3.1%-9.5% were males, which is significantly higher than in wild type (Figure 5A). This indicates that loss of mrg-1 leads to X chromosome non-disjunction. Moreover, approximately 8%-17% oocyte nuclei in mrg-1(lf) worms contained more than six DAPI-stained bodies, varying from 7 to 12 (Figure 5B and 5C), which probably resulted from defects in crossover formation.
Figure 5.
mrg-1 mutants are defective in meiosis. (A) Quantification of brood sizes, hatching and male percentages of N2, mrg-1(RNAi), mrg-1(qa6200), mrg-1(tm1227), ced-3(n717) and mrg-1(qa6200);ced-3(n717) worms. (B) mrg-1 mutant oocytes show > 6 DAPI-stained bodies. Bars, 5 μm. (C) Quantification of mrg-1 mutant oocytes with > 6 DAPI-stained bodies.
To determine the effect of apoptosis on meiosis in mrg-1(lf) animals, we analyzed the meiotic phenotype in double mutants of mrg-1(qa6200) with ced-3(n717), a strong loss-of-function allele of the ced-3 gene that is required for cell death in C. elegans. No germ cell death was observed in mrg-1(qa6200);ced-3(n717) double mutants (data not shown), but their brood size and survival of embryos were strongly decreased compared with that of mrg-1(qa6200) and ced-3(n717) single mutants (Figure 5A). Notably, up to 36.7% of the surviving progeny of mrg-1(qa6200);ced-3(n717) double mutants were males, which is more than three times as many as in mrg-1(qa6200) single mutants (Figure 5A). These findings suggest that apoptosis is required to eliminate cells that harbor meiotic defects resulting from loss of mrg-1 function.
Apoptosis caused by loss of mrg-1 function involves the synapsis checkpoint
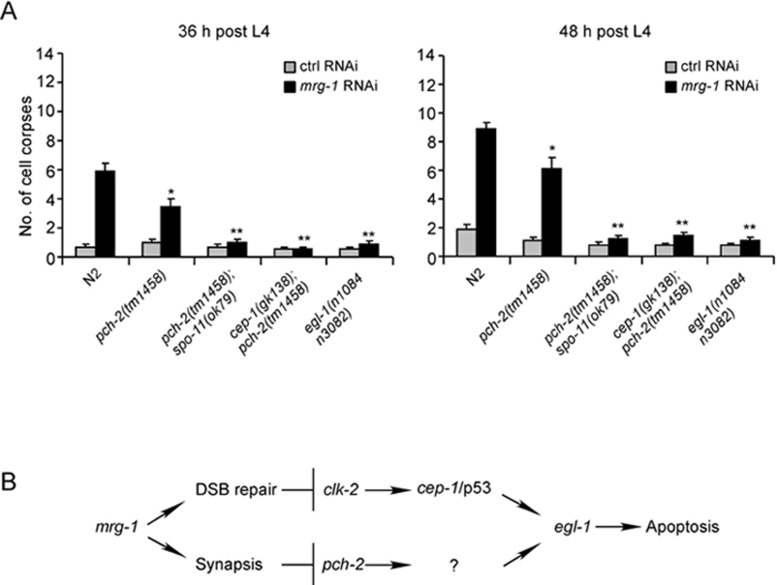
Our results indicate that mrg-1 is important for meiotic DSB repair. However, the elevated apoptosis in mrg-1(lf) germ lines is only partially dependent on the DNA damage checkpoint signaling pathway, suggesting that other checkpoint signals are also required. In the C. elegans germ line, defective chromosome synapsis in the prophase of meiosis I was found to trigger germ cell apoptosis 13. To test whether synapsis checkpoint signaling participates in germ cell apoptosis in mrg-1(lf) worms, we performed mrg-1 RNAi in pch-2(tm1458) deletion mutants that affect the synapsis checkpoint gene pch-2. As shown in Figure 6A, mrg-1 RNAi-induced germ cell apoptosis was partially but significantly reduced in pch-2(tm1458) mutants at all adult ages examined, suggesting that the synapsis checkpoint is involved in the increase of germ cell apoptosis in the absence of mrg-1. Next, we tested whether inactivation of both DNA damage and synapsis checkpoint pathways could abolish mrg-1 RNAi-induced germ cell apoptosis. In double mutants of pch-2(tm1458) with spo-11(ok79), mrg-1 RNAi did not cause an obvious increase in germ cell apoptosis compared with control RNAi (Figure 6A). Similarly, no increase in germ cell apoptosis was observed in cep-1(gk138);pch-2(tm1458) double mutants following mrg-1 RNAi treatment (Figure 6A). In addition, mrg-1 RNAi failed to induce obvious apoptosis in the egl-1(n1084n3082) strong loss-of-function mutants affecting the cell death initiator EGL-1 (Figure 6A). These results indicate that loss of mrg-1 function activated both synapsis and DNA damage checkpoint signaling pathways to trigger apoptosis (Figure 6B).
Figure 6.
The synapsis checkpoint gene pch-2 and DNA damage checkpoint genes are required for increased apoptosis in mrg-1(RNAi) worms. (A) Quantification of germ cell apoptosis in mrg-1 RNAi-treated N2, pch-2(tm1458), pch-2(tm1458);spo-11(ok79), cep-1(gk138);pch-2(tm1458), and egl-1(n1084n3082) worms. Germ cell corpses in RNAi-treated worms at adult ages of 36 h (left) and 48 h (right) post L4 stage were scored and analyzed as described in Materials and Methods. Comparisons were made between N2 and other mutants treated with mrg-1 RNAi. *P < 0.05, **P < 0.001. (B) A schematic summary of the signaling pathways leading to apoptosis in the absence of mrg-1.
mrg-1 mutant germ lines are defective in progression of meiotic prophase
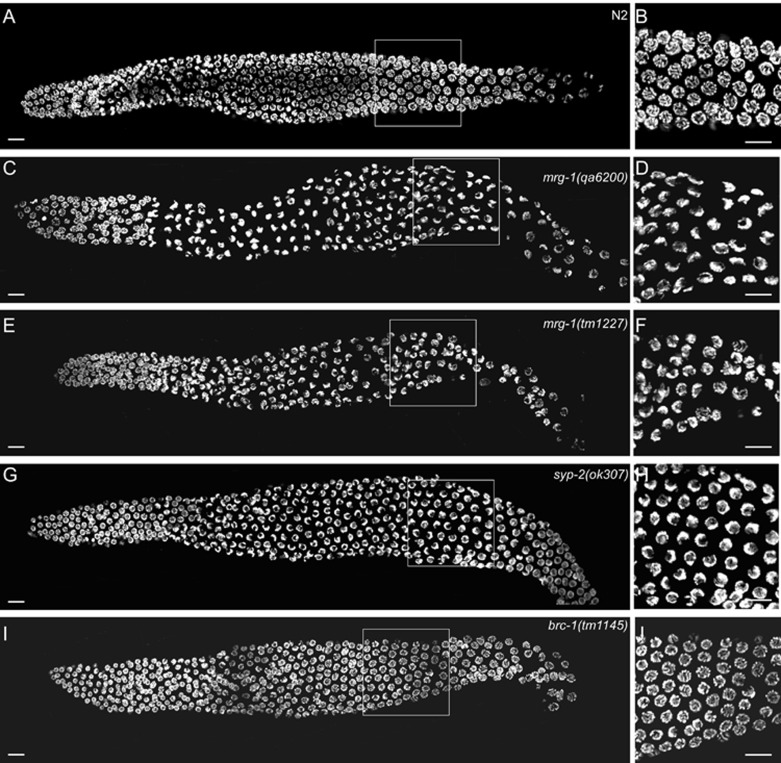
We next investigated whether chromosome synapsis is affected in mrg-1 mutants. In wild-type germ lines syncytial nuclei were spatially arranged according to the temporal progression of the prophase of meiosis I 3. Nuclei undergoing mitosis and premeiotic replication were in the distal most end of the gonad, which was followed by a transition zone where nuclei entered leptotene/zygotene and the chromatin polarized, adopting a crescent-like morphology. In pachytene, the chromatin redispersed and chromosomes synapsed, which could be visualized as parallel tracks when stained with DAPI. During diplotene and diakinesis, chromosomes further condensed and individualized, forming six bivalents (Figure 7A and 7B) 3. In mrg-1(qa6200) and mrg-1(tm1227) mutant germ lines, the mitotic nuclei entered the transition zone as in the wild type; however, leptotene/zygotene-nuclei with polarized crescent-like morphology were distributed in the germline regions corresponding to the mid to late pachytene regions in the wild-type gonad, indicative of a failure of the polarized chromatin to redisperse and align longitudinally (Figure 7C and 7E). Consistent with this, few nuclei with parallel tracks were observed in mrg-1(lf) germ lines (Figure 7D and 7F). Similarly, in syp-2(ok307) synapsis-defective mutants polarized chromosomes persisted from the transition zone through almost the whole pachytene region (Figure 7G and 7H) 33. In contrast, no obvious defects in nuclear progression from leptotene/zygotene to pachytene were found in brc-1(tm1145) mutants lacking the brc-1 gene that is essential for DSB repair through inter-sister recombination but dispensable for chromosome crossover (Figure 7I and 7J) 36. These findings indicate that the progression of germline nuclei from leptotene/zygotene to pachytene is defective in mrg-1(lf) germ lines, which may result from impaired synapsis and contribute to the activation of synapsis checkpoint.
Figure 7.
mrg-1 mutants are defective in progression of meiotic prophase. Representative images of DAPI-stained nuclei of whole-mount gonads from age-matched worms of the following strains are shown: (A, B) N2, (C, D) mrg-1(qa6200), (E, F) mrg-1(tm1227), (G, H) syp-2(ok307), and (I, J) brc-1(tm1145). Boxed regions in A, C, E, G and I representing mid to late pachytene stages are magnified and shown in B, D, F, H and J, respectively. ≥ 19 gonads for each strain were analyzed. Bars, 10 μm.
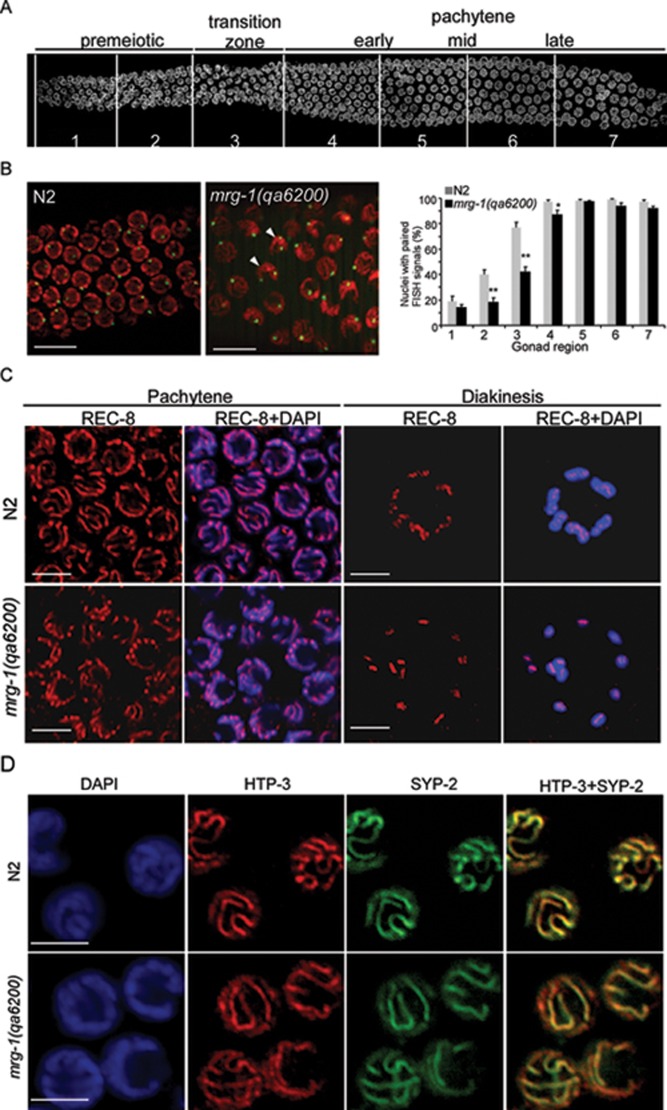
mrg-1 does not affect chromosomal loading of the SC
To evaluate the effect of mrg-1 on synapsis, we first used the 5S rDNA located on the non-pairing center region of the fifth chromosome as a probe to examine the pairing of chromosomes by fluorescence in situ hybridization (FISH) 9. Our results indicated that in mrg-1(qa6200) mutants chromosome pairing was reduced in gonadal regions corresponding to premeiotic to early pachytene zones (Figure 8A and 8B, gonad regions 2, 3 and 4) compared to the wild-type germ line, but no obvious defect was observed in the more proximal germline regions (Figure 8A and 8B), suggesting that chromosome pairing is slightly perturbed in the absence of mrg-1. Next, we performed immunostaining of REC-8 to examine whether mrg-1 mutants exhibited defects in sister chromatid cohesion. In wild type, REC-8 distributed along the axes of sister chromatids in pachytene stage nuclei and it persisted on condensed chromosomes at diakinesis (Figure 8C). Similar REC-8 staining was observed in the pachytene region in mrg-1(qa6200) mutants. In addition, REC-8 was also found on each chromosomal piece in mrg-1(qa6200) oocytes containing 6 or > 6 DAPI-stained bodies (Figure 8C). These results suggest that loss of mrg-1 function does not cause obvious defect in sister chromatid cohesion. Finally, we asked whether the chromosomal loading of the SC was affected in mrg-1 mutants by examining the co-existence of HTP-3 and SYP-2, a lateral element and a central element of the SC, respectively. We found that HTP-3 and SYP-2 colocalized well along the axes of chromosomes in mrg-1(qa6200) mutants, as observed in wild-type worms (Figure 8D), suggesting that loss of mrg-1 does not obviously affect chromosomal loading of the SC.
Figure 8.
Analysis of chromosome pairing, sister chromatid cohesion, and chromosomal loading of the SC in mrg-1 mutants. (A) Representative image of a wild-type germ line with regions of meiotic progression indicated. (B) Representative FISH images of mid pachytene nuclei in N2 (left) and mrg-1(qa6200) (middle) germ lines using 5S rDNA (green) on chromosome V. DAPI staining is shown in red. Arrowheads indicate nuclei with unpaired FISH signals. Bars, 10 μm. Quantification of nuclei with paired FISH signals in different germline regions is indicated in the right panel. For each strain, 7 gonads were examined and a total of ≥ 74 nuclei were analyzed for each zone. Comparisons were performed between N2 and mrg-1(qa6200) mutants using unpaired t-test. *P < 0.05, **P < 0.001. (C) Representative images of REC-8 antibody staining (red) in pachytene and diakinesis nuclei in N2 and mrg-1(qa6200) worms. DAPI staining is shown in blue. Bars, 5 μm. (D) Representative images of HTP-3 (red) and SYP-2 (green) antibody staining in mid pachytene nuclei in N2 and mrg-1(qa6200) germ lines. DAPI staining is indicated in blue. Bars, 5 μm.
mrg-1 mutants have increased germline H3K9me2, similar to mutants defective in synapsis
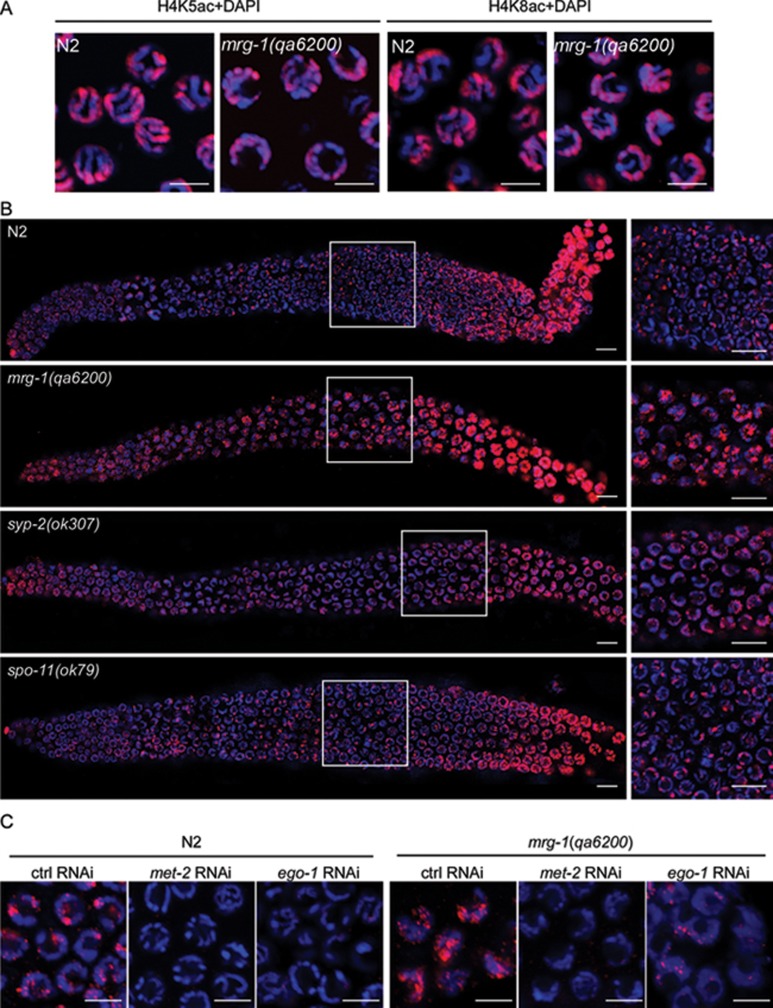
Because MRG-1 is homologous to yeast Eaf3/Alp13 and mammalian MRG15, which are subunits of the NuA4/Tip60 acetyltransferase complex, we sought to determine whether the acetylation of Histone H4 is affected in mrg-1 mutants using antibody staining. We did not observe an obvious change in H4K5 and H4K8 acetylation (H4K5ac and H4K8ac) between wild-type and mrg-1(qa6200) worms (Figure 9A). Nevertheless, it was previously reported that dimethylation of lysine 9 of Histone H3 (H3K9me2), which marks heterochromatin and transcriptionally inactive regions, is enriched on unpaired chromosomes and chromosome regions 37, 38, 39. Thus, we examined H3K9me2 in mrg-1 mutants. As described previously 40, in wild-type germ lines nuclei from the early to mid pachytene displayed small foci of H3K9me2. From the late pachytene to diplotene, H3K9me2 staining was strongly increased. The gradual increase in H3K9me2 may reflect the transition from a looser chromatin structure in the early pachytene stage to a more condensed structure in the diplotene stage (Figure 9B). In contrast, mrg-1(qa6200) germline nuclei maintained a much higher level of H3K9me2 from the early pachytene region to diplotene, suggesting that the chromatin adopts a more condensed structure (Figure 9B). A similar increase in H3K9me2 level was observed in syp-2(ok307) and him-3(e1256) synapsis-defective mutants (Figure 9B and data not shown) 39. By contrast, H3K9me2 in spo-11(ok79) mutants, in which meiotic recombination is disrupted but synapsis progression is not affected, was similar to that in wild type (Figure 9B). To assess the effect of H3K9me2 on meiotic progression, we used RNAi to deplete met-2 40 and ego-1 41, which encode a methyltransferase responsible for H3K9me2 and an RNA-dependent RNA polymerase required for H3K9me2 enrichment on unpaired chromosome regions during meiosis, respectively. In both wild type and mrg-1(qa6200) mutants RNAi of met-2 and ego-1 caused a strong decrease in H3K9me2 level; however, these RNAi treatments did not change the chromatin morphology compared with control RNAi (Figure 9C). In addition, met-2 RNAi did not inhibit the chromosome fragmentation in mrg-1(lf) mutants after exposure to γ-irradiation (Figure 4C). Taken together, these results suggest that the elevated H3K9me2 is not responsible for the meiotic progression defects and DSB repair defects in mrg-1 mutants. Instead, it is likely to be a mark of chromosome regions with impaired synapsis at pachytene stage, which further suggests that MRG-1 likely plays a role in regulating chromosome synapsis.
Figure 9.
mrg-1 and syp-2 mutants show increased H3K9me2 in the germ line. (A) Representative images of H4K5ac (left) and H4K8ac (right) of nuclei in the pachytene regions of N2 and mrg-1(qa6200) germ lines. Bars, 5 μm. (B) Representative images of H3K9me2 patterns in N2, mrg-1(qa6200), syp-2(ok307), and spo-11(ok79) germ lines. The boxed regions in the left panels are magnified and shown in the right panels. Bars, 10 μm. (C) Representative images of H3K9me2 in nuclei of late pachytene regions in N2 and mrg-1(qa6200) germ lines treated with RNAi of met-2 and ego-1. Bars, 5 μm. In all panels DAPI-stained chromatins are shown in blue and antibody staining is shown in red.
Discussion
In an effort to search for factors whose inactivation may cause abnormal germ cell apoptosis in C. elegans, we have identified the chromodomain-containing protein MRG-1 as an important factor for maintenance of genomic integrity during meiosis. We found that loss of mrg-1 function led to an elevation in germ cell apoptosis. Importantly, the increase in apoptosis in mrg-1(lf) germ lines was partially inhibited by mutations affecting either genes involved in the DNA damage signaling pathway or the synapsis checkpoint gene pch-2. These findings suggest that the elevation in germ cell apoptosis likely results from defective DSB repair and/or impaired synapsis during the prophase of meiosis I. In agreement with this hypothesis, inactivation of both DNA damage and chromosome synapsis checkpoints abrogated the elevated germ cell apoptosis. Thus the requirement of these signaling pathways in the absence of mrg-1 suggests that MRG-1 is important for proper DSB repair and synapsis in the prophase of meiosis I.
In support of the notion that MRG-1 is important for DSB repair, we found that mrg-1(lf) worms accumulated spo-11-dependent RAD-51 foci from mid to late pachytene stages in germ lines, suggesting that DSBs are generated normally but fail to be repaired in a timely fashion in meiosis. Such DSB repair defects likely lead to the activation of DNA damage checkpoint pathway for triggering apoptosis. In addition, mrg-1(lf) worms exhibited excessive apoptosis and more chromosome fragmentation following exposure to DNA-damaging agents, indicating that MRG-1 is also essential for repair of DNA lesions generated exogenously, which provides further evidence that MRG-1 plays an important role in DNA repair in C. elegans. On the other hand, we found that mrg-1 RNAi still induced obvious apoptosis in spo-11 mutants in which no DSB was generated, and mrg-1(lf)-caused apoptosis was partially dependent on the synapsis checkpoint gene pch-2, suggesting that MRG-1 likely plays a role in synapsis and its loss of function triggers the synapsis checkpoint. To figure out how MRG-1 may function in this respect, we examined several events leading to chromosome synapsis, including sister chromatid cohesion, homologous chromosome pairing, and chromosomal loading of the SC. To our surprise, we only detected slight delay in chromosome pairing in mrg-1(qa6200) mutants, whereas no obvious defects was found in sister chromatid cohesion and chromosomal loading of the SC. Thus the weak defect in chromosome pairing might result from impaired synapsis, but not a direct aftermath of failure in chromosome pairing. Importantly, we found that chromosomes polarized at leptotene/zygotene stages fail to redisperse and align along their entire lengths at the pachytene stage in mrg-1 mutants, which is analogous to the defective progression of meiosis caused by synapsis failure in mutants deficient in the SC, suggesting that synapsis is likely not fully completed in the absence of mrg-1 although chromosomal loading of the SC is not obviously affected. We reason that MRG-1 may be involved in the regulation of synapsis progression through some yet unknown mechanisms.
Eaf3/Alp13 and MRG15, the MRG-1 homologs in yeast and mammals, respectively, are components of the NuA4/Tip60 HAT complex that acts at multiple levels to affect DNA damage response (DDR) and DNA repair. NuA4/Tip60 is recruited to the chromatin surrounding DSBs to facilitate DNA repair through chromatin modifications such as acetylation of the tail of Histone H4, thus opening and relaxing the chromatin for access of DNA repair proteins 28, 42. Tip60 can directly acetylate the ATM (ataxia-telangiectasia mutated) kinase, promoting its activation in response to DNA damage 28. Moreover, the acetyltransferase activity of Tip60 is stimulated by binding of its chromodomain to H3K9me3 43. Whether MRG-1/MRG15 affects DSB repair by regulating the HAT activity of Tip60 remains to be explored, though it was found that histone acetylation and recruitment of initial DNA repair proteins such as 53BP1 were delayed in Mrg15-null mouse embryonic fibroblasts following γ-irradiation 27. In C. elegans, loss of mrg-1 caused accumulation of RAD-51 foci, suggesting that MRG-1 should play a role downstream of the formation of the nucleoprotein filament in the process of DSB repair. Nevertheless, we did not detect an obvious change in either H4K5ac or H4K8ac in mrg-1 mutant germ line. Thus it remains to be further investigated whether MRG-1 affects the HAT activity of the Tip60 complex on other sites. Interestingly, using the yeast two-hybrid assay, we found that MRG-1 directly interacts with the DNA helicase RUVB-2, a homolog of another component of the Tip60 complex, RVB1/2/Tip49 44 (data not shown). RVB1/2/Tip49 is important for the HAT activity of the Tip60 complex 42. Notably, the prokaryotic homologs of RVB1/2, RuvABC, promote branch migration and Holliday junction processing 45. Thus the interaction between MRG-1 and RUVB-2 may promote the recruitment of DNA repair proteins other than RAD-51 to DNA breaks, and/or facilitate the processing of Holliday junctions in HR-mediated DSB repair.
Recently, it was found that loss of spr-5, which encodes a histone H3K4me2 demethylase, led to elevated levels in germline H3K4me2, p53-dependent apoptosis, and meiotic RAD-51 foci, suggesting that histone modifications play a role in meiotic DSB repair in C. elegans 46. Intriguingly, we found that dimethylation of the lysine 9 of histone H3 was similarly increased from the early to late pachytene regions in mutants deficient in mrg-1, syp-2, and him-3, compared with wild-type germ lines. However, RNAi depletion of met-2 or ego-1, which are essential for generation of H3K9me2 and enrichment of H3K9me2 on unpaired chromosome regions, respectively 40, 41, failed to restore the progression of synapsis in mrg-1(qa6200) mutants. Thus the increase in H3K9me2 is likely to be a mark of chromosome regions with impaired synapsis instead of playing a causative role. In addition, whether the elevation in H3K9me2 has a direct effect on meiotic DSB repair needs to be further explored. As MRG-1 directly interacts with RUVB-2 and ZK1127.3 47, the homologs of the NuA4/Tip60 subunits RVB1 and Eaf7/MRGBP, respectively, it remains possible that these interactions may affect histone acetylation and/or methylation at certain sites during chromosome synapsis and/or DSB repair. Alternatively, MRG-1 may affect the progression of meiotic prophase by regulating the expression of genes involved in synapsis and DSB repair since MRG-1 was found to be required for germline gene silencing 18. Further elucidation of how MRG-1 functions will provide deep insights into the molecular mechanisms underlying DSB repair, synapsis, and maintenance of genomic integrity.
Materials and Methods
C. elegans strains and genetics
C. elegans strains were cultured at 20 °C and maintained with standard procedures 48. The Bristol N2 strain was used as wild type. Mutant and balancer strains used are listed by linkage groups:
LG I: cep-1(gk138), hus-1(op244); hT2[bli-4(e937) let-?(q782) qIs48] (I;III).
LG II: mrt-2(e2663), pch-2(tm1458), vps-18(tm1125).
LG III: brc-1(tm1145), clk-2(mn159), mrg-1(ok1262), mrg-1(qa6200), mrg-1(tm1227), qC1[dpy-19(e1259)glp-1(q339)qIs26(lag-2::GFP)](III;V).
LG IV: ced-3(n717), him-3(e1256), spo-11(ok79), nT1[unc(n754)let qIs50](IV;V).
LG V: syp-2(ok307), egl-1(n1084n3082).
Deletion strains were outcrossed with the N2 strain at least four times. Double mutants were constructed with standard protocols [49]. smIs34 is an integrated array expressing CED-1::GFP 24. M+Z- mrg-1 hermaphrodites (homozygous hermaphrodites from heterozygous mothers; M, maternal load; Z, zygotic synthesis) were used for all germline apoptosis and meiosis analyses.
RNA interference
RNAi clones of mrg-1, met-2, and ego-1 were obtained from the C. elegans RNAi library generated by the Ahringer laboratory and verified by sequencing. RNAi was performed at 20 °C using a feeding assay. Briefly, worms synchronized at early larval stage 4 (L4) with different genetic backgrounds were grown on plates seeded with the bacterial strain HT115 (DE3) expressing dsRNAs of mrg-1, met-2 and ego-1. The progeny were maintained on RNAi plates and worms with intact germ lines after reaching adult age were synchronized for germ cell corpse quantification and other experimental analyses (see below). The efficiency of mrg-1 RNAi was verified by examining the loss of germ lines in certain proportion of adult worms.
Germ cell apoptosis analysis and DNA damage treatment
Germ cell corpses were viewed by formation of disc-like structures under DIC optics. Alternatively, cell corpses were recognized by formation of CED-1::GFP rings in worms carrying the integrated array smIs34. For time course analysis of apoptosis, synchronized worms at 24, 36, 48, and 60 h post L4 stage were scored for germ cell corpses under DIC optics. Cell corpses of one gonad arm were scored for each worm and a total of ≥ 15 worms of each strain were examined at every time point. All experiments were repeated 2-3 times and the average number of germ cell corpses per gonad arm and the SEM were calculated from worms (≥ 30 totally) examined for each strain at every time point. For DNA damage treatment, worms at 12 h post L4 stage were irradiated with different dosages of γ-rays (30, 60, and 120 Gy) or treated with ENU at different concentrations (0, 2.5, and 5 mM) for 4 h. Worms were then recovered for 24 h and scored for germ cell corpses under DIC optics as described above. The experiments were repeated twice. Data comparisons were performed using the unpaired t-test.
To examine γ-irradiation-induced chromosome fragmentation, synchronized L4 worms were irradiated with 70 and 90 Gy, respectively, and back to culture for another 48 h. Gonads were then dissected out and diakinesis nuclei were stained with DAPI and observed under a fluorescence scope (Zeiss Axioimager M1).
Analysis of meiotic defects in mrg-1(lf) worms
The number of chromosomes in oocytes was examined by DAPI staining. Briefly, gonads of N2 and M+Z- mrg-1 hermaphrodites at the age of 48 h post L4 stage were dissected in phosphate buffer containing 0.25 mM levamisole and fixed with 2% formaldehyde. After extensive wash, gonads were stained with DAPI (2 μg/ml) and viewed under fluorescence scope. To quantify brood sizes and proportions of males, M+Z- mrg-1 mutants or mrg-1(RNAi) worms at L4 stage with intact germ lines were chosen to lay eggs on NGM plates. The total numbers of M-Z- mrg-1 eggs laid and hatched larvae were scored.
Antibodies and immunostaining
RAD-51 polyclonal antibody was generated in rabbits by injecting a GST-fused N-terminal peptide (amino acids 1-103) of RAD-51. SYP-2 polyclonal antibody was raised in mice by injecting a GST-fused C-terminal peptide (amino acids 101-213) of SYP-2. The RAD-51 and SYP-2 sera were affinity purified and used with 1:500 and 1:200 dilution, respectively. HTP-3 polyclonal antibody was generated in guinea pig by injecting His6-tagged HTP-3 (amino acids 414-607) protein and used with a dilution of 1:200. Rabbit polyclonal REC-8 antibody was purchased from Novus Biologicals (#29470002) and used with 1:500 diluton. Rabbit polyclonal antibodies against acetylated H4K5 and H4K8 were purchased from Upstate Biotechnology Inc. and used with 1:1 000 dilution. Mouse monoclonal antibody for H3K9me2 was purchased from Abcam (#ab1220) and used with 1:500 dilution. For immunostaining, worm gonads were dissected out from hermaphrodites at 21-24 h post L4 stage and fixed in 4% paraformaldehyde for 40 min at room temperature. Samples were then sequentially treated with methanol, methanol:acetone (1:1), and acetone for 5 min each at −20 °C. After being washed three times in phosphate buffer (1.1 mM KH2PO4, 153.8 mM NaCl, 3.0 mM Na2HPO4, pH 7.4) containing 1% Triton X-100, samples were sequentially incubated with primary antibodies for 16-18 h at 4 °C and FITC- or Cy3-conjugated secondary antibodies (1:200 and 1:4 000 dilution, respectively) for 2 h at room temperature. An antifade solution (Vector Laboratories) containing 2 μg/ml DAPI was added to samples before imaging under a confocal microscope (Olympus Fluoview FV1000, IX81 inverted laser scanning microscope).
Fluorescence in situ hybridization
5S rDNA 9 was amplified by polymerase chain reaction (PCR) and labeled with digoxigenin-16-dUTP (Roche) by nick translation. Worm gonads fixed on slides were denatured at 80 °C for 2 min in 2× SSC (0.3 mM NaCl, 0.03 mM Na3C6H5O7, pH7.0) containing 70% formamide and then dehydrated by sequentially immersing in a pre-cooled ethanol series (70%, 90%, and 100% for 5 min each). 50-100 ng probe labeled with digoxigenin was denatured and then incubated with worm gonads on slides at 37 °C overnight in a moist chamber. Slides were subsequently washed once in 2× SSC for 10 min at 42 °C and twice in 2× SSC for 5 min each at room temperature. Chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories).
Acknowledgments
We thank the C. elegans Genetic Center (CGC) and Dr Shohei Mitani (Tokyo Women's Medical Univeristy, Japan) for providing worm deletion strains; Dr Xiaochen Wang (National Institute of Biological Sciences, China) for helpful suggestions and Dr Isabel Hanson for proofreading of the manuscript. This research was supported by grants from the National Basic Research Program of China (2011CB910102 and 2007CB947201) and the National Natural Science Foundation of China (31025015 and 30871266). CY is supported by the Hundred Talents Program of the Chinese Academy of Sciences.
References
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Meiotic recombination in Caenorhabditis elegans. Chromosome Res. 2007;15:607–621. doi: 10.1007/s10577-007-1146-x. [DOI] [PubMed] [Google Scholar]
- Lemmens BB, Tijsterman M. DNA double-strand break repair in Caenorhabditis elegans. Chromosoma. 2011;120:1–21. doi: 10.1007/s00412-010-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP. The many facets of SC function during C. elegans meiosis. Chromosoma. 2006;115:195–211. doi: 10.1007/s00412-006-0061-9. [DOI] [PubMed] [Google Scholar]
- Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Schild-Prufert K, Colaiacovo MP. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 2009;5:e1000669. doi: 10.1371/journal.pgen.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Eizinger A, Schild-Prufert K, et al. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegansrad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. Germline survival and apoptosis. WormBook. 2008. pp. 1–20. [DOI] [PMC free article] [PubMed]
- Hofmann ER, Milstein S, Boulton SJ, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009;5:e1000514. doi: 10.1371/journal.pgen.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Liu Z, Habara Y, et al. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development. 2007;134:757–767. doi: 10.1242/dev.02771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Takasaki T, Nakajima N, Kawano T, Shimura Y, Sakamoto H. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech Dev. 2002;114:61–69. doi: 10.1016/s0925-4773(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Pena AN, Pereira-Smith OM. The role of the MORF/MRG family of genes in cell growth, differentiation, DNA repair, and thereby aging. Ann N Y Acad Sci. 2007;1100:299–305. doi: 10.1196/annals.1395.031. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, et al. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, et al. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Chen D, Fang Z, et al. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell. 2009;20:21–32. doi: 10.1091/mbc.E08-04-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xiao H, Zhang K, et al. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- Bertram MJ, Berube NG, Hang-Swanson X, et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Garcia SN, Kirtane BM, Podlutsky AJ, Pereira-Smith OM, Tominaga K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581:5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Stergiou L, Hengartner MO. Death and more: DNA damage response pathways in the nematode C. elegans. Cell Death Differ. 2004;11:21–28. doi: 10.1038/sj.cdd.4401340. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Bailly AP, Freeman A, Hall J, et al. The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet. 2010;6:e1001025. doi: 10.1371/journal.pgen.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, La Volpe A. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner CE, Kelly WG. Germline chromatin. WormBook. 2006. pp. 1–14. [DOI] [PMC free article] [PubMed]
- Kelly WG, Schaner CE, Dernburg AF, et al. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 2010;6:e1000830. doi: 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine EM, Hauth J, Ratliff T, Vought VE, She X, Kelly WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiacovo MP. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci USA. 2011;108:12805–12810. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]