Abstract
High-throughput sequencing and genotyping methods are dramatically increasing the number of observable genetic intraspecies differences that can be exploited as genetic markers. In addition, automated phenotyping platforms and “omics” profiling technologies further enlarge the set of quantifiable macroscopic and molecular traits at an ever-increasing pace. Combined, both lines of technological advances create unparalleled opportunities to identify candidate gene regions and, ideally, even single genes responsible for observed variations in a particular trait via association studies. However, as of yet, this new potential is not sufficiently matched by enabling software solutions to easily exploit this wealth of genotype/phenotype information. We have developed Matapax, a Web-based platform to address this need. Initially, we built the infrastructure to support association studies in Arabidopsis (Arabidopsis thaliana) based on several genotyping efforts covering up to 1,375 Arabidopsis accessions. Based on the user-supplied trait information, associated single-nucleotide polymorphism markers and single-nucleotide polymorphism-harboring or -neighboring genes are identified using both the GAPIT and EMMA libraries developed for R. Additional interrogation is facilitated by displaying candidate regions and genes in a genome browser and by providing relevant annotation information. In the future, we plan to broaden the scope of organisms to other plant species as more genotype/phenotype information becomes available. Matapax is freely available at http://matapax.mpimp-golm.mpg.de and can be accessed using any internet browser.
Genome-wide association studies (GWAS) are a powerful way to harness natural variation to detect the genomic causes for phenotypic variance by testing the relationship between sequence and phenotypic variation. Although GWAS are criticized for being a data-driven approach that potentially inflate type I error through the sheer number of tests performed, they have enjoyed several successes in many organisms through the identification of experimentally determined associations (Aranzana et al., 2005; Zhao et al., 2007) and the identification of associations that were subsequently experimentally confirmed (Klein et al., 2005; Sladek et al., 2007; Wellcome Trust Case Control Consortium, 2007). They also highlight many plausible novel associations (Atwell et al., 2010; Todesco et al., 2010).
The basic unit of GWAS is a statistical test of the association between the alleles of a genetic marker and the corresponding trait measurements. The number of these comparisons performed is set to rapidly rise due to two factors. (1) The advent of high-throughput macroscopic and molecular phenotyping platforms enabled the analysis of many more traits and accessions (i.e. organisms with a unique genetic makeup). For example, a single metabolomic study has the potential to produce measurements for hundreds to thousands of metabolites in as many accessions (Lisec et al., 2006; Sozzani and Benfey, 2011). (2) Novel de novo or resequencing technologies have yielded massively increased numbers of genetic markers at ever-falling costs. Thus, the density of genetic markers may now be high enough to potentially allow single-gene resolution of GWAS studies. These factors combined mean that a single GWAS may require over one million trait-marker comparisons. Although this allows the genetic causes for the trait variation to be much more tightly defined, a single study now demands significantly more computational resources to complete in a timely and statistically meaningful manner.
There are many tools available that perform GWAS (Aulchenko et al., 2007; Bradbury et al., 2007; Browning and Browning, 2007; Purcell et al., 2007; Muñiz-Fernandez et al., 2011), two of which deserve particular mention as they are popular choices in plant and human GWAS. TASSEL (Bradbury et al., 2007) is a Java-based tool, which can be either downloaded or run from the home page, that provides a plethora of different analyses such as the prefiltering of trait and marker data, phylogenetic analyses, and general linear model and mixed linear model analyses. TASSEL was developed for maize (Zea mays) but is applicable to any organism. PLINK (Purcell et al., 2007) provides a set of tools to perform GWAS and supporting analyses and is heavily used in the HapMap project (International HapMap Consortium, 2003). PLINK was developed for humans and is also applicable to any organism.
However, out of the box, none of the aforementioned tools appear capable of performing the number of comparisons needed in a timely manner without extensive modification or knowledge of concurrent/asynchronous programming. They also require extensive data formatting by the users to obtain the necessary input file formats. This poses a significant barrier to research groups that wish to perform GWAS but lack the necessary technical expertise or computing power.
Recently, the publication of three definitive Arabidopsis (Arabidopsis thaliana) accession resources (Atwell et al., 2010; Cao et al., 2011; Horton et al., 2012) has made it possible to develop a platform that provides GWAS for practically any Arabidopsis trait data. Combined, these resources provide high-density single-nucleotide polymorphism (SNP) marker maps for over 1,000 Arabidopsis accessions, which should be sufficient for many, if not most, Arabidopsis GWAS. However, these data require a prohibitive amount of preprocessing to assemble the data in a format that can be used by the aforementioned GWAS programs.
We seek to introduce a platform for the genome-wide analysis of trait-marker interaction that simultaneously addresses the issue of computation time, renders GWAS analysis more easily accessible to biologists, provides informative post hoc analyses of the results, and makes such studies widely accessible to the broader scientific community regardless of in-house technical capability or analytical expertise.
RESULTS
We developed the Matapax Web-based platform to initially support GWAS in Arabidopsis. It was implemented as a Web-based solution utilizing the R library EMMA (Kang et al., 2008) and GAPIT (Lipka et al., 2011). EMMA is the current tool of choice for GWAS on Arabidopsis and provides efficient computation of associations using mixed models and kinship matrices for population structure control. GAPIT enhances EMMA with algorithms designed to boost statistical power and further decrease computation time. Processing of user data is performed in parallel on the server using the Torque resource management system. For use, the user requires an internet browser and need only upload a matrix of trait measurements including trait and accession names. The developed platform was named MArker-Trait Association Platform And eXplorer (Matapax).
Initial Data Check
Matapax incorporates three marker sets from the Arabidopsis POLYmorphism DataBase (AtPolyDB; referred to in this paper as SNP-Set1 and SNP-Set2; Atwell et al., 2010; Horton et al., 2012) and the 1,001 Genomes Project marker database (SNP-Set3; Cao et al., 2011) into the pipeline with the option to choose either one or the other for association mapping. Both of these databases provide high-quality genotyping data for an extensive number of accessions and markers (SNPs) that have been produced using either SNP chips (SNP-Set1 and SNP-Set2) or resequencing techniques (SNP-Set3). Matapax currently utilizes 199, 1,307, and 91 accessions from SNP-Set1, SNP-Set2, and SNP-Set3, respectively. The number of genotyped accessions in SNP-Set3 is set to increase to 1,001 accessions once initial, prepublication use limitations are lifted by the data providers. Matapax uses the public marker information as provided by the respective data providers, thereby relying on their proper SNP-calling quality control. However, we have included an option here for users to filter out markers that do not meet a specified minimum allele frequency. In the future, this option will also be available for the Hardy-Weinberg equilibrium.
There is currently no naming standard for Arabidopsis accessions, so the three different marker databases that Matapax uses occasionally have slightly different names for the same accessions. Matapax attempts to automatically match user accessions to the database accessions; however, when there is no direct match, the option to interactively select the correct accession is offered. In the marker databases, some accessions have been genotyped more than once. In this case, the user is presented with an option to choose from the different unique ecotype identifiers that the marker databases provide. If the user’s accession is not present in the marker database, the option to ignore the accession is available. The user can also specify that an accession is a cross between two other homozygous accessions by writing the accession name in the form “<accession 1> x <accession 2>” (i.e. separating the two accessions with an “x”).
In cases where the distribution of trait measurements is skewed, we provide the option to perform a Box-Cox transformation, which lessens the influence of outliers and produces a more symmetric distribution. The fit of the trait data to a normal distribution is estimated using the Shapiro-Wilk test (Shapiro and Wilk, 1965), and a plot of the measurement distribution is provided.
RESULTS
Post hoc analysis is assisted through the use of a graphical genome browser (Fig. 1) and a detailed results table (Fig. 2) listing the P values for the association of each marker with each trait. The user is able to load and manipulate this table entirely within his or her own internet browser and can recall the results of previous studies by supplying a job identification number that is provided after job submission. The results can also be downloaded as a compressed table for local analysis.
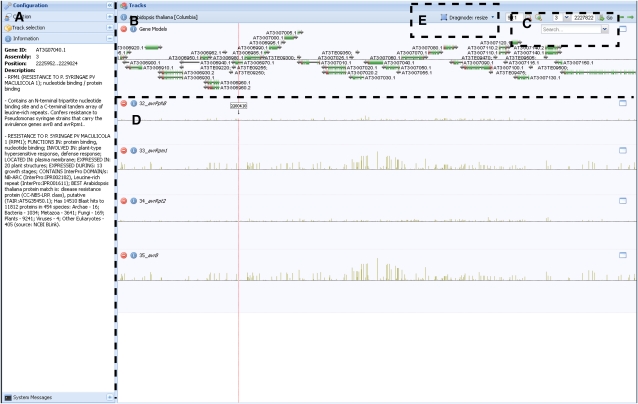
Figure 1.
A screen shot of the genome browser. Matapax allows users to view association P values in the context of the genome with the aid of the AnnoJ genome browser. This browser supports several features. A, The visible tracks can be selected in the side menu, and any information about the track source or gene annotation is also displayed here. B, The default TAIR genome annotation is the default track. The TAIR version is dependent on the marker database selected for association. Selecting the gene model names will display the model annotation in the side menu. C, The annotation track can be searched using the search tool. Searches can be made on both AGI codes and model annotation terms. D, The remaining tracks display the marker association strengths of each analyzed trait. Each analyzed trait is displayed in a separate track. The association P values are –log10 transformed, meaning the higher the bar, the stronger the trait-marker association. E, The genome browser enables browsing by “dragging” the tracks horizontally, scaling the values of the individual tracks as well as resizing the track height and zooming on areas of interest. Users can also go directly to positions of interest. The displayed figure shows high association between the avrRpm resistance phenotype and the rpm1 resistance gene, which has been shown to play a significant role in Pseudomonas syringae resistance (Grant et al., 1995). [See online article for color version of this figure.]
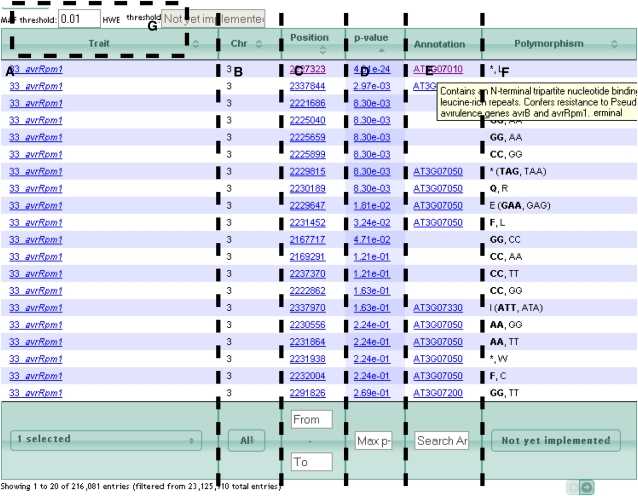
Figure 2.
A screen shot of the results table. The results displayed in the results table of Matapax are highly configurable to assist users in the interpretation of their results. A, The Trait column can be both sorted and filtered. Trait names link to quantile-quantile plots of the trait. B, The Chromosome column can be both sorted and filtered. C, The Position column can be both sorted and filtered on a range. Each position is linked to the genome browser, enabling the user to visualize the genomic context of the marker. D, The P Value column displays the association strength. Higher values have stronger association. This column can be both sorted and filtered with a minimum value. The association values link to a box plot of the marker-trait segregation. E, The Annotation column displays an AGI code if the marker can be found in a gene model. The AGI code is linked to the TAIR Web site, and holding the mouse over the AGI code will display the TAIR annotation. The TAIR version displayed is dependent on the marker data set version. The Annotation column can be filtered by both AGI codes and model annotation terms. F, The Polymorphism column displays the polymorphisms at the current position either as the nucleotide change from ecotype Columbia, if the marker is noncoding, or as the amino acid change, if the marker is coding. Currently, this column can be neither sorted nor filtered. G, It is also possible to filter the markers based on their minimum allele frequency (MAF). In the future, this filter will be extended to the Hardy-Weinberg equilibrium. [See online article for color version of this figure.]
The genome browser is based on AnnoJ (http://www.annoj.com), a system that utilizes Representational State Transfer principles. This allows AnnoJ to be customized toward our purposes and to draw data from many sources. The genome browser displays gene models provided by the Salk Institute (http://pbio.salk.edu/pbioe) along with the obtained –log10-transformed association P values and allows the user to browse, scale, zoom, and search the results.
The results table provides several features, including filter and multiple sort capabilities on the traits, chromosomes, positions, and P values. Searching of TAIR10 (for The Arabidopsis Information Resource) Arabidopsis annotation is enabled along with links to TAIR annotation (http://www.arabidopsis.org). Additionally, polymorphism information is provided as base changes in intergenic and intronic regions and amino acid changes in coding regions. Further information is provided as box plots of marker-trait segregation and quantile-quantile plots of the association values.
As the very nature of GWAS involves hundreds of thousands of accession-phenotype association tests, multiple testing correction needs to be addressed. To this end, the Benjamini-Hochberg method (Benjamini and Hochberg, 1995) is employed to correct the provided P values. Another approach is to visually observe how the most significant P values deviate from the expected distribution. Ideally, a higher than expected fraction of marker-associated P values will be skewed toward significant values. To assist in this form of analysis, quantile-quantile plots of the association values per trait are available. Such plots are a scatterplot of the expected and observed distributions that allow deviations to be clearly identified. Points that are noticeably different from the diagonal or deviate from the trend are potential candidates for further examination.
Matapax Run-Time
To assess Matapax processing times, we ran the pipeline while varying the numbers of traits and accessions independently. Increasing the number of traits submitted in a single job resulted in a linear increase in run-time, with jumps every 12 traits corresponding to the number of nodes on the server (Fig. 3A). There was also a gradual overall increase due to the overhead of querying the marker data and inserting the results data into a relational database management system.
Figure 3.
Computational run-time for Matapax. The run time of Matapax was tested for increasing numbers of traits and accessions. The results were plotted as a Lowess fit of the runtime as the number of traits and accessions increased, where the solid line is the fit and the dotted lines indicate the 95% confidence interval. A, The run-time over a number of traits increasing from two to 25. As 12 nodes were available on the server when the jobs were run, there is a rough doubling of computation time every 12 traits. There is also a slight extra cost for each trait in the postprocessing step of Matapax, as the results are inserted into a relational database. B, The run-time for both GAPIT and EMMA over increasing numbers of accessions: two, five, 10, 20, 50, 100, 200, and 1,000. EMMA was unable to complete running within a reasonable time when tested with 500 and 1,000 accessions. [See online article for color version of this figure.]
Increasing the number of accessions resulted in an exponential increase in run-time for both GAPIT and EMMA, although GAPIT performed much faster than EMMA (Fig. 3B). Due to the exponential computational complexity of EMMA, we were unable to test EMMA with more than 200 accessions. In the future, the next iteration of EMMA (EMMAx) will be used once it leaves the beta stage, providing a significant increase in computational speed. These results suggest that using GAPIT for any number of accessions over 200 is advisable. With GAPIT, it is possible to perform GWAS for 1,000 accessions in a little over 4 h per trait, implying that the use of all accessions in SNP-Set1, SNP-Set2, and SNP-Set3 is computationally feasible.
Matapax-Desktop Comparison
A simple test between the results of Matapax and a desktop computer was conducted using simulated phenotypes for all 199 accessions in SNP-Set1, resulting in a perfect correlation. The generated trait values, the association R script for a desktop computer, and the results for both Matapax and the desktop are available in Supplemental File S1. The marker data can be obtained from AtPolyDB (https://cynin.gmi.oeaw.ac.at/home/resources/atpolydb), and the EMMA R script can be obtained from the EMMA home page (http://mouse.cs.ucla.edu/emma).
Case Study
To test Matapax, we replicated an earlier association study (Atwell et al., 2010) that associated 107 Arabidopsis traits covering resistance, ionomics, flowering, and developmental phenotypes with the high-density SNP marker set SNP-Set1 using a kinship matrix for population structure correction. By comparing our results with those in that study, we were not only able to test our pipeline over several traits but also to assess how closely our method agrees with established results. Through Matapax, we associated the trait data with SNP-Set1 using EMMA to perform the associations and a K-matrix to correct for population structure.
The vast majority of Matapax results had an almost 1:1 correlation with the Atwell et al. (2010) data (Fig. 4; Supplemental Table S1). For these traits, observations could be drawn that are highly similar or identical to the original results concerning the location and significance of the association. Further information about the nature of the polymorphism was also provided, including whether it is intergenic, synonymous, or nonsynonymous.
Figure 4.
Correlation of published with Matapax association P values. The association P values of the published data were correlated with the association P values obtained by Matapax. For the vast majority of traits, Matapax is able to obtain identical results to the published data. However, Matapax is unable to reproduce the association results for a few traits. Possible reasons are discussed in the text.
There were a few traits, however, that remained difficult to reproduce, although the results clearly correlated. Due to the complicated nature of GWAS, there are several places where the discrepancies could have arisen. (1) Differences in the way the genomic data were calculated could alter how the measurements were compared with each other in the mixed linear model. Such differences could stem from the use of different marker or sequence data versions or differing missing value imputation methods. (2) Differences in the kinship matrix calculation would change the way the fixed effects are calculated in the model. (3) Other potential sources of differences are the algorithm version and exact parameters used.
DISCUSSION
By developing Matapax, we sought to create a platform that is easy to use, that makes the available rich genotypic information readily available, where the user requires little to no technical knowledge about GWAS or programming, and that assists the user in answering and further developing hypotheses from the results.
Although we are able to reproduce the results of the Atwell et al. (2010) study with high accuracy (87 out of 107 traits with a correlation greater than 0.80), there were seven traits that correlated rather poorly (correlation less than 0.60; Fig. 3). Due to the complex nature of a GWAS, it is difficult to test all the possible combinations of different parameters and data sets to try and tease out the work flow that the original study followed. This lack of reproducibility highlights the need for standards that could be easily provided by a platform such as Matapax.
As discussed by Zhao et al. (2007), the effectiveness of correction for population structure is widely debated, and top results require a significant degree of skepticism. This is hopefully mitigated with an informed and annotated review of the results and the application of the right population structure correction(s). Matapax provides two forms of correction (kinship matrix [K] and population structure matrix [P]), which can be newly calculated or uploaded by the user and which appear to be sufficient to reduce inflated P values in many traits (Zhao et al., 2007).
Aside from population structure correction, a great deal of manual inspection of the results is necessary to produce meaningful and biologically relevant results. To assist in manual inspection, we sought to include all relevant information about the results of a GWAS in a highly manipulable manner. With input from users, we hope to refine this aspect of Matapax further.
Matapax presents three high-coverage Arabidopsis marker database choices for association that provide whole-genome coverage of the Arabidopsis genome: AtPolyDB (SNP-Set1 and SNP-Set2; Atwell et al., 2010; Huang et al., 2011; Horton et al., 2012) and the 1,001 Genomes Project (SNP-Set3; Ossowski et al., 2008; Weigel and Mott, 2009), using different methods for genotyping. The choice of marker database for association can be informed by the differences between the three databases. SNP-Set1 uses a 250k SNP resequencing chip that interrogates 250,000 positions in the Arabidopsis genome that were chosen based upon earlier genotyping work (Kim et al., 2007). This allows the rapid genotyping of Arabidopsis accessions but suffers from incomplete coverage, as is evidenced by the presented case study. There are only 199 accessions available in this data set. SNP-Set2 uses the same experimental procedure for detecting SNPs as SNP-Set1 but uses a later calling method and has 1,307 accessions. The SNP-Set3 project markers are called using next-generation sequencing technology potentially identifying all markers in the sequenced accessions. This high-density genomic coverage comes at the cost of accession coverage. Currently, there are 447 accessions available in this data set, although only 91 are permitted for use.
In its first release, Matapax focuses on the model plant Arabidopsis. Evidently, the infrastructure can easily be expanded to include other species as well, pending increased availability of broad information on genetic variability in the respective species. Furthermore, upon positive reception of Matapax by the scientific community, creating a central repository for GWAS around the Matapax nucleus may also be worthwhile, especially in light of the increased need to report testable and reproducible GWAS study results.
CONCLUSION
We have developed a genome-wide association pipeline that performs all essential steps for basic GWAS, is capable of handling genotypic crosses as well as a large number of requests, and presents the results in an easy-to-use manner that assists in the development of hypotheses. The entry-level requirements to GWAS on Arabidopsis have been significantly lowered, as investigators no longer need to format the myriad data sets to fit the specifications of the particular tool they are using, nor are they required to maintain computational resources beyond a computer with internet browser capability. The results analysis is being assisted by the provision of necessary annotation and contextual information and the possibility to search the annotation using keywords.
We present Matapax with the core capabilities required for GWAS and with all initial development goals met. Future development of Matapax will require user input on functionality, and with enough interest, Matapax will be placed under active development, where we expect to work closely with users to plan many improvements and features that will be developed for future release. Matapax is freely available at http://matapax.mpimp-golm.mpg.de.
MATERIALS AND METHODS
Matapax Design
Matapax is a pipeline that ties together several technologies and publicly available tools and resources to achieve the desired goals and features.
We incorporate three genome-wide marker databases into the pipeline, all generated from Arabidopsis. SNP-Set1 and SNP-Set2 provide marker data for 199 and 1,307 accessions, respectively. The SNPs are determined using a 250k Affymetrix genotyping chip, thus providing a high-resolution map of genomic variation across a large number of accessions. Currently, SNP-Set3 provides marker data for 91 accessions. The SNPs are determined through resequencing, thus providing an extremely high-resolution map of genomic variation, although not as many accessions are available. However, in the future, this number will increase. The marker databases are formatted as SQLite (http://www.sqlite.org) relational databases for efficient storage, quick retrieval times, and scalability. At the time of submission, Matapax utilizes 91 accessions from SNP-Set3 because of prepublication use restrictions imposed by the original data providers. After publication by the original data providers, the entire sets will readily be made available via Matapax.
Association tests are performed in the R statistical computing environment (http://www.r-project.org) using the EMMA library (Kang et al., 2008). Through GAPIT and EMMA, we are able to provide efficient mixed-model association with optional corrections for kinship (K) and population structure (P). The K matrix is a distance matrix where the difference is defined as the mean of the similarities plus the differences between two accessions. In the current version of Matapax, the difference is calculated as a simple genetic distance:
where Gi and Gj are the accessions being compared and n is the number of markers. The values of G range between 0 and 1, typically falling on the values 0, 0.5, and 1, indicating a genotype with two major alleles, a heterozygous genotype, and a genotype with two minor alleles. Heterozygous genotypes are standardized before calculating the K and P matrices. If another form of kinship matrix is desired, then it is possible to upload custom matrices. In the future, Matapax will include other kinship matrices, such as the Loiselle kinship matrix. Matapax derives the P matrix by taking the first three principal components calculated from the genotypic data. To improve computational time, the NIPALS algorithm is used. However, power users should consider determining the number of principal components separately and uploading their own P matrix.
In addition to using efficient association algorithms, we implemented parallelization with the Torque Resource Manager (http://www.clusterresources.com), ensuring that all jobs are processed without conflict and that the pipeline remains scalable in the event that more nodes are added in the future. Currently, Matapax processes the traits of each submission in parallel, but in the future, each trait-marker comparison will be parallelized.
Result analysis is assisted with the implementation of the genome browser AnnoJ (http://www.annoj.org). Each trait is displayed as a separate track along with graphical and textual genome annotation. An association track can be scaled, zoomed, and browsed, allowing the user to peruse the results and see how well genomic regions associate with the given traits.
Results are also presented as a table where each row displays the association between a trait and a marker. The columns cover the tested Trait, the Chromosome of the marker, the Position of the marker on the chromosome, the P Value of the association, the Annotation of the marker as TAIR (http://www.arabidopsis.org) Arabidopsis Genome Initiative (AGI) codes, and the type of Polymorphisms that occur at that position. All columns except Annotation and Polymorphisms support multiple sorting and filtering to allow users to display the information they wish. The Annotation column supports searching allowing the user to search for genes or biological processes/molecular functions of interest. Filtering on the Polymorphism column will be implemented in a future release. Entries in the Trait column are hyperlinked to the quantile-quantile plot for that trait, entries in the Position column are hyperlinked to the genome browser, entries in the P Value column are hyperlinked to a box plot of the trait-marker segregation, and entries in the Annotation column are hyperlinked to the TAIR Web site.
Pipeline Flow
Only one input file is needed to start the pipeline, and there is a choice of marker database available for the user. Traits are submitted as a simple tab-delimited flat file where each row is a separate accession and each column shows the trait measurements. Accessions can be named by their native name or by the identifiers used by the groups that produced the marker databases. A basic normality check is performed on each trait, and a P value indicating how well the trait measurements fit a normal distribution is given for each trait, along with the option to perform a box-cox transformation that may improve the fit. A selection of population structure correction matrices is also presented to the user. Once all checks are complete, the markers for the matching accessions are extracted from preformatted SQL databases. Each trait is submitted individually to a cluster that is managed by the Torque Resource Manager (http://www.clusterresources.com), ensuring that all jobs are processed properly without resource conflicts.
Timing the Pipeline
To test the run-time of Matapax, we independently increased the number of traits and accessions over several submissions. The run-time response to increasing traits was tested for random values generated for 50 accessions using all SNPs from SNP-Set1, GAPIT, and a K matrix over increasing numbers of traits from two to 25. Each set of traits was submitted one at a time to ensure that all the nodes on the server were dedicated to a single submission. The run-time response to increasing accessions was tested for random values generated for a single trait using all SNPs from SNP-Seq1 and a K matrix over increasing numbers of accessions and for both GAPIT and EMMA. For reasons described previously, each set of accessions was submitted one at a time.
Result Comparisons
As an initial comparison, we compared the association P values of Matapax and a desktop computer. The trait data were created by generating random values for all 199 accessions in SNP-Set1. Matapax was run using a K matrix, and an R script was written to do the same on a desktop computer. The P values resulting from both were then correlated.
To test the pipeline on published data, we obtained phenotypic and genomic data from Atwell et al. (2010). These are publicly available from the AtPolyDB Web site. In addition, we obtained the association P values via direct communication. The phenotype data cover disease resistance, ionomics, flowering, and developmental phenotypes in Arabidopsis, and the accession data were SNP-Set1. The trait and marker data were fed into the Matapax pipeline, and the resulting association P values were correlated with the published association P values.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Trait names and correlation of Matapax with published data.
Supplemental File S1. Desktop R script, trait values, and desktop and Matapax results for the Matapax-desktop comparison.
Supplementary Material
Acknowledgments
We acknowledge Thomas Degenkolbe for his role in helping us test Matapax. We also acknowledge Uemit Seren of the Nordborg laboratory for providing the association values of the Atwell et al. (2010) article. We thank Detlef Weigel and colleagues for graciously allowing us the use of the generated genotyping information prior to publication. Finally, we acknowledge Reviewer 1 of our manuscript, whose detailed suggestions have helped shape and improve many aspects of Matapax.
References
- Aranzana MJ, Kim S, Zhao KY, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang CL, et al. (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81: 1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Horton M, Vilhjálmsson BJ, Seren U, Meng D, Meyer C, Ali Amer M, Borevitz JO, Bergelson J, Nordborg M. (May 23, 2011) Analysis and visualization of Arabidopsis thaliana GWAS using Web 2.0 technologies. Database (Oxford) 2011: http://dx.doi.org/10.1093/database/bar014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium (2003) The International HapMap Project. Nature 426: 789–796 [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. (2008) Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore M, Buckler ES, Zhang Z. (2011) User Manual of GAPIT: Genome Association and Prediction Integrated Tool. Buckler Lab for Maize Genetics and Diversity. http://www.maizegenetics.net/gapit (January 12, 2012) [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Muñiz-Fernandez F, Carreño-Torres A, Morcillo-Suarez C, Navarro A. (2011) Genome-wide association studies pipeline (GWASpi): a desktop application for genome-wide SNP analysis and management. Bioinformatics 27: 1871–1872 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. (1965) An analysis of variance test for normality (complete samples). Biometrika 52: 591–611 [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Benfey PN. (March 28, 2011) High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biol 12: http://dx.doi.org/10.1186/gb-2011-12-3-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Mott R. (May 27, 2009) The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol 10: http://dx.doi.org/10.1186/gb-2009-10-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, et al. (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.