Abstract
Plant Aldehyde Dehydrogenase10 (ALDH10) enzymes catalyze the oxidation of ω-primary or ω-quaternary aminoaldehydes, but, intriguingly, only some of them, such as the spinach (Spinacia oleracea) betaine aldehyde dehydrogenase (SoBADH), efficiently oxidize betaine aldehyde (BAL) forming the osmoprotectant glycine betaine (GB), which confers tolerance to osmotic stress. The crystal structure of SoBADH reported here shows tyrosine (Tyr)-160, tryptophan (Trp)-167, Trp-285, and Trp-456 in an arrangement suitable for cation-π interactions with the trimethylammonium group of BAL. Mutation of these residues to alanine (Ala) resulted in significant Km(BAL) increases and Vmax/Km(BAL) decreases, particularly in the Y160A mutant. Tyr-160 and Trp-456, strictly conserved in plant ALDH10s, form a pocket where the bulky trimethylammonium group binds. This space is reduced in ALDH10s with low BADH activity, because an isoleucine (Ile) pushes the Trp against the Tyr. Those with high BADH activity instead have Ala (Ala-441 in SoBADH) or cysteine, which allow enough room for binding of BAL. Accordingly, the mutation A441I decreased the Vmax/Km(BAL) of SoBADH approximately 200 times, while the mutation A441C had no effect. The kinetics with other ω-aminoaldehydes were not affected in the A441I or A441C mutant, demonstrating that the existence of an Ile in the second sphere of interaction of the aldehyde is critical for discriminating against BAL in some plant ALDH10s. A survey of the known sequences indicates that plants have two ALDH10 isoenzymes: those known to be GB accumulators have a high-BAL-affinity isoenzyme with Ala or cysteine in this critical position, while non GB accumulators have low-BAL-affinity isoenzymes containing Ile. Therefore, BADH activity appears to restrict GB synthesis in non-GB-accumulator plants.
Osmotic stress caused by drought, salinity, or low temperatures is a major limitation of agricultural production. Some plants synthesize and accumulate glycine betaine (GB), the most efficient osmoprotector known (Courtenay et al., 2000), when subjected to osmotic stress (Hanson and Wyse, 1982; Yancey et al., 1982; Weretilnyk et al., 1989; Valenzuela-Soto and Muñoz-Clares, 1994). It is generally accepted that GB is synthesized in the chloroplast stroma, as it is in spinach (Spinacia oleracea; Hanson et al., 1985), by a two-step oxidation of choline: first the alcohol group of choline is oxidized to the aldehyde group of betaine aldehyde (BAL) in a reaction catalyzed by choline monooxygenase (EC1.14.15.7; CMO), an enzyme unique to plants (Burnet et al., 1995); then the aldehyde group of BAL is oxidized to the acid group of GB in a reaction catalyzed by plant betaine aldehyde dehydrogenase [betaine aldehyde:NAD(P)+ oxidoreductase (EC 1.2.1.8); BADH; Hanson et al., 1985; Arakawa et al., 1987; Weretilnyk and Hanson, 1989; Valenzuela-Soto and Muñoz-Clares, 1994; Burnet et al., 1995; Hibino et al., 2001; Nakamura et al., 2001; Fujiwara et al., 2008; Kopěcný et al., 2011], an enzyme that belongs to the aldehyde dehydrogenase family10 (ALDH10; Vasiliou et al., 1999). Engineering the synthesis of GB in crops that naturally lack this ability has been a biotechnological goal for improving tolerance to osmotic stress (McNeil et al., 1999; Rontein et al., 2002; Waditee et al., 2007). The several attempts made so far have had limited success, stressing the need for a better understanding of the structural and functional properties of the enzymes involved in the GB biosynthetic pathway.
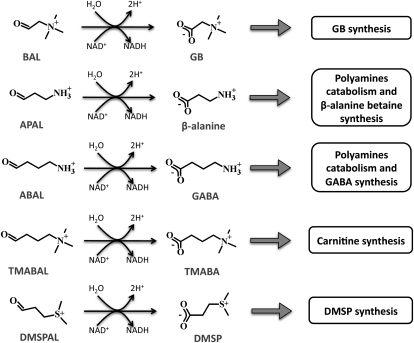
Most biochemically characterized plant ALDH10s appear to be ω-aminoaldehyde dehydrogenases (AMADHs) that can oxidize small aldehydes possessing an ω-primary amine group, such as 3-aminopropionaldehyde (APAL) and 4-aminobutyraldehyde (ABAL; Trossat et al., 1997; Vojtechová et al., 1997; Šebela et al., 2000; Livingstone et al., 2003; Oishi and Ebina, 2005; Bradbury et al., 2008; Fujiwara et al., 2008), or an ω-quaternary amino (trimethylammonium) group, as in BAL and 4-trimethylaminobutyraldehyde (TMABAL; Brauner et al., 2003; Fujiwara et al., 2008). In addition, unsurprisingly, given the structural similarity between the trimethylammonium and dimethylsulfonium groups, some plant BADHs can also use as substrate 3-dimethylsulfoniopropionaldehyde to produce the osmoprotectant 3-dimethylsulfoniopropionate (Trossat et al., 1997; Vojtechová et al., 1997; Fig. 1). Because of their relatively broad specificity, the plant ALDH10 enzymes may be involved not only in the synthesis of these two osmoprotectants but also in the synthesis of others, such as β-Ala betaine (Rathinasabapathi et al., 2000) and 4-aminobutyric acid (Bouché and Fromm, 2004), as well as in polyamine catabolism and the synthesis of carnitine (Fig. 1). To date, there are no other known plant ALDHs with AMADH or BADH activities other than the ALDH10 enzymes. With the exception of the two BADH isoenzymes from the mangrove plant Avicennia marina (Hibino et al., 2001), plant ALDH10s oxidize ω-primary aminoaldehydes with similar, or even higher, efficiency than they oxidize BAL, but some of these enzymes exhibit a very low activity with BAL (Šebela et al., 2000; Livingstone et al., 2003; Bradbury et al., 2008; Fujiwara et al., 2008), for as yet unknown reasons. On the basis of their affinity and activity with BAL, it has been proposed that plant BADHs constitute two subfamilies: true BADHs and high-BADH-homology aminoaldehyde dehydrogenases (Fitzgerald et al., 2009).
Figure 1.
Physiological roles of the reactions catalyzed by plant ALDH10 enzymes with ω-aminoaldehydes and 3-dimethylsulfoniopropionaldehyde as substrates. DMSP, 3-Dimethylsulfoniopropionate; DMSPAL, 3-dimethysulfoniopropionaldehyde; GABA, 4-aminobutyrate; TMBA, 4-trimethylaminobutyrate.
Neither how some ALDH10 enzymes discriminate against BAL nor the structural bases for BAL binding is known. Here, to our knowledge for the first time, we present the crystal structure of a chloroplastic ALDH10 with high BADH activity, the one from spinach (Spinacia oleracea; SoBADH), and report the results of site-directed mutagenesis of the residues involved in the binding of BAL as well as of the critical residue involved in discriminating BAL from other aminoaldehydes in those plant ALDH10s with poor BADH activity.
RESULTS
Overall Description of the SoBADH Three-Dimensional Structure
The structure of SoBADH in complex with NAD+ was determined at 2.3 Å resolution. The crystal belongs to the P1 space group and contains four subunits in the asymmetric unit describing two dimers, each of which corresponds to the biological unit of the enzyme (Valenzuela-Soto and Muñoz-Clares, 1994). The final model has an Rwork value of 21.3% and an Rfree value of 24.6%. Rwork is a measure of the agreement between the crystallographic model and the experimental x-ray diffraction data. Rwork = ∑|Fobs – Fcalc/Σ∑Ffobs, where Fobs and Fcalc are the observed and the calculated structure factors, respectively. Rfree is an R calculated using only 5% of reflections, which were randomly chosen and omitted from refinement. All data collection and refinement statistics are summarized in Supplemental Table S1. The folding (Supplemental Fig. S1) and topology (Supplemental Fig. S2) are very similar to those of every ALDH of known three-dimensional structure. The two K+-binding sites, one intrasubunit and the other intersubunit, which were previously found in the tetrameric BADH from Pseudomonas aeruginosa (González-Segura et al., 2009) also occur in the dimeric SoBADH.
Role of Aromatic Residues in the BAL-Binding Site of SoBADH
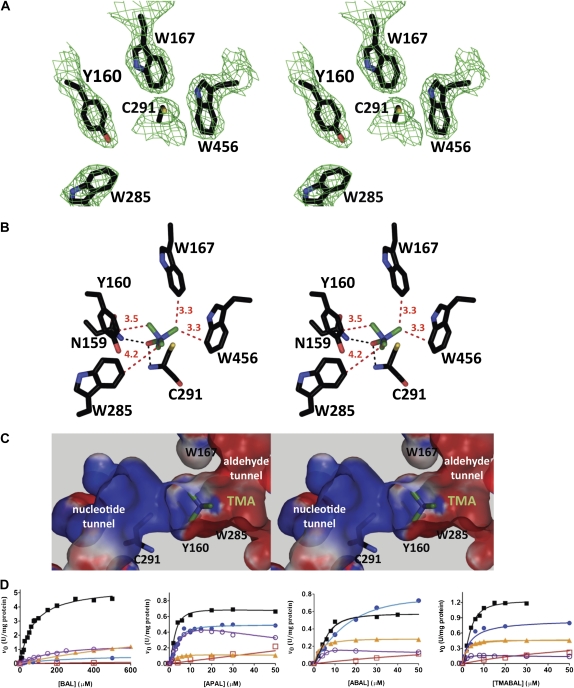
An inspection of the SoBADH crystal structure revealed four aromatic residues, Tyr-160, Trp-167, Trp-285, and Trp-456, in an arrangement suitable for the binding of the trimethylammonium group of BAL through cation-π interactions (Fig. 2A). As a first step to evaluate whether these residues play a role in substrate recognition and enzyme specificity, we constructed an energy-minimized model of the SoBADH active site with BAL productively bound, so that its carbonyl oxygen is inside the oxyanion hole (Muñoz-Clares et al., 2010), accepting two hydrogen bonds, one from the side chain amide nitrogen of Asn-159 and the other from the main-chain nitrogen of the catalytic Cys (Cys-291; SoBADH numbering), and its carbonyl carbon is in a position suitable for accepting the nucleophilic attack of the catalytic Cys. In the model, the trimethylammonium group is surrounded by the four aromatic residues at distances in the range of 3.2 to 4.2 Å, consistent with their involvement in BAL binding (Fig. 2B). The tunnel through which the aldehyde enters the active site has an irregular shape, widening in the vicinity of the catalytic Cys (Fig. 2C). The molecular electrostatic potential of the surface of this tunnel is clearly negative, mainly due to Glu and Asp residues near the tunnel entrance and to the aromatic rings of the four residues mentioned above, located where the trimethylammonium group binds (Fig. 2C). The side chain aromatic electrons contribute to the binding of the positively charged trimethylammonium group of BAL through cation-π interactions.
Figure 2.
Aromatic residues involved in binding of BAL to SoBADH. A, Stereoview of the 2Fo − Fc map contoured at 1 σ showing the aromatic residues in the aldehyde-binding site. B and C, Stereoviews of energy-minimized models of a BAL molecule docked into the active site showing the most favorable position of the trimethylammonium group when the carbonyl oxygen is kept inside the oxyanion hole and the carbonyl carbon is trigonal. In C, a section of the SoBADH active site shows the surface electrostatic potentials (−10 kT/e, red; 10 kT/e, blue) of the solvent-accessible molecular surface of the aldehyde and nucleotide entrance tunnels. Aromatic side chains are shown as sticks, with carbon atoms in black, oxygen in red, nitrogen in blue, and sulfur in yellow. The methyl groups are shown as green sticks. Distances of the methyl groups to the aromatic residues are given in angstroms and are depicted as red dashed lines. The hydrogen bonds between the carbonyl oxygen of the aldehyde and the oxyanion groups (i.e. the side chain amide nitrogen of Asn-159 and the main chain amide nitrogen of Cys-295) are depicted as black dashed lines. These panels were generated using PyMOL (DeLano, 2002). D, Saturation curves of wild-type SoBADH (black squares) and the mutants Y160A (red squares), W167A (blue circles), W285A (orange triangles), and W456A (violet circles) with BAL, APAL, ABAL, and TMABAL as substrates. Assays were carried out at pH 8.0 and fixed 0.2 mm NAD+. The points shown are experimentally determined values, and the lines drawn through these points are those calculated from the best fit of the data by nonlinear regression to Equation 1. Other experimental details are described in “Materials and Methods.”
To determine the relative contribution of these four aromatic residues to the binding of BAL, we individually mutated each of them to Ala to exclude the possibility for π-cation stabilization of the BAL trimethylammonium group. The kinetics of the pure mutant enzymes were studied with BAL, APAL, ABAL, and TMABAL as substrates. Steady-state kinetics studies were carried out at pH 8.0, which is close to the physiological intrachloroplastic pH value in spinach under light conditions (7.88; Werdan and Heldt, 1972), and at fixed 0.2 mm NAD+, which is the concentration estimated to exist in the stroma of spinach chloroplast (0.19; Heineke et al., 1991). This NAD+ concentration was found to be saturating or near saturating for both the wild-type and mutant enzymes in experiments in which the concentration of NAD+ was varied at a fixed concentration of BAL [a high but noninhibitory concentration that was at least four times the Km(BAL) value of each enzyme; Supplemental Table S2].
The wild-type SoBADH exhibited kinetic parameters for BAL, APAL, and ABAL (Table I; Fig. 2D) similar to those reported earlier for this enzyme (Incharoensakdi et al., 2000). The kinetics of TMABAL were comparable to those of APAL and ABAL. The Vmax and Km values determined using BAL as substrate were between four and nine times and between 12 and 26 times higher, respectively, than the values obtained using the other ω-aminoaldehydes, whose tighter binding to the enzyme appears to correlate with a slower catalysis. This results in a higher (between 1.2 and 4.3 times) catalytic efficiency (measured as Vmax/Km) for the other ω-aminoaldehydes tested than for BAL (Table I; Fig. 2D). The mutant enzymes exhibited significantly increased Km(BAL) and decreased Vmax/Km(BAL) values, particularly the Y160A mutant, which had 140 times higher Km(BAL) and 550 times lower Vmax/Km(BAL) than the wild-type SoBADH (Table I; Fig. 2D), indicating that the Tyr aromatic ring is of the utmost importance for binding of the aldehyde. On the basis of the observed changes in Km(BAL), Trp-285 also appears to be very important for the productive BAL binding, followed by Trp-167 and last by Trp-456. Although in our energy-minimized model Trp-285 is the farthest from the methyl groups of BAL, the significant effect of its mutation may be due in part to the loss of the van der Waals interactions of its indole ring carbon atoms CH2 and CZ3 with the phenol oxygen of Tyr-160. These interactions contribute to maintain the position of Tyr-160, which allows binding of the trimethylammonium group (see below). The mutant enzymes exhibited decreased Vmax values, but to a lesser extent than the increases in Km values. The exception was the W167A mutant, which had a greater effect on Vmax than on Km. Interestingly, the mutation of the four aromatic residues had a much smaller effect on the kinetics of SoBADH with APAL, ABAL, and TMABAL than on those with BAL (Table I; Fig. 2D). The mutant enzymes W167A, W285A, and W456A exhibited small changes in Km for APAL, ABAL, and TMABAL compared with the wild-type enzyme, indicating that their affinity for these aldehydes has not been importantly affected and, therefore, that these aromatic residues do not contribute to their binding. The Vmax values determined using these aldehydes as substrates were not importantly affected by the mutation of these three aromatic residues. Consequently, neither were the Vmax/Km values. The Y160A mutant, however, exhibited an 80 times increase in the Km value for APAL and around 20 times increases in the Km values for ABAL and TMABAL with respect to the wild-type SoBADH, resulting in importantly decreased Vmax/Km values. Multiple alignments of the known plant ALDH10 amino acid sequences indicated that Tyr-160, Trp-167, and Trp-456 are strictly conserved residues in these enzymes, whereas Trp-285 is a Phe or an Ala in some of them. On the basis of our results, it could be speculated that those enzymes with an Ala in the position equivalent to Trp-285 in SoBADH would use APAL, ABAL, and TMABAL as substrates preferentially to BAL.
Table I. Kinetic parameters of wild-type and mutant SoBADH enzymes using different ω-aminoaldehydes as substrates.
Initial velocities were obtained at 30°C, pH 8.0, and 0.2 mm NAD+. The kinetic parameters ± se were estimated by nonlinear regression of the experimental data to Equations 1 or 2, as appropriate. Vmax and Km values are given as units/mg protein and μm, respectively.
| Enzyme | Vmax | Km | Vmax/Km |
| BAL | |||
| Wild type | 5.4 ± 0.1 | 69 ± 5 | 7.8 ± 0.4 (×10−2) |
| Y160A | 1.4 ± 0.1b | 9.7 ± 1.2 (×103)b | 1.4 ± 0.8 (×10−4)b |
| W167A | 0.62 ± 0.01b | 2.9 ± 0.2 (×102)b | 2.1 ± 0.1 (×10−3)b |
| W285A | 3.4 ± 0.4 | 1.2 ± 0.3 (×103)b | 2.8 ± 0.3 (×10−3)b |
| W456A | 1.7 ± 0.2b | 2.4 ± 0.5 (×102)b | 7.0 ± 0.7 (×10−3)b |
| APAL | |||
| Wild type | 0.70 ± 0.04a | 2.6 ± 0.3a | 2.7 ± 0.4 (×10−1)a |
| Y160A | 0.86 ± 0.21 | 2.0 ± 0.2 (×102)b | 4.4 ± 0.4 (×10−3)b |
| W167A | 0.49 ± 0.01b | 3.6 ± 0.3 | 1.4 ± 0.1 (×10−1) |
| W285A | 0.14 ± 0.01b | 3.3 ± 0.8 | 4.2 ± 0.4 (×10−2)b |
| W456A | 0.52 ± 0.08 | 3.9 ± 0.8 | 1.3 ± 0.0 (×10−1) |
| ABAL | |||
| Wild type | 0.58 ± 0.03a | 5.5 ± 0.6a | 1.0 ± 0.2 (×10−1) |
| Y160A | 0.34 ± 0.02b | 1.2 ± 0.1 (×102)b | 2.8 ± 0.2 (×10−3)b |
| W167A | 1.4 ± 0.1b | 31 ± 6b | 4.5 ± 0.4 (×10−2)b |
| W285A | 0.30 ± 0.00b | 2.3 ± 0.2b | 1.3 ± 0.1 (×10−1) |
| W456A | 0.19 ± 0.03b | 4.3 ± 0.6 | 4.4 ± 0.0 (×10−2)b |
| TMABAL | |||
| Wild type | 1.2 ± 0.0a | 3.6 ± 0.2a | 3.4 ± 0.1 (×10−1)a |
| Y160A | 0.68 ± 0.02b | 86 ± 8b | 7.9 ± 0.6 (×10−3)b |
| W167A | 0.85 ± 0.03 | 3.7 ± 0.6 | 2.3 ± 0.3 (×10−1) |
| W285A | 0.47 ± 0.01b | 1.8 ± 0.2 | 2.6 ± 0.3 (×10−1) |
| W456A | 0.19 ± 0.02b | 1.5 ± 0.4b | 1.3 ± 0.2 (×10−1)b |
The estimated parameter is different to that determined for the wild-type enzyme with 99% confidence.
The estimated parameter is different to that determined with BAL with 99% confidence.
Residues Involved in Discriminating against the Binding of BAL in Plant ALDH10 Enzymes
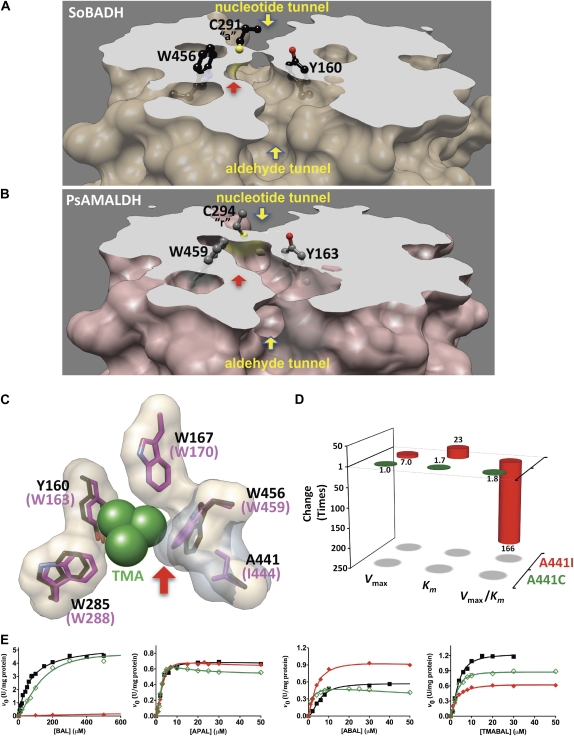
To date, there are only three plant ALDH10 enzymes whose three-dimensional structures are known: the SoBADH reported here (Protein Data Bank [PDB] code 4A0M) and two isoenzymes from pea (Pisum sativum [PsAMADH1 and PsAMADH2; PDB codes 3IWK and 3IWJ, respectively]; Tylichová et al., 2010). As none of the pea enzymes can use BAL as substrate (Šebela et al., 2000; Tylichová et al., 2010) whereas the spinach enzyme uses this aldehyde very efficiently, we compared the active sites of SoBADH and PsAMADH2 to find out the structural reasons for this important difference between them. We choose PsAMADH2 for this comparison because it has a Trp residue in the position of Trp-285 of SoBADH, whereas PsAMADH1 has a Phe. The superposition of the aldehyde-binding sites of the two enzymes shows that every residue lining the aldehyde entrance tunnel has a very similar conformation in both of them, with the exception of the side chain of the Trp residue equivalent to Trp-456 (Trp-459 in PsAMADH2), which in the pea enzyme is closer to the phenol group of the side chain of the Tyr residue equivalent to Tyr-160 (Tyr-163 in PsAMADH2) than in the spinach enzyme. This results in a narrower cavity in PsAMADH2 than in SoBADH at the place where the bulky trimethylammonium group of the BAL should be accommodated (Fig. 3, A and B). Energy-minimized models of the productively bound BAL molecule indicated that the trimethylammonium group can be bound in SoBADH but that it clashes with the Trp residue in PsAMADH2 (Fig. 3C). The reason for this is the different position of the Trp side chain in PsAMADH2, which is pushed toward the phenol group of the Tyr by the side chain of a nonactive site residue (an Ile [Ile-444; PsAMADH2 numbering]) that is behind and in close contact with the indole ring, at van der Waals distance (3.6 Å). Instead of Ile-444, SoBADH has Ala-441, whose much smaller side chain allows Trp-456 to be at a distance from Tyr-160 sufficient for accommodating the trimethylammonium group of BAL (Fig. 3C). To investigate whether this difference in a residue in the second sphere of interaction of the aldehyde could account for the differences in BAL specificity between the spinach and pea enzymes, we individually mutated Ala-441 in SoBADH for an Ile. As expected, the mutant A441I enzyme showed a greatly reduced affinity for BAL, indicated by a Km(BAL) value 23 times higher than that of the wild-type enzyme. It also exhibited an approximately seven times lower Vmax, which results in a decrease in the catalytic efficiency of the mutant A441I with BAL as substrate [Vmax/Km(BAL)] of around 160 times. On the contrary, the Km and Vmax values for the other ω-aminoaldehydes were very similar to those of the wild-type enzyme (Table II; Fig. 3, D and E). These results confirm our hypothesis of the critical importance of the side chain of the residue behind the indole ring of the Trp residue equivalent to Trp-456 for plant ALDH10s to discriminate against BAL.
Figure 3.
Structural determinants for BAL discrimination in ALDH10 enzymes. A to C, Comparison of the aldehyde-binding sites of SoBADH and PsAMADH2. A and B, Transverse sections of SoBADH and PsAMADH2 monomers (PDB codes 4A0M and 3IWJ, respectively) showing the catalytic tunnel and the narrowing of the active-site region where the trimethylammonium group of BAL binds due to the different position of Trp-459 (PsAMADH2) compared with that of Trp-456 (SoBADH), as signaled by the red arrow. Also shown is the catalytic Cys, which is in the attacking (“a”) conformation in SoBADH and in the resting (“r”) conformation in PsAMADH2. Amino acid side chains are shown as sticks, with carbon atoms in black (SoBADH) or gray (PsAMADH), oxygen in red, nitrogen in blue, and sulfur in yellow. C, Molecular surface of the aromatic residues of SoBADH (beige molecular surface, black carbon atoms) and PsAMADH (gray molecular surface, pink carbon atoms) showing the steric clash (marked by a red arrow) in the latter enzyme with the trimethylammonium group (TMA), shown as green balls, of a modeled BAL molecule. In PsAMADH, Ile-444 pushes Trp-459 toward Tyr-163, while in SoBADH, Ala-441 allows Trp-456 to be more distant from Tyr-160, thus leaving enough room between these two residues for a trimethylammonium group to bind. Images were generated using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen et al., 2004). D, Effects of mutation of residue Ala-441 to Ile or Cys on the kinetic parameters of SoBADH with BAL as variable substrate. Enzyme assays were carried out as described in “Materials and Methods,” and the data were analyzed as described in Figure 2D. E, Saturation curves of wild-type SoBADH (black squares) and the mutants A441I (red diamonds) and A441C (green diamonds) with BAL, APAL, ABAL, and TMABAL as substrates. Enzyme assays were carried out and the data analyzed as described in Figure 2D.
Table II. Kinetic parameters of wild- type and mutant SoBADH enzymes using different ω-aminoaldehydes as substrates.
Initial velocities were obtained at 30°C, pH 8.0, and 0.2 mm NAD+. The kinetic parameters ± se were estimated by nonlinear regression of the experimental data to Equations 1 or 2, as appropriate. Vmax and Km values are given as units/mg protein and μm, respectively.
| Enzyme | Vmax | Km | Vmax/Km |
| BAL | |||
| Wild type | 5.4 ± 0.1 | 69 ± 5 | 7.8 ± 0.4 (×10−2) |
| A441I | 0.77 ± 0.02a | 1.6 ± 0.2 (×103)a | 4.7 ± 0.4 (×10−4)a |
| A441C | 5.1 ± 0.6 | 120 ± 20 | 4.3 ± 0.7 (×10−2)a |
| APAL | |||
| Wild type | 0.70 ± 0.04 | 2.6 ± 0.3 | 2.7 ± 0.4 (×10−1) |
| A441I | 0.65 ± 0.01 | 2.5 ± 0.1 | 2.6 ± 0.8 (×10−1) |
| A441C | 0.78 ± 0.16 | 2.8 ± 0.6 | 2.7 ± 0.2 (×10−1) |
| ABAL | |||
| Wild type | 0.58 ± 0.03a | 5.5±0.6 | 1.0 ± 0.2 (×10−1) |
| A441I | 1.05 ± 0.06a | 3.8±0.5 | 2.7 ± 0.2 (×10−1) |
| A441C | 0.58 ± 0.05 | 1.8 ± 0.2 a | 3.2 ± 0.5 (×10−1) |
| TMABAL | |||
| Wild type | 1.2 ± 0.02 | 3.6 ± 0.2a | 3.4 ± 0.1 (×10−1) |
| A441I | 0.70 ± 0.2 a | 2.8 ± 0.3 | 2.5 ± 0.2 (×10−1) |
| A441C | 0.88 ± 0.22a | 2.7 ± 0.2 | 3.2 ± 0.2 (×10−1) |
The estimated parameter is different to that determined for the wild-type enzyme with 99% confidence.
The only other plant ALDH10 that so far has been found to have a very poor affinity for BAL is the barley (Hordeum vulgare) BADH isoenzyme called BBD1 by the authors (Fujiwara et al., 2008), which also has an Ile in this position. Interestingly, barley has another BADH isoenzyme with a high affinity for BAL, called BBD2, which possesses a Cys residue in the position of Ala-441 of SoBADH (Cys-439; BBD2 numbering). We anticipated that in this enzyme the steric impediment for the binding of the trimethylammonium group of BAL does not occur because of the small size of the Cys side chain. To prove this, we constructed the A441C mutant and confirmed that this change did not affect the Vmax and had a slight negative effect on Km(BAL), which was increased 1.7 times compared with the wild-type value (Table II; Fig. 3, D and E). These findings give additional support to our proposal of the critical importance of a small residue behind the indole group of the Trp residue for allowing BAL binding. The saturation kinetics with APAL, ABAL, and TMABAL were not affected in the A441C mutant, as was also expected.
DISCUSSION
The positive charge of the quaternary nitrogen of the trimethylammonium group of BAL suggests that negatively charged active-site residues should be involved in conferring substrate specificity to plant BADHs, by analogy with other enzymes that bind this group (Quaye et al., 2008). In a first study with SoBADH, Glu-103, which is strictly conserved in the known plant ALDH10 enzymes, was thought to be this residue; when mutated to Gln, however, there were no changes in the kinetics with BAL as substrate and only a small negative effect on those with APAL and ABAL (Incharoensakdi et al., 2000). The crystal structure of the spinach enzyme reported here explains these results: the side chain carboxylic group of Glu-103 is far from the aldehyde tunnel (Supplemental Fig. S3). ALDH10 enzymes also have two conserved Asp residues (Asp-107 and Asp-110; SoBADH numbering) whose carboxyl groups are exposed, or partially exposed in the case of Asp-107, to the solvent filling the aldehyde tunnel. The energy-minimized model of the productively bound BAL indicates that these carboxyl groups are too far away from the trimethylammonium group to directly interact with it. While this paper was in preparation, Kopečný et al. (2011) reported marked decreases in the affinity for APAL and ABAL of PsAMADH2 mutants in which these two Asp residues were changed to Ala. Our energy-minimized models with these aminoaldehydes productively bound (data not shown) showed that Asp-107 and Asp-110 are more than 7.5 Å away from the amino group of ABAL and APAL. Since the carboxyl of Asp-110 is relatively close to that of Asp-107, and therefore may influence its position, and the carboxyl group of Asp-107 is at an appropriate distance from Tyr-160 to electrostatically interact with the aromatic ring, the observed negative effects of the mutations D107A and D110A in PsAMADH2 may be, at least in part, due to the loss of this latter interaction, which may be relevant for the correct positioning of the tyrosyl residue equivalent to Tyr-160, a residue that is critical for the binding of the aldehydes (see below).
On the other hand, the trimethylammonium group of choline or GB has been shown to bind to proteins mainly through cation-π interactions with aromatic residues (Schiefner et al., 2004; Horn et al., 2006). The crystal structure of SoBADH showed four aromatic residues, Tyr-160, Trp-167, Trp-285, and Trp-456, in an arrangement suitable for cation-π interactions with the trimethylammonium group of BAL (Fig. 2). The residue equivalent to Trp-167 was suspected to participate in binding BAL in the BADH from cod liver (Johansson et al., 1998), which is an ALDH9, not an ALDH10, enzyme (Vasiliou et al., 1999), but this possibility was discarded on the basis that this residue is conserved in several other ALDHs that are not specific for BAL. The architecture of the BAL-binding site in SoBADH is similar to but not exactly the same as that of the GB-binding site in the GB transporter from Bacillus subtilis (Horn et al., 2006; PDB code 2B4L). While in the transporter, there are only three tryptophanyl residues forming a prism that perfectly accommodates and binds the trimethylammonium group in a fixed position, in SoBADH, there are four aromatic residues that could interact with the trimethylammonium group even if this group adopts different positions inside the active site. This allows a certain degree of flexibility in the binding of BAL that is needed for catalysis. The chemical mechanism of the ALDH-catalyzed reactions involves intermediates covalently attached to the catalytic Cys in which the trigonal carbonyl carbon of the aldehyde substrate changes to a tetrahedral carbon. First, the nucleophilic attack of the catalytic Cys on the carbonyl carbon results in the formation of a tetrahedral thiohemiacetal intermediate; second, the oxidation of this intermediate by the transfer of an hydride to NAD(P)+ forms a trigonal thioester intermediate; and then, the nucleophilic attack of the hydrolytic water molecule to the thioester forms a new tetrahedral intermediate that collapses into the trigonal acid product of the reaction. The oxygen atom remains in the oxyanion hole, but the rest of the molecule moves inside the active site as the geometry of the carbonyl carbon repeatedly changes (Muñoz-Clares et al., 2010). If the trimethylammonium group were bound in a fixed position, these movements would be hindered and catalysis negatively affected.
The kinetics of the SoBADH mutant enzymes in which the four aromatic residues were separately changed were consistent with the involvement of these residues in binding of the trimethylammonium group. Our results indicate that Tyr-160 is an important residue for binding the shortest ω-aminoaldehydes, BAL and APAL, particularly for BAL, but also for binding of ABAL and TMABAL, in spite of their amino group being farther from the aromatic ring. Interestingly, the Km for propionaldehyde, which lacks the amino group, is increased 50 times in the SoBADH Y160A mutant (data not shown). The energy-minimized model constructed by us suggests that the carbonyl oxygen of the aldehyde when bound into the oxyanion hole is at van der Waals distance from the CE1 carbon of the phenol ring and in the same plane. In this position, the carbonyl oxygen can electrostatically interact with the positive pole of the aromatic ring quadrupole. In this way, Tyr-160 may be involved in the binding of any aldehyde. Most known ALDH sequences have a Tyr or a Phe in this position (Julián-Sánchez et al., 2007), supporting the relevance of the aromatic ring for binding of the aldehyde group and probably also for its correct positioning for the following catalytic steps.
Our kinetic results with the mutant enzymes indicate that the binding of BAL to SoBADH depends on the interactions with the four aromatic residues much more than does the binding of APAL, ABAL, and TMABAL (i.e. the structural requirements for BAL binding are stricter than those for binding of the other ω-aminoaldehydes). This is because the architecture and size of the active-site pocket where the bulky trimethylammonium group binds should allow for cation-π interactions with the four aromatic rings and, at the same time, should prevent steric clashes between the active-site residues and the trimethylammonium group. Comparison of the aldehyde-binding sites of SoBADH and PsAMADH2 confirms this. Although an almost identical arrangement of the four aromatic residues can be observed in both, there is a subtle but critical difference between the spinach and pea enzymes responsible for the low affinity for BAL of the latter one: the size of the pocket formed by the residues equivalent to Tyr-160 and Trp-456 (SoBADH numbering), where the trimethylammonium group of BAL binds (Fig. 3). The residue behind the indole group of the Trp, an Ile in the pea enzyme and an Ala in SoBADH, determines the size of this pocket. The bulkier Ile pushes the Trp against the Tyr, thus hindering the binding of BAL, as the kinetics of the SoBADH mutant A441I demonstrated. Our findings are consistent with the increase in BADH activity of the Y163A mutant of PsAMADH2 relative to the wild-type enzyme (Kopěcný et al., 2011). Sequence alignments show that plant ALDH10 enzymes have only one of three different residues in the position of Ala-441, an Ala, a Cys, or an Ile. On the basis of the structural and biochemical studies reported here, we propose that those ALDH10 enzymes that have an Ala or a Cys at the position equivalent to Ala-441 of SoBADH are high-BAL-affinity isoenzymes, which presumably are involved in the synthesis of GB, while those ALDH10 enzymes that have an Ile in this position are low-BAL-affinity isoenzymes, which are likely involved in any of the other physiological functions of the plant ALDH10 enzymes (Fig. 1). The biochemical characterization of the ALDH10 isoenzymes from amaranth (Amaranthus hypochondriacus), spinach, barley, rice (Oryza sativa), pea, maize (Zea mays), and Arabidopsis (Arabidopsis thaliana) supports our proposal that those having Ala or Cys exhibit a high activity with BAL while those having Ile have a poor activity with this substrate (Supplemental Table S3). The exception is the Ile-containing isoenzyme from mangrove, which was reported to have a high affinity for BAL and to be unable to use APAL and ABAL as substrates (Hibino et al., 2001). Interestingly, all known ALDH10s that we propose as high-BAL-affinity isoenzymes contain a Trp in the position equivalent to Trp-285 of SoBADH, while several low-BAL-affinity isoenzymes have an Ala, Phe, Pro, or Ser instead. This is consistent with our results of the kinetics of the W285A mutant.
Different ALDH10 genes have been reported to exist in the genomes of several plants (McCue and Hanson, 1992; Ishitani et al., 1995; Wood et al., 1996; Legaria et al., 1998; Hibino et al., 2001; Bradbury et al., 2005), and the complete sequencing of several plant genomes has confirmed this trend. Some of them were considered alleles, given their high similarity at the nucleotide and amino acid sequences (McCue and Hanson, 1992), but others may be true isoenzymes, not only because they might be located in different loci but mainly because they may perform different physiological functions (i.e. some may be involved in the synthesis of GB from BAL and others in oxidizing other ω-aminoaldehydes). To clarify this, we searched all available ALDH10 sequences in the nonredundant protein sequences database of the National Center for Biotechnology Information (NCBI) using SoBADH as a query. After eliminating duplicates, we found that many species have two distinct ALDH10 sequences (Supplemental Table S3), which may correspond to two different isoenzymes. An examination of these sequences under the criterion of the residue occupying the position of Ala-441 in SoBADH indicated that some plants, such as amaranth, mangrove, barley, sorghum (Sorghum bicolor), Leymus chinensis, maize, and Zoysia tenuifolia, have the two kinds of isoenzymes: one with Ala or Cys in the position equivalent to Ala-441 of SoBADH and the other with Ile. Moreover, the known plant genomes have two ALDH10 genes that code for two isoenzymes; although there are other sequences deposited in the NCBI protein database, they are allelic variants or sequencing errors of these two. In some plants, such as maize, Zoysia, and sorghum, one of the two is the high-BAL-affinity isoenzyme and the other is the low-BAL-affinity isoenzyme. In other plant species whose genomes are known, such as Arabidopsis, Arabidopsis lyrata, soybean (Glycine max), rice, Populus trichocarpa, and potato (Solanum tuberosum), the two ALDH10 isoenzymes contain Ile and therefore are of the low-BAL-affinity kind. Phylogenetically, however, these two isoenzymes do not constitute two subfamilies within the ALDH10 family. It appears that they are the result of independent gene duplications that took place in different plant families.
Our phylogenetic analysis, described in ”Materials and Methods,” showed that plant ALDH10 sequences form a highly supported monophyletic clade, to the exclusion of bacterial ALDH10 and other plant ALDH sequences (bootstrap support for the monophyly of plant ALDH10, 100%; support for the monophyly of bacterial ALDH10, 99%; support for the monophyly of other plant ALDHs, 100%). Since these three groups of sequences formed well-supported separate clades, either the bacterial ALDH10s or the plant ALDHs can be used to root the plant ALDH10 clade, thus revealing its branching order, which was the same regardless of whether the rooting was done with the bacterial ALDH10s or the plant ALDHs, and a sequence of mutations from the hypothetical ancestral sequence toward contemporary plant ALDH10s. The mapping of amino acid residues for the position corresponding to Ala-441 in SoBADH indicates that Ile is the more common and ancient residue and that Ala tends to show up in gene duplicates in which the sister gene, if available, bears an Ile, whereas there are duplicates in which both copies bear an Ile. Furthermore, the bacterial ALDH10 sequences bear an Ile in the discussed position. Taken together, these observations strongly suggest that the ancestral plant ALDH10 gene coded for an Ile at the position homologous to Ala-441 in SoBADH. A functional specialization seems to have occurred when, in one of the two copies of the gene, the Ile mutated into an Ala or a Cys (or in the case of Vitis vinifera and potato, into a Val).
It has been suggested that the main reason why some plants do no accumulate GB is the lack of a functional CMO or an inadequate supply of choline to the chloroplast (Nuccio et al., 1998). Our results indicate that the absence of the high-BAL-affinity ALDH10 isoenzyme may be a major limitation for GB biosynthesis in plants. This is supported by our finding that the plant species in which one of the two isoenzymes is a high-BAL-affinity enzyme according to our criterion of possessing Ala or Cys in the position of Ala-441 of SoBADH, as well as other plants in which there have been found so far only one isoenzyme with Ala in this position, such as spinach and sugar beet (Beta vulgaris), or with Cys, such as wheat (Triticum aestivum), have been reported as being GB accumulators, whereas those plants that only have low-BAL-affinity isoenzymes are reported as lacking the ability to accumulate GB (Supplemental Table S3). Moreover, a functional CMO has only been found in species of Amaranthaceae that have the high-BAL-affinity isoenzyme, such as amaranth (Russell et al., 1998; Meng et al., 2001), orache (Atriplex hortensis; Shen et al., 2002), spinach (Rathinasabapathi et al., 1997), and sugar beet (Russell et al., 1998), whereas a nonfunctional gene was found in rice (Luo et al., 2007) and the recombinant CMO protein from Arabidopsis has no activity (Hibino et al., 2002). There are other CMO sequences deposited in GenBank, but it is not yet known whether the CMO proteins in these plants are functional or not.
Although there are no experimental data concerning the subcellular locations of most of the ALDH10 enzymes, the presence of Ala or Cys in position 441 of SoBADH correlates with a chloroplastic location in some of them while the presence of an Ile correlates with a peroxisomal location in others, but this is not a general rule (Supplemental Table S3). Thus, in Amaranthaceae, the high-BAL-affinity isoenzymes lack the C-terminal tripeptide SKL that has been considered as a signal for transport into peroxisomes (Gould et al., 1988) and the chloroplastic location of the spinach enzyme has been experimentally determined (Weigel et al., 1986), consistent with CMO being also chloroplastic (Burnet et al., 1995). According to their C-terminal signal, the low-BAL-affinity isoenzymes, but not the high-BAL-affinity ones, from mangrove, L. chinensis, and wheat are predicted to be peroxisomal isoenzymes, but both kinds of isoenzymes from Zoysia, sorghum, and maize have the peroxisomal signal. Interestingly, the high-BAL-affinity isoenzyme from barley, BBD2, was found to be cytosolic (Fujiwara et al., 2008) and the barley CMO peroxisomal (Mitsuya et al, 2011), suggesting that the subcellular location of GB synthesis in this plant is not chloroplastic but probably cytosolic. On the other hand, some low-BAL-affinity isoenzymes lack the peroxisomal signal. They may be located in leucoplasts, as was recently found for one of the two low-BAL-affinity isoenzymes from Arabidopsis (Missihoun et al., 2011). The rest of the low-BAL-affinity isoenzymes have an SKL or SKL-like C-terminal tripeptide. The peroxisomal location has been proved for the low-BAL-affinity isoenzymes from barley (Nakamura et al., 1997) and from the second Arabidopsis isoenzyme (Missihoun et al., 2011), both of which have the C-terminal SKL tripeptide.
CONCLUSION
The first crystal structure of a plant BADH, that from spinach, together with site-directed mutagenesis studies provide, to our knowledge for the first time, experimental evidence of the aromatic residues involved in binding of the trimethylammonium group of BAL in plant ALDH10 enzymes and, importantly, of the main structural feature determining whether they accept BAL as substrate: a non-active-site amino acid residue located in the second sphere of interaction of the aldehyde bound inside the active site. If this is a small residue, Ala or Cys, the enzyme will be a true BADH, whereas if this residue is an Ile, it pushes an active-site residue so that the cavity where the bulky trimethylammonium group of BAL binds is narrowed and the binding of BAL is prevented. Consequently, the activity with BAL will be low and the enzyme can be described as an AMADH. This conclusion is confirmed by the previously reported biochemical characterization of some of these enzymes. A survey of the known plant ALDH10 sequences indicates that the presence or absence of the high-BAL-affinity ALDH10 isoenzyme in plants correlates with them being a GB accumulator or a non GB accumulator, respectively. Therefore, the lack of the high-BAL-affinity ALDH10 isoenzyme appears to be a major limitation for GB biosynthesis in plants.
MATERIALS AND METHODS
Chemicals and Biochemicals
BAL chloride, the diethylacetals of APAL and ABAL, and NAD+ were obtained from Sigma-Aldrich. The diethylacetal of TMABAL was synthesized following the described method (Vaz et al., 2000). APAL, ABAL, and TMABAL were prepared freshly, hydrolyzing the corresponding diethylacetal forms following the method described by Flores and Filner (1985). The exact concentration of the resulting free aldehydes was determined in each experiment by determining the amount of NADH produced after their complete oxidation in the reaction catalyzed by SoBADH in the presence of an excess of NAD+.
Construction of the Expression Plasmid
The cDNA for SoBADH, a kind gift from Dr. Andrew D. Hanson, was the template to obtain the His-tagged enzyme by PCR, using the forward primer 5′-AGCATATGGCGTTCCAATTCC-3′, which contains the starting codon and an NdeI restriction site, and the reverse primer 5′-CTCGAGAGGAGACTTGTACC-3′, which corresponds to the 3′ end of the gene and contains the restriction site XhoI. The amplified DNA fragments were ligated into the pGEM-T Easy vector (Promega) and selected on Luria-Bertani agar plates. Plasmids were purified and digested with NdeI and XhoI restriction enzymes, and the fragment corresponding to the spinach (Spinacia oleracea) badh gene was purified and ligated into the corresponding sites of the pET28b+ vector (Invitrogen). The resulting plasmid, pET28-SoBADH, was used for the expression of full-length, N-terminal, His-tagged SoBADH.
For the expression of the recombinant proteins, cells of Escherichia coli BL21(DE3) (Agilent) were grown at 37°C in 400 mL of Luria-Bertani broth, added with 50 mg mL−1 kanamycin, until the optical density at 600 nm reached 0.6. At this point, protein expression was induced by the addition of 0.1 mm isopropyl thio-β-d-galactoside. The cells were allowed to grow for 4 h and harvested by centrifugation at 3,800g for 10 min. The pellet was suspended in 10 mL of 50 mm HEPES-KOH buffer, pH 7.5, containing 10 mm 2-mercaptoethanol and 10% (v/v) glycerol (buffer A) and sonicated for 20 min. Cell debris was removed by centrifugation at 15,000 rpm for 20 min, and the supernatant was applied to a Q-Sepharose Fast Flow column (GE Healthcare) equilibrated with buffer A. The column was washed with 10 volumes of the same buffer, and the enzyme was eluted with 300 mL of a linear KCl gradient from 0 to 350 mm in buffer A. His-tagged enzymes were applied into a column packed with Protino Ni-(tris(carboxymethyl)ethylene diamine) resin (Macherey-Nagel) equilibrated with 50 mm HEPES-KOH buffer, pH 6.5, containing 10% (v/v) glycerol and 10 mm imidazole (buffer B). The column was washed with the same buffer, and the enzymes were eluted by applying a linear gradient of imidazole from 10 to 250 mm in buffer B. Imidazole excess was removed by centrifugal concentration using Amicon Ultra 30 (Millipore), while buffer B was replaced by buffer A. To get rid of small contaminants, in the crystallization experiments a final step of purification through a Mono Q HR5 column (GE Healthcare) connected to a HPLC system (Waters) was included.
Site-Directed Mutagenesis
The plasmid pET28-SoBADH, containing the full sequence of the spinach badh gene and an N-terminal His tag, was used as a template for site-directed mutagenesis, which was performed via PCR using the Quick Change XL-II Site Directed Mutagenesis system (Agilent) and the following mutagenic primers: Y160A, 5′-GATTAATATCCCCATGGAATGCCCCACTTCTAATGGCTAC-3′ (forward) and 5′-AGCCATTAGAAGTGGGGCATTCCATGGGGATATTAATCC-3′ (reverse); W167A, 5′-CCACTTCTAATGGCTACAGCGAAAATTGCTCCAGCACTTGC-3′ (forward) and 5′-AAGTGCTGGAGCAATTTTCGCTGTAGCCATTAGAAGTGGG-3′ (reverse); W285A, 5′-ACTATTTTTGGCTGTTTCGCGACAAATGGTCAAATATGTAGTGC-3′ (forward) and 5′-ACATATTTGACCATTTGTCGCGAAACAGCCAAAAATAGTCC-3′ (reverse); W456A, 5′-TTTGTTCAAGCTCCTGCGGGAGGCATCAAGCGTAGTGG-3′ (forward) and 5′-CTACGCTTGATGCCTCCCGCAGGAGCTTGAACAAAGCATG-3′ (reverse); A441I, 5′-GAAGGCTCTAGAAGTTGGAATTGTTTGGGTTAATTGCTCAC-3′ (forward) and 5′-TTGTGAGCAATTAACCCAAACAATTCCAACTTCTAGAGCC-3′ (reverse); A441C, 5′-GAAGGCTCTAGAAGTTGGATGTGTTTGGGTTAATTGCTCAC-3′ (forward) and 5′-TTGTGAGCAATTAACCCAAACACATCCAACTTCTAGAGCC-3′ (reverse). The underlined portion of the oligonucleotides is the noncomplementary mutagenic region. Mutagenesis was confirmed by DNA sequencing. The level of production of functional recombinant protein in E. coli was similar for the wild-type SoBADH and its mutants.
Activity Assay and Kinetic Characterization of the Wild-Type and Mutant SoBADH Enzymes
The specific dehydrogenase activities of wild-type SoBADH and its mutants were measured spectrophotometrically at 30°C by monitoring the increase in A340 (ε = 6,220 m−1 cm−1) in a mixture (0.5 mL) consisting of 50 mm HEPES-KOH buffer, pH 8.0, 1 mm EDTA, and 0.2 mm NAD+ and variable concentrations of the aldehydes or saturating concentrations of the aldehydes and variable NAD+. The exact concentration of the aldehydes was determined by end-point assays using SoBADH and the standard assay conditions described below, and the exact concentration of NAD+ was determined by its A260 using a molar absorptivity of 18,000 m−1 cm−1 (Dawson et al., 1986). All assays were initiated by addition of the enzyme. Each saturation curve was determined at least in duplicate using enzymes from different purification batches. One unit of activity is defined as the amount of enzyme that catalyzes the formation of 1 μmol of NADPH per minute under our assay conditions.
Kinetic data were analyzed by nonlinear regression calculations using a Michaelis-Menten equation that includes terms in the numerator and denominator to account for partial substrate inhibition:
where v is the experimentally determined initial velocity, Vmax is the maximal velocity, [S] is the concentration of the variable substrate, Km is the concentration of substrate at half-maximal velocity, KIS is the substrate inhibition constant, and β is the factor that describes the effect of substrate inhibition on Vmax. For the estimation of Vmax/Km values and their associated se, a modified form of Equation 1 was used:
where KA is Vmax/Km. In those cases where no substrate inhibition was observed within the substrate concentration range used in the experiment, the data were fitted to the Michaelis-Menten equation.
Two estimated values for a given kinetic parameter determined for the wild-type enzyme and a mutant enzyme, or for two different amino aldehyde substrates, were considered significantly different at 99% confidence levels when the estimated values had confidence intervals that did not overlap. Confidence intervals were computed by multiplying the se of the estimated value by a factor that depends on the confidence level chosen and on the number of degrees of freedom, which equals the number of data points minus the number of parameters that were fit.
Protein concentrations were determined spectrophotometrically using the molar absorptivity at 280 nm deduced from the amino acid sequence by the method of Gill and von Hippel (1989): 43,200 m−1 cm−1 for the wild-type and A441I and A441C enzymes; 40,450 m−1 cm−1 for the single Trp mutant enzymes (W167A, W285A, and W456A); and 42,455 m−1 cm−1 for the Y160A mutant enzyme.
Crystallization, Structure Solution, and Refinement
Crystals of wild-type SoBADH were obtained by the hanging-drop technique under anaerobic conditions. Prior to crystallization, wild-type SoBADH at a concentration of 20 mg mL−1 was incubated with 2 mm NAD+ for 15 min at room temperature. Subsequently, 2 μL of protein solution was mixed with 2 μL of reservoir solution, which contained 85 mm Tris-HCl, pH 8.5, 0.17 m sodium acetate trihydrate, 25.5% (w/v) polyethylene glycol 4000, and 15% (v/v) glycerol (solution 22 of the crystal screen Cryo of Hampton Research). Bar-shaped crystals appeared at 18°C after 48 h and grew to their final dimensions within 1 week. The crystals were cooled under a nitrogen stream at 100 K during data collection. Synchrotron data were collected at the National Synchrotron Light Source on beamline X6a. The data were indexed with Mosflm (Leslie, 1992), integrated using XDS (Kabsch, 2010), and scaled and truncated with programs from the CCP4 suite (Collaborative Computational Project Number 4, 1994).
Before phases were obtained, SoBADH diffraction data analysis suggests a triclinic symmetry, which was probed after phases were determined with molecular replacement. A clear solution for the molecular replacement was obtained with the program Phaser (McCoy et al., 2007) using the coordinates of the BADH from Staphylococcus aureus (SaBADH; PDB code 3ED6) as a starting model. Two dimers were found in the asymmetric unit; however, the low data-parameters ratio, because of the resolution of the diffraction, compromises a free refinement of all the components in the asymmetric unit. To avoid an overrefinement, noncrystallographic symmetries (NCS) were applied, starting with constrained NCS, moving to tight restrained NCS and loose restrained NCS, and finishing the refinement releasing the NCS. Once the NCS were retired, alternating cycles of automatic and manual refinement were carried out with the standard protocols of Phenix (Adams et al., 2002) monitoring the Rwork and Rfree split during the whole process. The program Coot (Emsley and Cowtan, 2004) was used to analyze the electron density. Water molecules were automatically localized using Phenix (Adams et al., 2002) and Coot. Structural alignments were performed with Coot and PyMOL (DeLano, 2002).
Docking and Surface Electrostatic Potential Calculations
Aldehyde molecules were rigidly docked into the active site of the SoBADH and PsAMADH2 three-dimensional structure, so that the carbonyl oxygen makes the known interactions inside the oxyanion hole, using the PyMOL building mode and then energy minimized using the GROMOS 96 force field potential (van Gunsteren et al., 1996) of the Swiss PDB Viewer software (Guex and Peitsch, 1997). The convergence criterion was a value of 0.05 kJ mol−1 for the averaged derivative. The trimethylammonium group of BAL was nonrigidly docked into the SoBADH after removing NAD+ and glycerol molecules from the crystallographic structure using the PatchDock server (Schneidman-Duhovny et al., 2005) and then further refined with FireDock (Mashiach et al., 2008). The solution with the highest geometric shape complementarity score was used.
Surface electrostatics calculations of SoBADH were carried out with the Adaptive Poisson-Boltzmann Solver (Baker et al., 2001) using the PDB 2PQR Web portal (http://kryptonite.nbcr.net/pdb2pqr/). The PQR (for per-atom charge and radius) file was generated using the PARSE force field. The PROPKA program (Li et al., 2005) was used to assign the protonation state of SoBADH at pH 7.5. The rendered electrostatic potential was visualized using the plug-in Adaptive Poisson-Boltzmann Solver of the PyMOL software (DeLano, 2002).
Retrieval and Phylogenetic Analysis of ALDH10 Orthologs
To obtain a phylogenetically wide sampling of ALDH10 orthologs from plants, we performed a BLASTP search in the nonredundant protein sequences database of the NCBI using the amino acid sequence of SoBADH as query. The maximum number of target sequences was set at 100, and in order not to exclude any candidate plant ALDH10, the expected threshold was set at 10; the scoring matrix used was BLOSUM62, with gap-opening and gap-extension costs of 11 and 1, respectively. All sequences retrieved belong to either Viridiplantae or Eubacteria. In order to determine whether different hits coming from a given species correspond to different isoenzymes, we performed a series of BLASTP searches on the protein databases of the following completely sequenced and annotated species, maize (Zea mays), rice (Oryza sativa japonica), and potato (Solanum tuberosum), with the same parameters as indicated previously. Only hits with an E-value higher than 10e-3 were excluded. The resulting sequences were added to the first set and aligned using MAFFT (Katoh et al., 2002). A maximum likelihood phylogeny was constructed using RAxML (Stamatakis, 2006) on the CIPRES Web portal (Miller et al., 2010) under the JTT+G substitution model. The resulting tree was rooted at the bacterial sequences, showing a clade containing all the plant ALDH10 isoenzymes and a second clade including other plant ALDHs. This phylogeny was used as a guide, and the following criteria were applied to exclude sequences that could represent allelic variants or sequencing errors: (1) all sequences with associated activity information were kept; (2) when chromosome-mapping information was available, a representative of every locus was kept; and (3) for sequences that were 99% identical or that differed only by single position mutations, only one representative was kept.
The coordinates and the structure factors for the structure of SoBADH in complex with NAD+ have been deposited in the Protein Data Bank (www.rcsb.org) with the accession code 4A0M.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fold and secondary structure elements of the dimeric SoBADH.
Supplemental Figure S2. Topology diagram of the SoBADH subunit.
Supplemental Figure S3. Section of a SoBADH monomer showing the aldehyde tunnel and the positions of Glu-103, Asp-107, and Asp-110.
Supplemental Table S1. Data collection and refinement statistics for the SoBADH crystal.
Supplemental Table S2. Kinetic parameters of wild-type and mutant SoBADH enzymes using NAD+ as a variable substrate.
Supplemental Table S3. Plant isoenzymes of the ALDH10 family.
Supplementary Material
Acknowledgments
We thank Dr. Andrew D. Hanson (University of Florida, Gainesville) for providing the spinach BADH cDNA and Patricia Demare and Dr. Ignacio Regla (Facultad de Estudios Superiores Zaragoza, Universidad Nacional Autónoma de México) for the synthesis of TMABAL. We acknowledge the use of the X6a beamline of the National Synchrotron Light Source, Brookhaven National Laboratory.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Arakawa K, Takabe T, Sugiyama T, Akazawa T. (1987) Purification of betaine-aldehyde dehydrogenase from spinach leaves and preparation of its antibody. J Biochem 101: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H. (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Bradbury LMT, Fitzgerald TL, Henry RJ, Jin Q, Waters DLE. (2005) The gene for fragrance in rice. Plant Biotechnol J 3: 363–370 [DOI] [PubMed] [Google Scholar]
- Bradbury LMT, Gillies SA, Brushett DJ, Waters DLE, Henry RJ. (2008) Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol 68: 439–449 [DOI] [PubMed] [Google Scholar]
- Brauner F, Šebela M, Snégaroff J, Peč P, Meunier J-C. (2003) Pea seedling aminoaldehyde dehydrogenase: primary structure and active site residues. Plant Physiol Biochem 41: 1–10 [Google Scholar]
- Burnet M, Lafontaine PJ, Hanson AD. (1995) Assay, purification, and partial characterization of choline monooxygenase from spinach. Plant Physiol 108: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Courtenay ES, Capp MW, Anderson CF, Record MT., Jr (2000) Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 39: 4455–4471 [DOI] [PubMed] [Google Scholar]
- Dawson RM, Elliot DC, Elliot WH, Jones KM. (1986) Data for Biochemical Research, Ed 3. Oxford University Press, New York [Google Scholar]
- DeLano WL. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fitzgerald TL, Waters DLE, Henry RJ. (2009) Betaine aldehyde dehydrogenase in plants. Plant Biol (Stuttg) 11: 119–130 [DOI] [PubMed] [Google Scholar]
- Flores HE, Filner P. (1985) Polyamine catabolism in higher plants: characterization of pyrroline dehydrogenase. Plant Growth Regul 3: 277–291 [Google Scholar]
- Fujiwara T, Hori K, Ozaki K, Yokota Y, Mitsuya S, Ichiyanagi T, Hattori T, Takabe T. (2008) Enzymatic characterization of peroxisomal and cytosolic betaine aldehyde dehydrogenases in barley. Physiol Plant 134: 22–30 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- González-Segura L, Rudiño-Piñera E, Muñoz-Clares RA, Horjales E. (2009) The crystal structure of a ternary complex of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa provides new insight into the reaction mechanism and shows a novel binding mode of the 2′-phosphate of NADP+ and a novel cation binding site. J Mol Biol 385: 542–557 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Subramani S. (1988) Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J Cell Biol 107: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hanson AD, May AM, Grumet R, Bode J, Jamieson GC, Rhodes D. (1985) Betaine synthesis in chenopods: localization in chloroplasts. Proc Natl Acad Sci USA 82: 3678–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Wyse R. (1982) Biosynthesis, translocation, and accumulation of betaine in sugar beet and its progenitors in relation to salinity. Plant Physiol 70: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Peter U, Flügge UI, Heldt HW. (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Meng YL, Kawamitsu Y, Uehara N, Matsuda N, Tanaka Y, Ishikawa H, Baba S, Takabe T, Wada K, et al. (2001) Molecular cloning and functional characterization of two kinds of betaine-aldehyde dehydrogenase in betaine-accumulating mangrove Avicennia marina (Forsk.) Vierh. Plant Mol Biol 45: 353–363 [DOI] [PubMed] [Google Scholar]
- Hibino T, Waditee R, Araki E, Ishikawa H, Aoki K, Tanaka Y, Takabe T. (2002) Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J Biol Chem 277: 41352–41360 [DOI] [PubMed] [Google Scholar]
- Horn C, Sohn-Bösser L, Breed J, Welte W, Schmitt L, Bremer E. (2006) Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J Mol Biol 357: 592–606 [DOI] [PubMed] [Google Scholar]
- Incharoensakdi A, Matsuda N, Hibino T, Meng Y-L, Ishikawa H, Hara A, Funaguma T, Takabe T, Takabe T. (2000) Overproduction of spinach betaine aldehyde dehydrogenase in Escherichia coli: structural and functional properties of wild-type, mutants and E. coli enzymes. Eur J Biochem 267: 7015–7023 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Nakamura T, Han SY, Takabe T. (1995) Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol Biol 27: 307–315 [DOI] [PubMed] [Google Scholar]
- Johansson K, El-Ahmad M, Ramaswamy S, Hjelmqvist L, Jörnvall H, Eklund H. (1998) Structure of betaine aldehyde dehydrogenase at 2.1 A resolution. Protein Sci 7: 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julián-Sánchez A, Rivero-Rosas H, Martínez-Castilla LP, Velasco-García R, Muñoz-Clares RA. (2007) Phylogenetic and structural relationships of the betaine aldehyde dehydrogenase. In H Weiner, B Plapp, R Lindahl, E Maser, eds, Enzymology and Molecular Biology of Carbonyl Metabolism, Vol 13. Purdue University Press, West Lafayette, IN, pp 64–76 [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečný D, Tylichová M, Snegaroff J, Popelková H, Šebela M. (2011) Carboxylate and aromatic active-site residues are determinants of high-affinity binding of ω-aminoaldehydes to plant aminoaldehyde dehydrogenases. FEBS J 278: 3130–3139 [DOI] [PubMed] [Google Scholar]
- Legaria J, Rajsbaum R, Muñoz-Clares RA, Villegas-Sepúlveda N, Simpson J, Iturriaga G. (1998) Molecular characterization of two genes encoding betaine aldehyde dehydrogenase from amaranth: expression in leaves under short-term exposure to osmotic stress or abscisic acid. Gene 218: 69–76 [DOI] [PubMed] [Google Scholar]
- Leslie AGW. (1992) Recent changes to the MOSFLM package for processing film and image plate data. In Joint CCP4 and EACMB Newsletter Protein Crystallography 26. Daresbury Laboratory, Warrington, UK, pp 22–33 [Google Scholar]
- Li H, Robertson AD, Jensen JH. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61: 704–721 [DOI] [PubMed] [Google Scholar]
- Livingstone JR, Maruo T, Yoshida I, Tarui Y, Hirooka K, Yamamoto Y, Tsutui N, Hirasawa E. (2003) Purification and properties of betaine aldehyde dehydrogenase from Avena sativa. J Plant Res 116: 133–140 [DOI] [PubMed] [Google Scholar]
- Luo D, Niu X, Wang Y, Zheng W, Chang L, Wang Q, Wei X, Yu G, Lu B-R, Liu Y. (2007) Functional defect at the rice choline monooxygenase locus from an unusual post-transcriptional processing is associated with the sequence elements of short-direct repeats. New Phytol 175: 439–447 [DOI] [PubMed] [Google Scholar]
- Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. (2008) FireDock: a Web server for fast interaction refinement in molecular docking. Nucleic Acids Res 36: W229–W232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue KF, Hanson AD. (1992) Salt-inducible betaine aldehyde dehydrogenase from sugar beet: cDNA cloning and expression. Plant Mol Biol 18: 1–11 [DOI] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Hanson AD. (1999) Betaines and related osmoprotectants: targets for metabolic engineering of stress resistance. Plant Physiol 120: 945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YL, Wang YM, Zhang B, Nii N. (2001) Isolation of a choline monooxygenase cDNA clone from Amaranthus tricolor and its expressions under stress conditions. Cell Res 11: 187–193 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop. Institute of Electrical and Electronics Engineers, Inc., Piscataway, NY, pp 1–8 [Google Scholar]
- Missihoun TD, Schmitz J, Klug R, Kirch H-H, Bartels D. (2011) Betaine aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 233: 369–382 [DOI] [PubMed] [Google Scholar]
- Mitsuya S, Kuwahara J, Ozaki K, Saeki E, Fujiwara T, Takabe T. (2011) Isolation and characterization of a novel peroxisomal choline monooxygenase in barley. Planta 234: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Muñoz-Clares RA, Díaz-Sánchez AG, González-Segura L, Montiel C. (2010) Kinetic and structural features of betaine aldehyde dehydrogenases: mechanistic and regulatory implications. Arch Biochem Biophys 493: 71–81 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nomura M, Mori H, Jagendorf AT, Ueda A, Takabe T. (2001) An isozyme of betaine aldehyde dehydrogenase in barley. Plant Cell Physiol 42: 1088–1092 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T. (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11: 1115–1120 [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD. (1998) The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J 16: 487–496 [DOI] [PubMed] [Google Scholar]
- Oishi H, Ebina M. (2005) Isolation of cDNA and enzymatic properties of betaine aldehyde dehydrogenase from Zoysia tenuifolia. J Plant Physiol 162: 1077–1086 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Quaye O, Lountos GT, Fan F, Orville AM, Gadda G. (2008) Role of Glu312 in binding and positioning of the substrate for the hydride transfer reaction in choline oxidase. Biochemistry 47: 243–256 [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B, Burnet M, Russell BL, Gage DA, Liao P-C, Nye GJ, Scott P, Golbeck JH, Hanson AD. (1997) Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci USA 94: 3454–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B, Sigua C, Ho J, Douglas AG. (2000) Osmoprotectant β-alanine betaine synthesis in the Plumbaginaceae: S-adenosyl-L-methionine dependent N-methylation of β-alanine to its betaine is via N-methyl and N,N-dimethyl β-alanines. Physiol Plant 109: 225–231 [Google Scholar]
- Rontein D, Basset G, Hanson AD. (2002) Metabolic engineering of osmoprotectant accumulation in plants. Metab Eng 4: 49–56 [DOI] [PubMed] [Google Scholar]
- Russell BL, Rathinasabapathi B, Hanson AD. (1998) Osmotic stress induces expression of choline monooxygenase in sugar beet and amaranth. Plant Physiol 116: 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefner A, Breed J, Bösser L, Kneip S, Gade J, Holtmann G, Diederichs K, Welte W, Bremer E. (2004) Cation-π interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J Biol Chem 279: 5588–5596 [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šebela M, Brauner F, Radová A, Jacobsen S, Havliš J, Galuszka P, Peč P. (2000) Characterisation of a homogeneous plant aminoaldehyde dehydrogenase. Biochim Biophys Acta 1480: 329–341 [DOI] [PubMed] [Google Scholar]
- Shen Y-G, Du B-X, Zhang W-K, Zhang J-S, Chen S-Y. (2002) AhCMO, regulated by stresses in Atriplex hortensis, can improve drought tolerance in transgenic tobacco. Theor Appl Genet 105: 815–821 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Trossat C, Rathinasabapathi B, Hanson AD. (1997) Transgenically expressed betaine aldehyde dehydrogenase efficiently catalyzes oxidation of dimethylsulfoniopropionaldehyde and ω-aminoaldehydes. Plant Physiol 113: 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylichová M, Kopecný D, Moréra S, Briozzo P, Lenobel R, Snégaroff J, Šebela M. (2010) Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes. J Mol Biol 396: 870–882 [DOI] [PubMed] [Google Scholar]
- Valenzuela-Soto E, Muñoz-Clares RA. (1994) Purification and properties of betaine aldehyde dehydrogenase extracted from detached leaves of Amaranthus hypochondriacus L. subjected to water deficit. J Plant Physiol 143: 145–152 [Google Scholar]
- van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. (1996) Biomolecular Simulation: The GROMOS96 Manual and User Guide. Verlag der Fachvereine, Zurich [Google Scholar]
- Vasiliou V, Bairoch A, Tipton KF, Nebert DW. (1999) Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9: 421–434 [PubMed] [Google Scholar]
- Vaz FM, Fouchier SW, Ofman R, Sommer M, Wanders RJA. (2000) Molecular and biochemical characterization of rat γ-trimethylaminobutyraldehyde dehydrogenase and evidence for the involvement of human aldehyde dehydrogenase 9 in carnitine biosynthesis. J Biol Chem 275: 7390–7394 [DOI] [PubMed] [Google Scholar]
- Vojtechová M, Hanson AD, Muñoz-Clares RA. (1997) Betaine-aldehyde dehydrogenase from amaranth leaves efficiently catalyzes the NAD-dependent oxidation of dimethylsulfoniopropionaldehyde to dimethylsulfoniopropionate. Arch Biochem Biophys 337: 81–88 [DOI] [PubMed] [Google Scholar]
- Waditee R, Bhuiyan NH, Hirata E, Hibino T, Tanaka Y, Shikata M, Takabe T. (2007) Metabolic engineering for betaine accumulation in microbes and plants. J Biol Chem 282: 34185–34193 [DOI] [PubMed] [Google Scholar]
- Weigel P, Weretilnyk EA, Hanson AD. (1986) Betaine aldehyde oxidation by spinach chloroplasts. Plant Physiol 82: 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werdan K, Heldt HW. (1972) Accumulation of bicarbonate in intact chloroplasts following a pH gradient. Biochim Biophys Acta 283: 430–441 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Bednarek S, McCue KE, Rhodes D, Hanson AD. (1989) Comparative biochemical and immunological studies of the glycine betaine synthesis pathway in diverse families of dicotyledons. Planta 178: 342–352 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Hanson AD. (1989) Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity. Arch Biochem Biophys 271: 56–63 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Saneoka H, Rhodes D, Joly RJ, Goldsbrough PB. (1996) Betaine aldehyde dehydrogenase in sorghum. Plant Physiol 110: 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. (1982) Living with water stress: evolution of osmolyte systems. Science 217: 1214–1222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.