Abstract
Chickpea is a very important crop legume plant, which provides a protein-rich supplement to cereal-based diets and has the ability to fix atmospheric nitrogen. Despite its economic importance, the functional genomic resources for chickpea are very limited. Recently, we reported the complete transcriptome of chickpea using next generation sequencing technologies. We analyzed the tissue-specific expression of chickpea transcripts based on RNA-seq data. In addition, we identified two sets of lineage-specific genes, including a total of 3,632 chickpea-specific and 741 as legume-specific transcripts based on sequence comparison with other species within plant kingdom. The study of lineage-specific genes provides insights into the species-/lineage-specific functions and evolutionary processes. In this study, we further analyze the expression profiles of legume- and chickpea-specific transcripts in various tissue samples. Several legume- and chickpea-specific transcripts showed preferential and/or specific expression in the tissue samples analyzed. Our analysis provides evidence for the role of legume- and chickpea-specific transcripts in various tissues and opens an important area of future research to elucidate the exact role of these genes.
Keywords: chickpea, legumes, lineage-specific transcripts; RNA-seq; tissue-specific expression
Despite being a very important food legume crop, very less genomic information is available for chickpea. It is only recently that genomic information, including expressed sequence tags, molecular markers, bacterial artificial chromosome sequences and physical map, has started accumulating about chickpea. Recently, we reported the complete transcriptome sequence of chickpea using next generation sequencing (NGS) technologies.1,2 The optimized de novo assembly of the chickpea transcriptome resulted in a total of 34,760 transcripts representing genes involved in diverse cellular processes.1 Further, the expression analysis resulted in the identification of several tissue-preferential and tissue-specific transcripts in chickpea. A web resource, Chickpea Transcriptome Database (www.nipgr.res.in/ctdb.html), has also been made publicly available.1
Lineage-specific genes are present exclusively in only one group of related species and do not have any significant similarity to the genes from other taxonomic groups. Due to the availability of several genomic resources, lineage-specific genes have been identified in plant species, including rice, Arabidopsis and legumes.3-6 Recently, based on the comparison of chickpea transcriptome sequence data with genome and unigene sequences available for other plant species, we identified a total of 3,632 and 741 transcripts as chickpea- and legume-specific, respectively.1 Among these, only a few of the transcripts could be assigned a putative function based on the presence of a conserved domain sequence or similarity with other known proteins in legumes. However, most of the genes remain uncharacterized due to lack of similarity with other known proteins and domains. In this study, we have analyzed the expression profiles of legume- and chickpea-specific genes to reveal their putative role in various tissue samples.
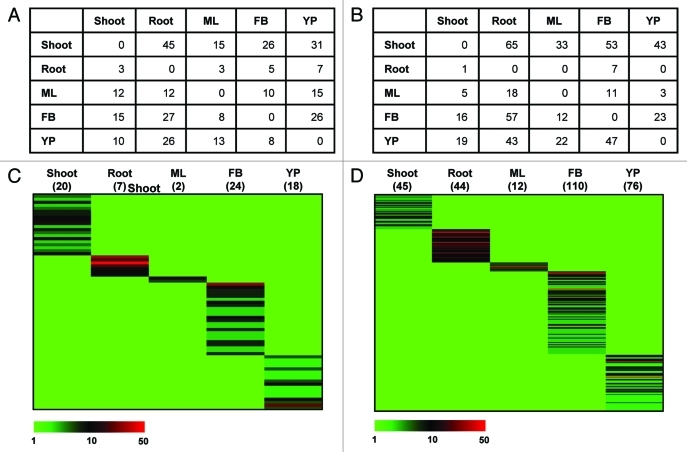
The use of sequence data from NGS technologies is considered a powerful tool for differential gene expression analysis.7-9 We also used the pyrosequencing data from five different tissue samples of chickpea, including shoot, root, mature leaf, flower bud and young pod,1 for expression profiling of legume- and chickpea-specific transcripts. Further, the differential expression of a few representative tissue-specific transcripts was validated by real-time PCR analysis, which correlated well with the RNA-seq results. We mapped all the high-quality 454 reads from different tissue samples on legume- and chickpea-specific transcripts using CLC Genomics Workbench software. A minimum of 90% coverage of the total length was used as a criterion for mapping of the reads. The number of uniquely mapped reads corresponding to each legume- and chickpea-specific transcripts in all the tissue samples was retrieved. Among the 741 legume-specific and 3632 chickpea-specific transcripts, no 454 read could be mapped on 470 and 2856 transcripts, respectively, using the above criterion. This indicated that most of the lineage-specific transcripts are expressed at very low level and are not represented in the 454 read data set. Hence, a very high depth of sequencing might be required to detect their expression. The normalized expression data (reads per kilobase per million; rpkm) for other lineage-specific transcripts (271 legume-specific and 776 chickpea-specific transcripts) showed large quantitative differences (very low expression represented by < 3 rpkm to very high expression represented by > 100 rpkm) in their expression profiles within a tissue sample. Further, the differential gene expression profiling based on normalized reads per million (rpm) data via tissue-by-tissue comparison identified several transcripts, which were expressed at significantly higher level in various tissue samples as compared with each other (Fig. 1A,B). We also identified putative legume- and chickpea-specific transcripts expressed specifically in a particular tissue sample only (the transcripts represented by ≥ 3 rpm in the tissue of interest and zero rpm in other tissues). The number of legume- and chickpea-specific transcripts specifically expressed in a tissue sample varied greatly. Overall, a total of 71 legume-specific and 287 chickpea-specific transcripts exhibited tissue-specific expression based on the above criteria. The heatmap depicting tissue-specific expression of these transcripts is shown in Figure 1C,D. In both the sets, largest number of transcripts were specifically expressed in flower bud. At least 18 legume-transcripts and 76 chickpea-specific were specifically expressed in young pod. The list of legume- and chickpea-specific transcripts showing tissue-specific expression is available as Tables S1 and S2, respectively. Only a few of these transcripts were predicted to contain a known putative conserved domain/motif, including extensin, amelogenin, serpentine-type 7TM GPCR chemoreceptor, WRKY, F-box and plant self-incompatibility response protein etc.
Figure 1.
Differential and tissue-specific expression analysis of legume- and chickpea-specific transcripts. (a,b) Number of legume- (A) and chickpea- (B) specific transcripts preferentially expressed in each tissue sample as compared with others in tissue-by-tissue comparison. The transcripts represented by ≥ 2-fold rpm as compared with other tissue sample are given. For the transcripts in each cell, there is preferential expression in the column tissue than the row tissue sample. (c,d) Heatmap of the legume- (C) and chickpea- (D) specific transcripts specifically expressed in various tissue samples. The number of transcripts showing specific expression in different tissue samples is also indicated. The color scale represents rpm value. The chickpea-specific transcripts with > 50 rpm (very highly expressed) are not distinguished by color scale (they are represented by the same color as that of transcripts with 50 rpm) to differentiate the genes with low to high expression. ML, mature leaf; FB, flower bud; YP, young pod.
The study of lineage-specific genes is very important because it provides insights into the lineage-specific functions and evolutionary processes such as speciation and adaptation.10 Although several hypotheses have been proposed for the origin of lineage-specific genes, the exact mechanism still remains unknown.11,12 Some of the lineage-specific genes identified in chickpea might represent the distant relatives of the genes present in other plant species and may perform similar functions. However, many of these may have got diversified to acquire the novel functions to perform lineage- or species-specific functions. Legumes represent a large family including several thousands of species inhabiting a wide range of environmental niches and possess unique developmental and physiological characteristics. The functional diversification of lineage-specific genes may be responsible for the unique features of the legumes and individual species. The most important feature of legumes is the nodulation and capacity to fix atmospheric nitrogen in association with nitrogen-fixing bacteria. The lineage-specific genes expressed specifically in roots might be involved in these legume-specific processes. Although several flowering genes were found to be conserved in legumes,13 our results clearly show that a set of genes specific to legumes also exist, which may be responsible for the specific biological processes occurring in legumes during flower and seed/pod development. Previously, only a few legume-specific genes from Medicago truncatula were shown to exhibit tissue-specific expression.5 However, our study provides evidence for the tissue-specific expression of quite a large number of legume- and chickpea-specific transcripts, suggesting their role in biology of corresponding chickpea tissues. To find out the exact role of these transcripts further experimental investigations are required. Further, as the expression data from diverse tissue samples representing different stages of tissue/organ development in chickpea become available, the identification of more number of tissue-specific lineage-specific transcripts will be possible.
Taken together, our results suggest that lineage-specific genes in chickpea may be involved in the regulation of cellular processes in specific tissue samples. Some of these may be responsible for lineage-specific functions also. The lineage-specific genes exhibiting tissue-specific expression are excellent candidates for further functional analysis.
Supplementary Material
Supplementary materials can be found at: www.landesbioscience.com/journals/psb/article/17879
Acknowledgment
The work is financially supported by the Department of Biotechnology, Government of India, New Delhi. We are thankful to Prof. Akhilesh Tyagi, NIPGR for reading of the MS and providing research facilities to RG.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/17879
References
- 1.Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, et al. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011;156:1661–78. doi: 10.1104/pp.111.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garg R, Patel RK, Tyagi AK, Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell MA, Zhu W, Jiang N, Lin H, Ouyang S, Childs KL, et al. Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol. 2007;145:1311–22. doi: 10.1104/pp.107.104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, et al. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol. 2010;10:41. doi: 10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham MA, Silverstein KA, Cannon SB, VandenBosch KA. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004;135:1179–97. doi: 10.1104/pp.104.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 7.Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–57. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, et al. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–95. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domazet-Loso T, Tautz D. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 2003;13:2213–9. doi: 10.1101/gr.1311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai JJ, Woo PC, Lau SK, Smith DK, Yuen KY. Accelerated evolutionary rate may be responsible for the emergence of lineage-specific genes in Ascomycota. J Mol Evol. 2006;63:1–11. doi: 10.1007/s00239-004-0372-5. [DOI] [PubMed] [Google Scholar]
- 12.Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 2006;103:9935–9. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht V, Foucher F, Ferrandiz C, Macknight R, Navarro C, Morin J, et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005;137:1420–34. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials can be found at: www.landesbioscience.com/journals/psb/article/17879