Abstract
The term contact inhibition (CI) encompasses the cellular changes that result in cessation of cell migration and of proliferation due to signals transduced when one cell comes into physical contact with another cell. Cancer cells, however, do not contact inhibit. A molecular understanding of the loss of CI in cancer cells is important for understanding tumor progression. In this Perspective, we propose that the loss of CI observed in cancer cells is the result of extracellular proteolysis of transmembrane cell-cell cell adhesion molecules (CAMs) in the tumor microenvironment. Proteolysis of homophilic cell-cell CAMs results in a shed extracellular fragment and released cytoplasmic fragment(s) that disrupts adhesion and induces signals that promote proliferation and/or migration. The importance of this observation in tumor progression is supported by the presence of the shed extracellular fragments of homophilic cell-cell CAMs in serum and tumor tissue of cancer patients suggesting that instead of acting as tumor suppressors, the shed CAM extracellular and cytoplasmic fragments actually function as oncogenes. The study of cell-cell CAM cleavage will provide important and novel means of diagnosing, imaging and treating tumor progression.
Keywords: cell-cell interactions, Receptors: structure and function, cell motility and migration, cadherin, receptor protein tyrosine phosphatase, cell-cell adhesion
Introduction
Normal cells grown in culture are inhibited or restricted from migration and/or proliferation by adhering to neighboring cells creating monolayers in culture (1). This phenomenon, known as contact inhibition (CI), was first observed in cells taken from chick embryo ventricles in 1953 (2). As early as 1957, it was recognized that cells derived from tumor tissue did not contact inhibit (3). CI of migration (CIM) describes the cessation of cell movement when a cell comes into contact with another cell. CI of proliferation (CIP) is a separate phenomenon that describes the cessation of cell division following cell-cell contact (1). The observation that tumor-derived cells do not contact inhibit (neither CIP nor CIM) led to the hypothesis that one step in tumor progression is the loss of cell-cell adhesion. Since loss of CI is one of the first observable steps in cancer progression, it is likely to be a key event. In the years since these observations, a number of proteins involved in promoting cell adhesion, known as cell adhesion molecules (CAMs; ref. 1) have been identified.
CAMs are subdivided into four superfamilies: cadherin, immunoglobulin (Ig), integrin, and selectin. Members of all CAM superfamilies can mediate cell-cell interactions, while integrins alone mediate cell-matrix interactions. CAMs bind to either the exact same proteins, known as homophilic binding, or different proteins, called heterophilic binding.
Many CAMs, including E-cadherin, cell-cell adhesion molecule 1 (C-CAM1) and deleted in colorectal cancer (DCC) are putative tumor suppressor genes (4). Genetic loss or DNA hypermethylation results in reduced CAM expression, and is one possible explanation for the loss of CI in cancer (4).
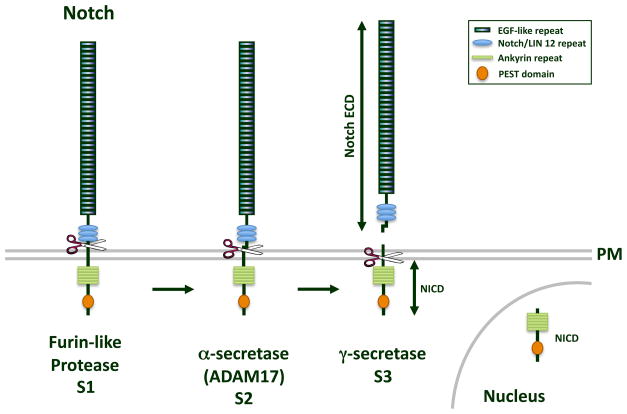
Post-translational changes of CAMs, such as proteolysis, has been observed and may also result in the loss of CI observed in cancer. Cleavage of many CAMs resembles that of the Notch receptor following ligand binding (5). Three cleavage events occur in the processing of Notch, identified as S1, S2, and S3 (Figure 1). S1 cleavage is attributed to a furin-like convertase in the trans-Golgi. S2 occurs in response to ligand binding and is mediated by a disintegrin and metalloprotease (ADAM). ADAM cleavage results in the shedding of the extracellular domain (ECD) of the Notch receptor. S3 is mediated by the γ-secretase complex and cleaves Notch within its transmembrane domain, thereby releasing its cytoplasmic fragment to translocate to the nucleus (5). γ-secretase consists of four proteins at any one time: the protease, either presenilin-1 (PS-1) or presenilin-2 (PS-2), and three other essential members of the complex, nicastrin, Aph-1, and Pen-2 (5). This same cleavage paradigm has been observed for other transmembrane proteins, including cell-cell CAMs (6).
Figure 1.
Notch cleavage. The Notch receptor is cleaved in three-step process to yield a functional signaling protein. S1 cleavage occurs in the Golgi by a furin-like protease. S2, or α-secretase, cleavage by ADAM17, occurs on the extracellular face of the plasma membrane (PM), and produces the shed ECD of Notch. S3 cleavage by the γ-secretase complex occurs in the transmembrane domain that releases the Notch ICD (NICD), which is able to translocate to the nucleus.
We suggest four potential mechanisms whereby cleavage of CAMs could alter their biological function and result in the loss of CI. First, the cleavage and shedding of the ectodomain (ECD) of cell-cell CAMs can reduce cell-cell adhesion and promote proliferation and/or migration due to its release from the plasma membrane, potentially antagonizing cell-cell adhesion by “occupying” the transmembrane receptor (7). Second, the shed fragment may associate with integrins and/or components of the extracellular matrix and form a new molecular substrate for cell migration (8, 9). Third, the shed ECD may bind to different receptors on other cells and activate distinct signals that regulate either proliferation or migration (9, 10). Finally, the released cytoplasmic fragment (ICD) may not associate with its normal signaling partners and may instead activate novel signals that regulate either proliferation or migration or potentiate normal signals when not anchored to the plasma membrane (11).
We propose a new theory to explain the loss of CI in tumor progression, in which cleavage of homophilic cell-cell CAMs deregulates CI by interfering with stable cell-cell adhesion and/or activating signals that promote cell proliferation and/or migration. We only consider homophilic cell-cell CAMs in this Perspective since cell-extracellular matrix CAMs are not involved in mediating inhibition of growth and migration initiated by cell-cell contact. We will review the four best examples of homophilic cell-cell CAM cleavage in cancer, E-cadherin, N-cadherin, EpCAM and PTPμ-subfamily receptor protein tyrosine phosphatases (RPTPs). Table 1 lists what is known about the cleavage and shedding of other homophilic cell-cell CAMs that may also disrupt CI. When considered altogether, there is a large body of evidence that supports our theory that disruption of stable cell-cell adhesion and adhesive signaling through homophilic cell-cell CAM proteolysis may be the key to explaining the loss of CI observed in cancer.
Table 1.
Summary of Cell-Cell CAM Cleavage in Cancer
| Cell-Cell CAM | ECD Protease | ICD Protease | Location of ICD | Evidence for role of cleavage in cancer | References |
|---|---|---|---|---|---|
| E-cadherin | Plasmin, MMP- 3, MMP-7, ADAM10, ADAM17 | PS 1/2 (presenilin) | Cytoplasm, nucleus | High levels in serum and urine of cancer patients; cleavage promotes cell migration | (7, 10–25) |
| N-cadherin | ADAM 10, MT1- MMP, MT5- MMP | PS 1 | Cytoplasm, not nucleus | High levels in serum of cancer patients; cleavage promotes migration of GBM cells, stimulates angiogenesis in vivo | (26–33) |
| P-cadherin | MMP-1, MMP-2 | Unknown | Unknown | Stimulation with sP-cad promotes breast cancer cell invasion | (42) |
| Protocadherin-γ3 | MMP | PS 1/2 | Cytoplasm, nucleus | Shedding reduces cell-cell aggregation | (43, 44) |
| PTPμ | ADAM/MMP | PS 1/2 | Cytoplasm, nucleus | Present in glioblastoma multiforme tissue not in normal brain tissue; PTPμ-ICD promotes cell migration, growth factor independent survival and anchorage independent cell growth | (39, 40) |
| PTPκ | ADAM10 | PS 1 | Cytoplasm, nucleus | Cleaved in breast cancer cells, results in dissolution of Adherens junctions | (38) |
| LAR | ADAM17 | PS 1/2 | Cytoplasm, nucleus | Shed in breast cancer cells, disrupts Adherens junctions | (45,46) |
| L1 | ADAM10 | PS 1 | Cytoplasm, nucleus | Soluble L1 in uterine and ovarian carcinoma tissue; shed L1 promotes adhesion to ECM and migration | (8,47,48) |
| EpCAM | ADAM17 | PS 2 | Cytoplasm, nucleus | EpCAM-ICD promotes cell proliferation, subcutaneously injected HEK293 cells expressing EpCAM-ICD induces tumor formation in mouse flank model | (36) |
| NrCAM | ADAM/MMP | Unknown | Unknown | NrCAM-Fc promotes proliferation and migration; NrCAM ECD is capable of inducing tumor formation in mouse flank model. | (9) |
Abbreviations used in the table. CAM: cell adhesion molecule; ECD: extracellular domain; ICD: intracellular domain; MMP; metalloprotease; ADAM: a disintegrin and metalloproteases; PS: presenilin;
E-cadherin
Cadherins are calcium dependent cell-cell adhesion molecules. All classical or type I cadherins share the same domain structure: 5 extracellular cadherin (EC) domains linked to conserved cytoplasmic tails. Like other classical cadherins, E-cadherin stabilizes cell-cell adhesions following homotypic binding by interacting with catenins and the actin cytoskeleton.
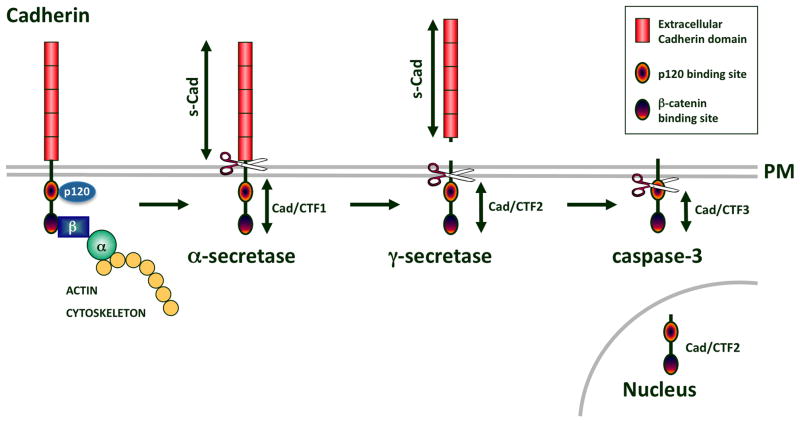
A cleaved soluble form of the ECD of E-cadherin, termed soluble E-cadherin (sE-cad) is generated by an α-secretase cleavage mechanism on the extracellular face of the plasma membrane (Figure 2). A handful of proteases are capable of mediating this cleavage event, including plasmin, MMP-3, MMP-7, ADAM-10, and ADAM-15 (7, 12–16). At least three cytoplasmic fragments of E-cadherin are also generated by separate proteolytic events (Figure 2) (11, 17–19). The membrane associated fragment, E-cad/CTF1, is produced by the α-secretase cleavage that generates sE-cad (15, 17, 19). The 33-kDa E-cad/CTF2 is produced by γ-secretase cleavage. E-cad/CTF2 is released into the cytosol (17, 19). E-cad/CTF3 is produced by caspase-3 cleavage to yield a 29-kDa fragment that is also found in the cytoplasm (17, 18).
Figure 2.
Cadherin cleavage. Classical cadherins are linked to the actin cytoskeleton via β-catenin (β) and α-catenin (α). E- and N-cadherin cleavage by an α-secretase produces the shed s-Cad subunit and a membrane tethered Cad/CTF1. Subsequent γ-secretase cleavage produces Cad/CTF2 that is released from the plasma membrane (PM) and is capable of translocating to the nucleus. Caspase 3 cleavage generates an even smaller fragment of E-cadherin, Cad/CTF3.
Cleavage of E-cadherin to yield sE-cad produces a fragment capable of homophilic binding that is no longer associated with the membrane. This fragment prevents cell-cell aggregation (7, 12, 20), induces cell invasion (7, 12, 14, 21), and promotes epithelial to mesenchymal transition (14). Stimulation of cells with an sE-cadherin-Fc chimera and/or the presence of sE-cad may promote cell proliferation (10, 15). The mechanism by which sE-cad is capable of disrupting cell-cell adhesion may be due to antagonizing cell-surface bound E-cadherin in a paracrine manner (7, 20). Recent data suggests that sE-cad may also antagonize CIM and CIP by stimulating the phosphorylation of ERK through heterodimerization and activation of the ErbB receptors HER2 and HER3 (10). Increased ERK activity may account for the increased motility and proliferation observed in cells with elevated sE-cad. In addition, stimulation of cells with sE-cad conditioned media can also induce MMP expression in the same cells (22), thereby promoting migration. To summarize, the literature supports our assertion that sE-cad can interfere with CIM and CIP.
Cleavage of E-cadherin also results in the dissolution of cadherin-based adherens junctions and, in the case of E-cad/CTF1 and CTF2 production, nuclear translocation of β-catenin (15, 17, 19), thereby potentiating Wnt signaling. Unlike full-length E-cadherin, E-cad/CTF2 itself is capable of translocating to the nucleus in association with p120 (11). p120 binds Kaiso and blocks its ability to repress transcription. When in a complex with p120, E-cad/CTF2 enhances the ability of p120 to block Kaiso-mediated gene repression (11). Enhanced transcription of the matrilysin (MMP7) gene results from the nuclear translocation of the p120-Ecad/CTF2 complex, which can further promote E-cadherin cleavage. Nuclear translocation of p120-E-cad/CTF2 also prevents staurosporine-induced apoptosis (11). Therefore, the sum of the literature on E-cadherin proteolysis suggests that E-cadherin cleavage may interfere with CIM by antagonizing stable cell-cell adhesion via its shed sE-cad (7, 12, 20) and by the dissolution of cadherin-catenin based adherens junction as a result of E-cad/CTF fragment generation. E-cadherin cleavage may also disrupt CIP via the activation of signals downstream of receptor tyrosine kinases, ERK, and β-catenin.
The presence of sE-cad in human tumor cell lines and in the serum of cancer patients has been frequently observed (for example see (23, 24). Circulating sE-cad is correlated with the development of distant metastases (25). Although the presence of sE-cad in human serum has not yet been shown to be causative in antagonizing CIM, it is supportive of our hypothesis that increased cell-cell CAM cleavage occurs during tumor progression. Cytoplasmic fragments of E-cadherin have also been observed in breast, colon, esophageal and epidermal carcinoma cell lines (11, 18, 19).
N-cadherin
As with E-cadherin, N-cadherin is first cleaved by an extracellular metalloprotease to yield a 95-kDa soluble N-cadherin fragment (sN-cad) and a 40-kDa membrane-tethered N-cad/CTF1 fragment (Figure 2) (26). Proteases capable of sN-cad production are ADAM10 (27), ADAM15 (28), MT1-MMP (29), and/or MT5-MMP (30). Ectodomain shedding and PS-1 activity are required to produce the 35-kDa N-cad/CTF2 fragment (26). N-cad/CTF2 binds to the CREB transcription factor and promotes its degradation, thereby decreasing CREB-mediated transcription (26). In addition, the cleavage of the cytoplasmic domain of N-cadherin increases β-catenin mediated transcription, which could be linked to CIP (27).
Studies of N-cadherin proteolysis suggest that the shed form of N-cadherin may also antagonize CIM. Using either a peptide containing the N-cadherin HAV motif necessary for N-cadherin-mediated adhesion or purified sN-cad, Derycke and colleagues demonstrated that sN-cad stimulates the migration of human endothelial pSV-1 cells and stimulates angiogenesis in vivo (31). Using siRNA directed against ADAM-10 or a cleavage site mutant of N-cadherin, Kohutek et al observed a significant reduction in the transwell migration of human glioblastoma cells (32). Clinical significance to N-cadherin cleavage in cancer is supported by the high levels of sN-cad found in the serum of cancer patients compared to normal controls (33).
EpCAM
Epithelial cell adhesion molecule (EpCAM) is an intriguing homophilic cell-cell CAM, as it mediates homophilic cell-cell aggregation, but can also interfere with cadherin-mediated adhesion (34). Structurally, EpCAM is a type I membrane protein composed of an extracellular epidermal growth factor (EGF)-like domain and a thyroglobulin-like domain fused to a short cytoplasmic domain with an NPXY internalization motif and two α-actinin binding sites (35). It does not fit into a canonical CAM superfamily.
EpCAM is a marker of various carcinomas (35). Recently, it has been suggested that proteolysis of EpCAM may be responsible for its ability to promote cell proliferation (36). Like the cadherins, EpCAM is cleaved by an ADAM, ADAM17, to yield Ep-ECD, and then is cleaved by the γ-secretase/PS-2 complex to yield Ep-ICD (36). Importantly, Ep-ICD is localized to the cytoplasm and nucleus in colon carcinoma tissue and cells, whereas full-length EpCAM is localized to the plasma membrane at cell junctions (36). Ep-ICD, but not full-length EpCAM, promotes cell proliferation, as assayed by cell counts and Ki67 labeling. This likely occurs through the interaction of Ep-ICD in the nucleus with ‘four and a half LIM domain’ protein 2 (FHL2), the Lef1 transcription factor and β-catenin to induce transcription of the c-myc gene. siRNA of EpCAM reduces the levels of c-myc protein and cell proliferation. C-myc expression and cell proliferation can be fully rescued by the re-expression of Ep-ICD. Finally, subcutaneous injection of HEK293 cells expressing EpCAM-YFP or Ep-ICD-YFP into mouse flank is capable of inducing tumor formation (36). Together, these data demonstrate that Ep-ICD can promote cell proliferation and tumorigenesis and may thus be involved in the deregulation of CIP in tumor progression.
PTPμ-subfamily of RPTPs
RPTPs differ from most CAMs in that they have catalytic tyrosine phosphatase domains in their intracellular segments in addition to extracellular segments capable of mediating cell-cell adhesion. RPTPs are classified based on their extracellular domain structure (37). The greatest evidence exists for the proteolysis of the type IIb PTPμ subfamily members in the loss of CI, although there is evidence for cleavage and shedding of the type IIa subfamily member, LAR, in cancer progression [Table 1].
The PTPμ-family of homophilic cell-cell CAMs is characterized by a meprin- A5 (neuropilin)-mu (MAM) domain, 1 Ig domain, and four fibronectin III (FNIII) repeats in its ectodomain. ECD shedding and ICD cleavage has been observed for PTPκ (38) and PTPμ (39). Like the other CAMs discussed (Figure 2), the ECDs of these RPTPs are cut by ADAM-like metalloproteases before being cleaved by the γ-secretase complex (38, 39). Both PTPκ- and PTPμ-ICDs translocate to the nucleus (38, 39). While both full-length PTPκ and PTPκ-ICD dephosphorylate β-catenin, dephosphorylation of β-catenin by PTPκ-ICD in the nucleus promotes TCF-mediated transcription, whereas full-length, transmembrane PTPκ does not (38). By promoting β-catenin/TCF signaling, we suggest that cleavage of PTPκ may lead to the loss of CIP.
Cleavage of PTPμ occurs preferentially in tumor-derived tissue and cancer cell lines (39, 40) and S.M.B-K., unpublished observations). For example, full-length PTPμ is down-regulated in glioblastoma multiforme (GBM) (41) via proteolysis (39, 40). ECD and ICD fragments of PTPμ are present only in GBM tissue compared to the surrounding normal tissue. Notably, PTPμ cleavage to yield PTPμ-ICD promotes GBM cell migration, growth factor independent survival and anchorage independent growth (39). The ability of PTPμ-ICD to promote GBM cell migration depends upon tyrosine phosphatase activity of the ICD fragment (39). Furthermore, studies of PTPμ cleavage demonstrate that PTPμ proteolysis and the presence of the PTPμ-ICD fragment directly result in more migratory cells, i.e. the loss of CIM.
Importantly, the shed ECD of PTPμ remains in the vicinity of the GBM tumor (40). Fluorescently tagged peptides that bind to PTPμ-ECD injected intravenously in mouse xenograft tumor models were effective at demarcating the main tumor and its edges in both mouse flank tumors and intracranial glioma tumors (40). This demonstrates that shedding of PTPμ-ECD occurs preferentially in the tumor microenvironment and can be utilized as a novel means of identifying the tumor and its margin.
Conclusions
The ability of cells to recognize one another and cease growth and/or migration as a result of cell-cell contact is an important cellular mechanism, and the loss of CI of proliferation and migration is a primary step in cancer progression. Transcriptional changes in CAMs that mediate cell-cell adhesion may be one means of overcoming CI. We propose that proteolysis of homophilic cell-cell CAMs also contributes to the loss of CIM and CIP that promotes tumorigenesis. The upregulation of ADAM proteases in tumor tissue (6) is one explanation for increased cell-cell CAM cleavage observed in cancer. We suggest that this has been overlooked because proteolysis can only be detected by examining protein size on immunoblots, and not through large scale screening techniques. Although this method is labor intensive, evidence is accumulating that proteolysis of many cell-cell CAMs results in the disruption of cellular adhesion, growth and migration (see Table 1). Cleavage of Nr-CAM (9) has been shown to promote cell proliferation, while proteolysis of P-cadherin (42) and L1 (8) increases cell migration.
Cell-cell CAM proteolysis results in increased motility of cells, increased cell proliferation, and resistance to apoptosis. How the cleaved cell-cell CAMs are able to affect these processes is not altogether understood. We know that the shed ECDs are found in the tumor microenvironment and/or circulating in bodily fluids. These shed ECDs are capable of antagonizing the adhesion of cell-surface bound CAMs (20), stimulating CAM cleavage (36), activating additional receptors [such as HER2 and HER3, in the case of sE-cad (10); or integrin receptors, in the case of L1 and NrCAM (8, 9)], and stimulating MMP expression (11, 22). In each of these situations, cell migration and/or proliferation can be stimulated.
The cleaved ICDs potentiate Wnt and Tcf/Lef signaling pathways. For example, the PTPκ-ICD, E-cad/CTF1, and N-cad/CTF2 all promote β-catenin nuclear translocation. Furthermore, PTPκ-ICD and Ep-ICD stimulate TCF- and Lef1-mediated transcription (36, 38). Ultimately, increased expression of c-myc (36), cyclin-D1 (15), and/or MMP-7 (11) has been observed following the generation of cell-cell CAM ICDs. These proteins can regulate signaling pathways that affect both cell proliferation and migration.
The recognition that tumor cells overcome CI by proteolyzing cell-cell CAMs provides a target for therapeutic intervention. While the use of ADAM/MMP inhibitors has been employed in treatment of cancer before (6), we suggest that they must be used in concert with γ-secretase inhibitors to limit the function of both the extracellular and intracellular fragments generated by cell-cell CAM cleavage. Given that the presenilin protease that cleaves cell-cell CAMs may differ (Table 1), it is possible that additional specificity in tumor treatment may be achieved using specific MMP and presenilin inhibitors in combination therapy.
Acknowledgments
Financial support: This research was supported by the following NIH grants: RO1-NS051520, R01-NS063971, and P30-CA043703.
We would like to thank Dr. Polly Phillips-Mason for her help editing this manuscript. This work is dedicated to Tabitha Yee-May Lou whose endowment supports research in the Brady-Kalnay lab.
References
- 1.Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–28. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res. 1953;5:111–31. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- 3.Abercrombie M, Heaysman JE, Karthauser HM. Social behaviour of cells in tissue culture. III. Mutual influence of sarcoma cells and fibroblasts. Exp Cell Res. 1957;13:276–91. doi: 10.1016/0014-4827(57)90007-1. [DOI] [PubMed] [Google Scholar]
- 4.Okegawa T, Li Y, Pong RC, Hsieh JT. Cell adhesion proteins as tumor suppressors. J Urol. 2002;167:1836–43. [PubMed] [Google Scholar]
- 5.Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3:673–84. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 6.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, Steelant W, et al. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J Cell Sci. 2001;114:111–8. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- 8.Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol. 2001;155:661–73. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conacci-Sorrell M, Kaplan A, Raveh S, Gavert N, Sakurai T, Ben-Ze’ev A. The shed ectodomain of Nr-CAM stimulates cell proliferation and motility, and confers cell transformation. Cancer Res. 2005;65:11605–12. doi: 10.1158/0008-5472.CAN-05-2647. [DOI] [PubMed] [Google Scholar]
- 10.Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283:18393–401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, et al. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol Chem. 2008;283:12691–700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryniers F, Stove C, Goethals M, Brackenier L, Noe V, Bracke M, et al. Plasmin produces an E-cadherin fragment that stimulates cancer cell invasion. Biol Chem. 2002;383:159–65. doi: 10.1515/BC.2002.016. [DOI] [PubMed] [Google Scholar]
- 13.Davies G, Jiang WG, Mason MD. Matrilysin mediates extracellular cleavage of E-cadherin from prostate cancer cells: a key mechanism in hepatocyte growth factor/scatter factor-induced cell-cell dissociation and in vitro invasion. Clin Cancer Res. 2001;7:3289–97. [PubMed] [Google Scholar]
- 14.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–73. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci U S A. 2005;102:9182–7. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KH, Choi EY, Hyun MS, Jang BI, Kim TN, Kim SW, et al. Association of extracellular cleavage of E-cadherin mediated by MMP-7 with HGF-induced in vitro invasion in human stomach cancer cells. Eur Surg Res. 2007;39:208–15. doi: 10.1159/000101452. [DOI] [PubMed] [Google Scholar]
- 17.Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–56. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–80. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Okamoto I, Araki N, Kawano Y, Nakao M, Fujiyama S, et al. Calcium influx triggers the sequential proteolysis of extracellular and cytoplasmic domains of E-cadherin, leading to loss of beta-catenin from cell-cell contacts. Oncogene. 1999;18:7080–90. doi: 10.1038/sj.onc.1203191. [DOI] [PubMed] [Google Scholar]
- 20.Wheelock MJ, Buck CA, Bechtol KB, Damsky CH. Soluble 80--kDa fragment of cell-CAM 120/80 disrupts cell-cell adhesion. J Cell Biochem. 1987;34:187–202. doi: 10.1002/jcb.240340305. [DOI] [PubMed] [Google Scholar]
- 21.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–72. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawrocki-Raby B, Gilles C, Polette M, Bruyneel E, Laronze JY, Bonnet N, et al. Upregulation of MMPs by soluble E-cadherin in human lung tumor cells. Int J Cancer. 2003;105:790–5. doi: 10.1002/ijc.11168. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths TR, Brotherick I, Bishop RI, White MD, McKenna DM, Horne CH, et al. Cell adhesion molecules in bladder cancer: soluble serum E-cadherin correlates with predictors of recurrence. Br J Cancer. 1996;74:579–84. doi: 10.1038/bjc.1996.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama M, Hirai S, Kamihagi K, Nakagawa K, Yasumoto M, Kato I. Soluble E-cadherin fragments increased in circulation of cancer patients. Br J Cancer. 1994;69:580–5. doi: 10.1038/bjc.1994.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charalabopoulos K, Gogali A, Dalavaga Y, Daskalopoulos G, Vassiliou M, Bablekos G, et al. The clinical significance of soluble E-cadherin in nonsmall cell lung cancer. Exp Oncol. 2006;28:83–5. [PubMed] [Google Scholar]
- 26.Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, et al. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–45. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, et al. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–52. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Najy AJ, Day KC, Day ML. ADAM15 supports prostate cancer metastasis by modulating tumor cell-endothelial cell interaction. Cancer Res. 2008;68:1092–9. doi: 10.1158/0008-5472.CAN-07-2432. [DOI] [PubMed] [Google Scholar]
- 29.Covington MD, Burghardt RC, Parrish AR. Ischemia-induced cleavage of cadherins in NRK cells requires MT1-MMP (MMP-14) Am J Physiol Renal Physiol. 2006;290:F43–51. doi: 10.1152/ajprenal.00179.2005. [DOI] [PubMed] [Google Scholar]
- 30.Monea S, Jordan BA, Srivastava S, DeSouza S, Ziff EB. Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J Neurosci. 2006;26:2300–12. doi: 10.1523/JNEUROSCI.3521-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derycke L, Morbidelli L, Ziche M, De Wever O, Bracke M, Van Aken E. Soluble N-cadherin fragment promotes angiogenesis. Clin Exp Metastasis. 2006;23:187–201. doi: 10.1007/s10585-006-9029-7. [DOI] [PubMed] [Google Scholar]
- 32.Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci. 2009;29:4605–15. doi: 10.1523/JNEUROSCI.5126-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derycke L, De Wever O, Stove V, Vanhoecke B, Delanghe J, Depypere H, et al. Soluble N-cadherin in human biological fluids. Int J Cancer. 2006;119:2895–900. doi: 10.1002/ijc.22219. [DOI] [PubMed] [Google Scholar]
- 34.Carpenter G, Red Brewer M. EpCAM: another surface-to-nucleus missile. Cancer Cell. 2009;15:165–6. doi: 10.1016/j.ccr.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Trzpis M, McLaughlin PM, de Leij LM, Harmsen MC. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–95. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 37.Ensslen-Craig SE, Brady-Kalnay SM. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev Biol. 2004;275:12–22. doi: 10.1016/j.ydbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, et al. Furin-, ADAM 10-, and gamma-Secretase-Mediated Cleavage of a Receptor Tyrosine Phosphatase and Regulation of beta-Catenin’s Transcriptional Activity. Mol Cell Biol. 2006;26:3917–34. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, et al. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69:6960–8. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, et al. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12:305–16. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, et al. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009 doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro AS, Albergaria A, Sousa B, Correia AL, Bracke M, Seruca R, et al. Extracellular cleavage and shedding of P-cadherin: a mechanism underlying the invasive behaviour of breast cancer cells. Oncogene. 2010;29:392–402. doi: 10.1038/onc.2009.338. [DOI] [PubMed] [Google Scholar]
- 43.Haas IG, Frank M, Veron N, Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. J Biol Chem. 2005;280:9313–9. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- 44.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, et al. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–52. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhe JE, Streit S, Hart S, Ullrich A. EGFR signaling leads to downregulation of PTP-LAR via TACE-mediated proteolytic processing. Cell Signal. 2006;18:1515–27. doi: 10.1016/j.cellsig.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Haapasalo A, Kim DY, Carey BW, Turunen MK, Pettingell WH, Kovacs DM. Preseniliin/gamma-Secretase-mediated Cleavage Regulates Association of Leukocyte-Common Antigen-related (LAR) Receptor Tyrosine Phosphatase with beta-Catenin. J Biol Chem. 2007;282:9063–72. doi: 10.1074/jbc.M611324200. [DOI] [PubMed] [Google Scholar]
- 47.Riedle S, Kiefel H, Gast D, Bondong S, Wolterink S, Gutwein P, et al. Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem J. 2009;420:391–402. doi: 10.1042/BJ20081625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A, et al. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003;362:869–75. doi: 10.1016/S0140-6736(03)14342-5. [DOI] [PubMed] [Google Scholar]