Abstract
22q11.2 deletion syndrome is mainly characterized by conotruncal congenital heart defects, velopharyngeal insufficiency, hypocalcemia and a characteristic craniofacial appearance. The etiology in the majority of patients is a 3-Mb recurrent deletion in region 22q11.2. Nevertheless, recently some cases of infrequent deletions with various sizes have been reported with a different phenotype. We report on a patient with congenital heart disease (truncus arteriosus type 2) in whom a de novo 1.3-Mb 22q11.2 deletion was detected by array comparative genomic hybridization. The deletion described corresponds to an atypical and distal deletion which spans low copy repeat (LCR) 4 and is associated with breakpoint sites that do not correspond to known LCRs of 22q11.2. We examine the clinical phenotype of our case and compare our findings with those published in the literature. The most prevalent clinical features in this type of deletion are a history of prematurity, pre-natal and post-natal growth retardation, slight facial dysmorphic features, microcephaly and developmental delay, with a speech defect in particular. These are clearly different from those found in the classic 22q11.2 deletion syndrome, and we believe that the main differential diagnosis should be with Silver-Russel syndrome. In our case we observe the cardiac phenotype with truncus arteriosus communis usually seen in the classic 22q11.2 deletion syndrome, and so far associated with the TBX1 gene. Significantly, however, TBX1 is not included in our patient's deletion. The possible roles of a position effect or other genes are discussed.
Key Words: 22q11.2 distal deletion syndrome, Pre- and post-natal growth retardation, TBX1, Truncus arteriosus
Introduction
Interstitial deletions of chromosome 22q11.2 are the most commonly seen microdeletions observed in humans, with a frequency of approximately 1:4,000–1:8,000 live births. The main clinical phenotypes associated with the 22q11.2 deletion syndrome are the velocardiofacial syndrome (OMIM 192430) and DiGeorge syndrome (OMIM 188400). Patients with the 22q11.2 deletion present various combinations of the following clinical features: congenital heart disease, in particular conotruncal heart anomalies, typical facial phenotype, palatal abnormalities with velopharyngeal insufficiency, congenital hypocalcemia and immune deficiency. The vast majority of patients with the 22q11.2 microdeletion exhibit a common ∼3-Mb deletion while 7% of the individuals have a smaller 1.5-Mb deletion [Carlson et al., 1997]. It was discovered that the 3-Mb as well as the 1.5-Mb deletions occur as a result of nonallelic homologous recombination [Stankiewicz and Lupski, 2002]. The majority of deletions were located within the common ∼3-Mb deletion region or were found to overlap with it. ‘Atypical deletions’ correspond to deletions located in the typical deletion region with varied breakpoints, while ‘distal deletions’ are located distal to the 3-Mb common deletion region.
So far, only a small number of ‘atypical deletions’ [Kurahashi et al., 1996, 1997; Garcia-Minaur et al., 2002; Rauch et al., 2005; D'Angelo et al., 2007; Fernandez et al., 2009] and ‘distal deletions’ [Rauch et al., 1999, 2005; Saitta et al., 1999; Mikhail et al., 2007; Ben-Shachar et al., 2008; Rodningen et al., 2008; Xu et al., 2008; Bruce et al., 2009; Lafay-Cousin et al., 2009] were discovered. Here, we describe the case of a 5-year-old girl with an atypical and distal 22q11.2 deletion slightly overlapping with the 3-Mb recurrent deletion and compare the clinical features of our case with those reported in the literature.
Materials and Methods
DNA Methylation Analysis
DNA methylation of KCNQ1OT1 and H19 imprinting control regions of 11p15.5 was analyzed in DNA derived from peripheral blood leukocytes by combined bisulfite restriction analysis, as previously described [Sparago et al., 2007]. Two micrograms of genomic DNA were treated with sodium bisulfite, amplified by polymerase chain reaction (PCR), the PCR product digested with a restriction enzyme containing a CpG dinucleotide in its target sequence and the fragments separated on a polyacrylamide gel.
Microsatellite Analysis
11p15 microsatellite markers TH, D11S4046 and D11S922 and chromosome 7 microsatellite markers D7S1818, D7S484, D7S1830, D7S686, D7S1824 and D7S669 were amplified using FAM-labeled and HEX-labeled primers and detected using an ABI 3100 capillary electrophoresis instrument. Data were analyzed using GeneMapper Software. Primers and PCR conditions for the microsatellite markers were obtained from the NCBI UniSTS Database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unists).
Array Comparative Genomic Hybridization
Single nucleotide polymorphism (SNP) array analysis was performed using the GeneChip Human Mapping 250K NspI Array (Affymetrix, Santa Clara, Calif., USA), which contains a 25-mer oligonucleotide representing a total of 262,264 SNPs with an average resolution of ∼12 kb. Labeling and hybridization were performed in accordance with the manufacturer's protocol while SNP copy number was assessed using the program Genotyping Console Software 4.0 (Affymetrix) by a standard Hidden Markov Model method.
Clinical Case and Results
The girl is the second of 2 children of healthy, nonconsanguineous parents (40-year-old mother and 41-year-old father). She has a healthy brother. Family history was otherwise unremarkable. Pre-natal ultrasound at 18 weeks’ gestation showed congenital heart disease (truncus arteriosus). The fetal movements were normal. The karyotype analysis at 400 band resolution on amniocytes was normal, 46,XX. She was born at 35 weeks’ gestation with Caesarean section, a birth weight of 1,680 g (<3rd centile), length of 41 cm (<3rd centile) and head circumference of 29 cm (<3rd centile). APGAR scores were 9 and 9 at 1 and 5 min, respectively. She was transferred from the neonatal service to the pediatric cardiac surgery at the age of 32 days and underwent surgery for truncus arteriosus type 2. The psychomotor development was delayed both on the mental and motor scales. She was able to sit at 6 months, started walking at 18 months and produced her first words at 18 months. A developmental assessment during the preschool years revealed mild mental retardation. She showed a harmonic cognitive profile with impairments in verbal and performance tasks. Delays in expressive language were worse than those expected by cognitive level, and were not always related to structural anomalies of the palate. Within the verbal domain, receptive language seemed to be the strongest component. Executive function and attention tests showed impairments of several types.
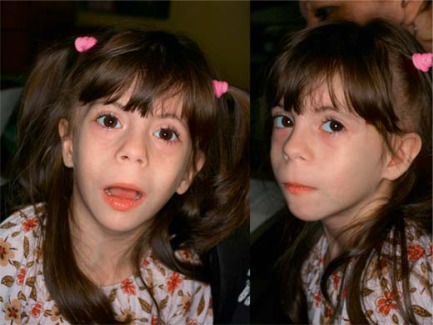
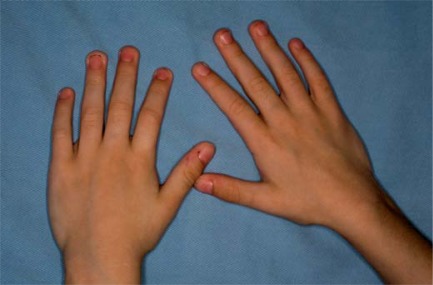
Because of the growth retardation, she was treated with growth hormone, and is also being treated for hypothyroidism. On examination at the age of 2 years and 3 months and prior to the beginning of the treatment with growth hormone, her head circumference was 43 cm (<−2 SD), height 77 cm (<<3rd centile) and weight 7.050 kg (<<3rd centile). At 4 years and 11 months of age, her head circumference was 46.5 cm (<−2 SD), height 99 cm (3rd–10th centile) and weight 12 kg (<<3th centile). She had microcephaly, triangular face, high forehead, frontal bossing, horizontal palpebral fissures, epicanthic folds, broad nose, long and flat philtrum, thin upper lip, downturned mouth, micrognathia, pointed chin, posteriorly rotated ears and long fingers (fig. 1, 2). Bone age at 1 year 11 months corresponded to that of 1 year 6 months. Renal ultrasound was normal.
Fig. 1.
Craniofacial phenotype of the proposita.
Fig. 2.
Hands of the patient. Note the long fingers.
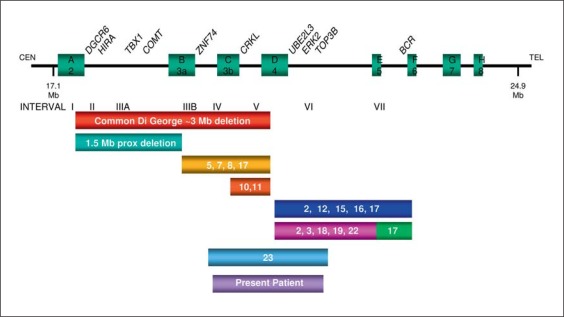
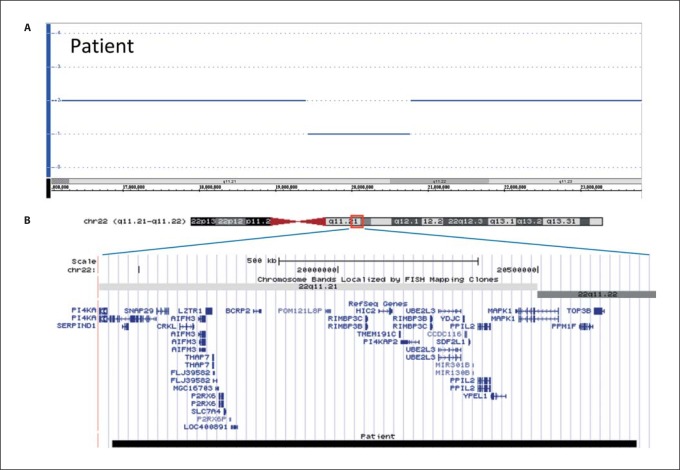
Concerning the molecular investigations performed on this case, microsatellite and combined bisulfite restriction analyses excluded the typical defects of Silver-Russel syndrome (SRS), e.g. chromosome 7 UPD and 11p15.5 copy number or DNA methylation abnormalities, while FISH analysis (probe D22S75) excluded the classical 22q11.2 deletion (data not shown). The DNAs of the proposita and her parents were then analyzed by array comparative genomic hybridization using the GeneChip Human Mapping 250K Nsp I Array. This study disclosed a de novo 1.3-Mb 22q11.2 distal deletion from SNP A-2255904 (19,433,551 bp) to SNP A-2110718 (20,747,829 bp) probes, based on UCSC Genome Browser (http://genome.ucsc.edu/; March 2006 release), in the proposita. The deletion marginally and distally overlapped the common 22q11.2 deletion (fig. 3, 4).
Fig. 3.
Schematic overview of the 22q11.2 deletion region [modified from Rauch et al., 2005]. Low copy repeats are depicted as green squares and labeled with A2 to H8, according to Edelmann et al. [1999], McDermid and Morrow [2002], and Shaikh et al. [2000]. Bars below the map depict deletions found in this study and in reported cases. The exact deletion borders are sometimes only estimated in reported cases.
Fig. 4.
Result of Affymetrix GeneChip Human Mapping 250K NspI Array. A Copy number state of each probe is drawn along chromosome 22 from 16,000,000–23,800,000 bp. The upper panel (blue line) represents the copy number state of the proband. Values of the y-axis indicate the inferred copy number according to probe intensity. B Map of RefSeq genes of the deleted regions (http://genome.ucsc.edu; hg18) in the patient. Black box indicates the deletion of 22q11.21–q11.22 region between 19,433,551 and 20,747,829 bp in the patient.
Discussion
The recent diffusion of genome-wide analytical methods for the identification of copy number variations resulted in an increasing number of reports on rare deletions differing from the more commonly observed ones and those associated with variant phenotypes.
In this report, we describe a patient with atypical and distal 22q11.2 deletion and congenital heart disease (truncus arteriosus type 2) and compare the molecular data and clinical phenotype with those previously reported on this genomic region [Kurahashi et al., 1996, 1997; Rauch et al., 1999, 2005; Saitta et al., 1999; Garcia-Minaur et al., 2002; D'Angelo et al., 2007; Mikhail et al., 2007; Ben-Shachar et al., 2008; Rodningen et al., 2008; Xu et al., 2008; Bruce et al., 2009; Fernandez et al., 2009; Lafay-Cousin et al., 2009; Ogilvie et al., 2009].
Apparently, this is the second case, after the first one described by Ogilvie et al. [2009], with an atypical and distal deletion spanning LCR4, the distal point of the common deleted region and the proximal point of many of the distal deletions. Thus, this present deletion, spanning atypical and distal deletions, represents a new variant deletion, associated with breakpoint sites that do not correspond to known LCRs of 22q11.2. Both deletions are novel deletions and are overlapping some of the reported atypical or distal deletions. In both cases, location of the breakpoints does not coincide with known LCRs, indicating alternative mechanisms in the origin of these deletions, which did not utilize LCRs as recombination substrates.
The most common clinical features associated with the atypical and the distal 22q11.2 deletions are a history of prematurity, pre-natal and post-natal growth retardation as well as developmental delay (tables 1, 2, 3, 4). This clinical aspect was already described by Ben-Shachar et al. [2008] with pre- and post-natal growth restriction found in 83% of his patients. In contrast, pre-natal growth retardation is not a typical feature of the classic 22q11.2 deletion syndrome and post-natal growth retardation has been noted in only 36% of the cases [Ben-Shachar et al., 2008].
Table 1.
Clinical features and review of the literature (interval VI-VII)
| Saitta et al, 1999 | Xu et al, 2008 | Ben-Shachar et al, 2008 | Rodningen et al., 2008 | Bruce et al., 2009 | Rauch et al., 2005 | Lafay-Cousin et al., 2009 | Rauch et al., 1999 | Rauch et al., 2005 | Mikhail et al., 2007 | Ben-Shachar et al., 2008 | Patient | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deleted interval and genes (see fig. 4) | VI IGLV | VI IGLV | VI IGLV Pat 3 | VI IGLV Pat 4 | VI IGLV Pat 5 | VI IGLV Pat 6 | VI Patl | VI Pat 2 | VI | VII GNAZ | VII GNAZ | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII Patl | VI+VII Pat 2 | V+VI | |
| 22qll.2 size deletion Mb | ? | 1.12 | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 | 1.1 | 0.6 | 0.6 | 2.7 | 2 | 2 | 2 | 2.1 | 1.55–1.92 | 2.1 | 2.1 | 1.3 | |
| Parental analysis for deletion | P-NL M-NL | P-NL M-NL | P-NL M-NL | P? M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | Pdel | P? M? | P-NL M-NL | Mdel | P-NL M-NL | P-NL M del | M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | |
| Gender Karyotype | m 46,XY | m 46,XY | m 46,XY | m 46,XY | f 46,XX | m 46,XY | f 46,XX | m 46,XY | f 46,XX | f | m | m | f 46,XX | f | f | f 46,XX | m 46,XY | m 47.XYY | m 46,XY | f 46,XX | llm/9f |
| Age, years | 2 | 11/12 | 6 | 5 | 3 | 4 | 7 | 7 | 12 | 8/12 | 11/12 | 2/12 | 31 | 12 | 7 | 15 | 11 | 3 | 4 | ||
| Premature death | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | 1/20 | |
| Gestational age, weeks | 35 | 37 | 38 | 36 | 35 | 37 | 38 | 36 | 36 | ? | ? | at term | ? | ? | ? | ? | 35 | 32 | 33 | 35 | mean 35 |
| Birth weight, kg centile | ? | 1.800 <3rd | ? | 2.210 3rd-10th | 1.250 <3rd | 1.700 <3rd | 2.100 <3rd | 2.100 3rd | 1.930 <3rd | ? | ? | ? | ? | ? | ? | ? | 2.450 | 1.050 3rd | 1.360 3rd-10th | 1.680 <3rd | 1.784 |
| Postnatal growth retardation | + | ? | + | − | + | + | − | ± | + | ? | ? | + | NA | − | − | − | − | + | + | 9/16 | |
| Weight at presentation (<3rd centile) | ? | ? | «3rd | 60th | «3rd | «3rd | 2.5th-10th | 5th | <3rd | ? | ? | ? | ? | ? | ? | ? | 98th | 97th | <5th | «3rd | 7/11 |
| Height at presentation (<3rd centile) | ? | ? | ? | 60th | «3rd | «3rd | 2.5th-10th | 5th | <3rd | ? | ? | ? | ? | 10th | 10th | ? | 10th | 20th | <5th | 3rd-10th | 5/12 |
| Head circumference (<3rd centile/<-2 SD) | ?MC | ?MC | ? | ? | ? | ? | <2.5th | <2.5th | −2SD (first year) | ? | ? | ? | ? | <3rd | 25th | ? | >95th | ? | ? | <3rd | 7/9 |
| Developmental delay | +sd | + | +sd | − | ± | +sd | +sd | +sd | motor | − | − | +sd | NA | ± | + | ± | + | +sd | +sd | +sd | 16/19 |
| Behavioural problems | ? | NA | − | + | − | − | − | ± | − | NA | ? | NA | NA | − | − | − | + | + | − | − | 4/14 |
| Congenital heart defect | +TAC2 VSD | VSD | +BAV | − | − | − | ? | − | VSD | PAS VSD PDA | − | − | IAAB TACA4 | − | − | VSD | − | ? | +TAC | +TAC2 | 9/18 |
| Hypocalcaemia | + | ? | ? | ? | ? | ? | ? | − | ? | ? | ? | ? | ? | ? | ? | − | ? | ? | ? | − | 1/3 |
| Urogenital anomalies | +HYSP | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | − | ? | ? | ? | − | ? | ? | ? | − | 1/4 |
| Tracheomalacia/laryngomalacia | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/20 |
| Immunodeficiency | − | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | + | − | ? | − | ? | ? | ? | − | 1/5 |
| Recurrent infections | − | NA | − | − | − | − | − | + | − | NA | ? | NA | + | + | + | + | − | − | − | − | 5/16 |
B AV = Bicuspid aortic valve; del = deletion; f = female; HYSP = hypospadias; IAAB = interrupted aortic arch type B; m = male; M = maternal; MC = microcephaly; NA = not applicable; NL = normal; P = paternal; PAS = pulmonary artery stenosis; Pat = patient; PDA = persistent ductus arteriosus Botalli; sd = speech delay; TAC = truncus arteriosus communis; TAC2/A4 = truncus arteriosus communis type 2 or A4; VSD = ventricular septal defect
Table 2.
Clinical features and review of the literature (interval VI-VII)
| Saitta et al., 1999 | Xu et al., 2008 | Ben-Shachar et al., 2008 | Rodningen et al., 2008 | Bruce et al., 2009 | Rauch et al., 2005 | Lafay-Cousin et al., 2009 | Rauch et al., 1999 | Rauch et al., 2005 | Mikhail et al., 2007 | Ben-Shachar et al., 2008 | Patient Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deleted interval and genes (see fig. 4) | VI IGLV | VI IGLV | VI IGLV | VI IGLV | VI IGLV | VI IGLV | VI | VI | VI | VII GNAZ | VII GNAZ | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | VI+VII | V+VI | |
| Pat 3 | Pat 4 | Pat 5 | Pat 6 | Pat l | Pat 2 | Pat 1 | Pat 2 | ||||||||||||||
| 22qll.2 size deletion Mb | ? | 1.12 | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 | 1.1 | 0.6 | 0.6 | 2.7 | 2 | 2 | 2 | 2.1 | 1.55–1.92 | 2.1 | 2.1 | 1.3 | |
| Parental analysis for deletion | P-NL M-NL | P-NL M-NL | P-NL M-NL | P? M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | P-NL M-NL | Pdel | P? M? | P-NL M-NL | Mdel | P-NL M-NL | Mdel | P-NL M-NL | P-NL M-NL | P-NL | P-NL M-NL M-NL | P-NL M-NL | |
| Gender Karyotype | m 46,XY | m 46,XY | m 46,XY | m 46,XY | f 46,XX | m 46,XY | f 46,XX | m 46,XY | f 46,XX | f | m | m | f 46,XX | f | f 46,XX | f | m 46,XY | m | m 47.XYY 46,XY | f 46,XX | llm/9f |
| Age, years | 2 | 11/12 | 6 | 5 | 3 | 4 | 7 | 7 | 12 | 8/12 | 11/12 | 2/12 | 31 | 12 | 7 | 15 | 11 | 3 | 4 | ||
| Craniofacial | |||||||||||||||||||||
| Arched eye brows | − | ? | − | + | + | + | − | − | ? | − | + | + | − | − | − | − | + | + | + | 7/18 | |
| Hypertelorism | + | ? | − | − | − | − | ? | − | − | − | + | + | − | − | + | − | + | − | 4/17 | ||
| Deep-set eyes | + | ? | + | − | + | + | ? | ? | ? | − | + | − | − | − | + | + | − | − | 7/16 | ||
| Iris coloboma | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/20 | |
| Choanal stenosis/atresia | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | 2/20 | |
| Broad nose | + | + | − | − | − | + | ? | ? | ? | − | + | + | ? | − | + | + | + | + | + | + | 11/16 |
| Hypoplastic alae nasi | + | + | + | − | + | + | + | + | + | − | − | − | + | − | − | − | + | − | 10/19 | ||
| Smooth philtrum | + | + | + | + | + | + | + | + | ? | − | − | + | - | − | ± | − | − | + | + | + | 13/19 |
| High arched palate | ? | ? | − | − | − | − | − | − | ? | ? | ? | ? | ? | + | + | ? | − | + | − | 3/12 | |
| Velopharyngeal insufficiency | − | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | − | ? | ? | ? | − | ? | + | ? | − | 1/5 |
| Cleft palate/bifid uvula | +BU | + | − | − | − | − | − | ? | − | − | − | − | − | +nick in the uvula | +nick in the uvula | − | − | +CP | − | − | 5/19 |
| Micrognathia | + | + | ? | ? | − | − | ? | ? | + | − | − | − | + | + | + | − | − | − | + | 7/16 | |
| Down-turned mouth | + | − | ? | ? | − | − | ? | ? | ? | − | + | − | − | − | − | − | + | + | + | 5/15 | |
| Pointed chin | − | − | + | − | + | + | ? | ? | ? | − | − | − | − | − | − | − | + | + | + | 6/17 | |
| Skeletal | ? | + | ? | ? | − | ? | short 5th finger | ? | − | 4/6 | |||||||||||
| 5th finger clinodactyly/short 5th finger | PT SEAC OAVS | SA | ptosis | SA | SA HL | SA | SA | PT HM HL EDC OAVS IATRT | hypoplastic toes | HL PT ST | SA | ||||||||||
BU = Bifid uvula; CP = cleft palate; del = deletion; EDC = epibulbar dermoid cyst; f = female; HL = hearing loss; HM = hemifacial microsomia; IATRT = intracranial atypical teratoid rhabdoid tumor; m = male; M = maternal; NL = normal; OAVS = oculo-auricolo-vertebral spectrum; P = paternal; Pat = patient; PT = preauricular tag; SA = various skeletal anomalies (1 scoliosis, 1 lumbar lordosis, 1 upper limb asymmetry, 1 anomaly in upper sacrum, 1 coxa valga and bowed ulnae, 1 short and broad fingernails and bilateral fifth finger clinodactyly, 2 hip dysplasia, 2 hips rotated in valgus, 1 poorly formed long bones, 1 brachydactyly and contractures, 2 flat feet); SEAC = small external auditory canals; ST = strabism.
Table 3.
Clinical features and review of the literature (interval IV-V)
| Garcia-Minaur et al., 2002 | Rauch et al., 2005 | D'Angelo et al., 2007 | Fernandez et al., 2009 | Kurahashi et al.,1996, 1997 | Ogilvie et al., 2009 | Patient | Total | ||
|---|---|---|---|---|---|---|---|---|---|
| Deleted interval and gene (see fig. 4) | IV+V | IV+V | IV+V | IV+V | IV+V | V | V+VI | V+VI | |
| CRKL | |||||||||
| 22ql 1.2 size deletion, Mh | ? | ? | 1 | 1 | 1.1 | ? | 1.5 | 1.3 | |
| Parental analysis for deletion | P del | P-NL | P? | P-NL | ? | P-NL | P-NL | ||
| M-NL | M-NL | Mdel | M-NL | M-NL | M-NL | ||||
| Gender | m | m | m | f | m | ? | f | f | 4m/3f |
| Karyotype | 46,XX | 46,XX | |||||||
| Age, years | 5/12 | ? | 6 | 4 | 7/12 | ? | 16/12 | 4 | |
| Premature death | − | − | − | − | − | − | − | − | 0/8 |
| Gestational age, weeks | 39 | ? | ? | at term | 37 | ? | 36 | 35 | mean 37 |
| Birth weight, kg | 3.250 | ? | ? | 2.900 | 2.450 | ? | 1.760 | 1.680 | mean 2.408 |
| Centile | 25th | 10th-25th | <3rd | ||||||
| Postnatal growth retardation | − | − | − | − | + | ? | + | + | 3/7 |
| Weight at presentation (<3rd centile) | <3rd | ? | ? | >97th | <3rd | ? | <3rd | «3rd | 4/5 |
| Height at presentation (<3rd centile) | 10th-25th | ? | ? | 25th-50th | <5th | ? | <3rd | 3rd-10th | 3/5 |
| Head circumference (<3rd centile/<-2 SD) | <3rd | ? | ? | 50th-98th | 5th-10th | ? | <3rd | <3rd | 3/5 |
| Developmental delay | − | + | +sd | +sd | + | ? | ? | +sd | 5/6 |
| Behavioural problems | NA | ? | + | + | NA | ? | NA | − | 2/3 |
| Congenital heart defect | ToF | ? | − | − | ASD, VSD | PA, ToF | ASD, VSD | +TAC2 | 4/6 |
| Hypocalcaemia | − | ? | − | − | − | − | ? | − | 0/6 |
| Urogenital Anomalies | ? | ? | − | − | ? | ? | − | 0/3 | |
| Tracheomalacia/laryngomalacia | − | − | − | − | − | ? | ? | − | 0/6 |
| Immunodeficiency | ? | ? | − | − | ? | ? | ? | − | 0/3 |
| Recurrent infections | NA | ? | + | ? | NA | ? | ? | _ | 1/2 |
ASD = Atrial septal defect; del = deletion; f = female; m = male; M = maternal; NA = not applicable; NL = normal; P = paternal; PA = pulmonary atresia; sd = speech delay; TAC2 = truncus arteriosus communis type 2; ToF = tetralogy of Fallot; VSD = ventricular septal defect.
Table 4.
Clinical features and review of the literature (interval IV-V)
| Garcia-Minaur et al., 2002 | Rauch et al., 2005 | D'Angelo et al., 2007 | Fernandez et al., 2009 | Kurahashi et al., 1996,1997 | Ogilvie etal., 2009 | Patient | Total | ||
|---|---|---|---|---|---|---|---|---|---|
| Deleted interval and gene (see fig. 4) | IV+V | IV+V | IV+V | IV+V | IV+V | V | V+VI | V+VI | |
| CRKL | |||||||||
| 22qll.2 size deletion, Mb | ? | ? | 1 | 1 | 1.1 | ? | 1.5 | 1.3 | |
| Parental analysis for deletion | P del | P-NL | p? | P-NL | ? | P-NL | P-NL | ||
| Gender | M-NL | M-NL | Mdel | M-NL | M-NL | M-NL | |||
| Karyotype | m | m | m | f | m | ? | f | f | 4 m |
| 46,XX | 46,XX | 3f | |||||||
| Age, years | 5/12 | ? | 6 | 4 | 7/12 | ? | 16/12 | 4 | |
| Craniofacial | |||||||||
| Arched eye brows | − | _ | _ | _ | + | ? | ? | + | 2/6 |
| Hyperte lor ism | + | − | − | − | − | − | ? | − | 1/7 |
| Deep-set eyes | − | − | − | + | − | ? | ? | − | 1/6 |
| Iris coloboma | − | − | − | − | − | ? | − | − | 0/6 |
| Choanal stenosis/atresia | − | − | − | − | − | ? | − | − | 0/6 |
| Broad nose | − | + | − | + | + | ? | ? | + | 4/6 |
| Hypoplastic alae nasi | − | − | + | − | − | ? | ? | − | 1/6 |
| Smooth philtrum | − | − | − | − | − | ? | ? | + | 1/6 |
| High arched palate | − | ? | + | − | ? | − | 1/4 | ||
| Velopharyngeal insufficiency | − | _ | _ | _ | _ | _ | ? | _ | 0/7 |
| Cleft palate/bifid uvula | − | ? | − | − | ? | − | 0/4 | ||
| Micrognathia | − | + | + | + | + | − | ? | + | 5/7 |
| Down-turned mouth | + | − | − | − | − | ? | ? | + | 2/6 |
| Pointed chin | − | _ | _ | _ | _ | ? | ? | + | 1/6 |
| Skeletal | |||||||||
| 5th finger clinodactyly | ? | ? | ? | ? | ? | ? | + | − | ½ |
| Other clinical features | pes planus | imperforate | |||||||
f = Female; m = male; M = maternal; NL = normal; P = paternal; OB = obesity.
We subdivided the reported cases with atypical and distal 22q11.2 deletions into 2 groups: those associated with a ‘distal deletion’ in the more telomeric region (interval VI–VII, fig. 3 and tables 1, 2) [Rauch et al., 1999, 2005; Saitta et al., 1999; Mikhail et al., 2007; Ben-Shachar et al., 2008; Rodningen et al., 2008; Xu et al., 2008; Bruce et al., 2009; Lafay-Cousin et al., 2009] and those with an ‘atypical deletion’ in the more centromeric region (interval IV–V, fig. 3 and tables 3, 4) [Kurahashi et al., 1996, 1997; Garcia-Minaur et al., 2002; Rauch et al., 2005; D'Angelo et al., 2007; Fernandez et al., 2009]. The case reported by Ogilvie et al. [2009] is very similar to the present patient, with an ‘atypical’ and ‘distal’ 22q11.2 deletion, with the same breakpoints, but it is difficult to do a comparison between the facial phenotypes because these clinical aspects have not been described in detail by Ogilvie et al. We did not consider in our review the 5 patients reported by Jackson et al. [2007], with germline 22q11.2 deletion, because these children had a more distal deletion including the INI1 gene and predisposing to development of malignant rhabdoid tumors.
In each of the 2 groups, the clinical features and morphologic phenotypes are similar, although the sample of the second group is too small to allow statistically significant conclusions. In both groups, we observe the presence of prematurity (13 out of 18 patients), with an average of 35 weeks gestation, and pre-natal growth retardation (8 out of 11 newborns). Post-natal growth retardation is present in 11 out of 20 cases.
The facial phenotype of the atypical and distal 22q11.2 deletion patients is characterized by slight dysmorphic features with arched eyebrows, deep-set eyes, broad nose, hypoplastic alae nasi, smooth philtrum, down-turned mouth, micrognathia and pointed chin. In our experience, the phenotype of the atypical and distal 22q11.2 deletion patients is not too evident and tricky to suspect, but clearly distinguishable from that of the classic 22q11.2 deletion syndrome. Instead, the main differential diagnosis is with SRS. Indeed, we and others [Bruce et al., 2009] had initially suspected SRS in distal 22q11.2 cases. However, the molecular analyses for chromosome 7 UPD and 11p15.5 copy number or methylation defects have not revealed any abnormality. Moreover, patients with SRS do not present with microcephaly, like children with the atypical and the distal 22q11.2 deletions, but, on the contrary, they have a pseudohydrocephalic appearance and normal head circumference.
We also noted that 2 of the reported patients with distal 22q11.2 deletion had clinical features of the oculo-auriculo-vertebral spectrum [Xu et al., 2008; Lafay-Cousin et al., 2009].
Consistent with the observations by Rauch et al. [2005], mental retardation is apparently more serious in patients with deletions encompassing the CRKL gene, including our case, as opposed to cases with deletions in the proximal area, especially those with proximal 1.5-Mb nested 22q11.2 deletion.
Developmental delay is very frequent in patients with atypical and distal 22q11.2 deletions: this is also confirmed in our 2 groups and is found in 20 out of 25 cases, more than in the classic 22q11.2 deletion. Although the number of patients is not high, it is interesting to note that a speech defect is described in 12 out of 25 of our children, but it is difficult to make a comparison between the 2 groups owing to the fact that the numbers involved are so low. In our case, we observed the presence of truncus arteriosus, a heart defect usually seen in the classic 22q11.2 deletion syndrome. A similar cardiac phenotype was found in 3 other cases with 22q11.2 distal deletion [Rauch et al., 1999; Saitta et al., 1999; Ben-Shachar et al., 2008]; moreover, varied congenital heart defects have also been reported in other cases. Since a key role of Tbx1 in heart development has been demonstrated in the mouse, this 22q11.2 gene was also involved in the pathogenesis of the associated human heart defects [Lindsay et al., 1999; Baldini, 2002]. TBX1 is not included in our as well as other distal 22p11.2 deletion cases (fig. 3). However, genes distal to and functionally interacting with TBX1 such as CRKL and ERK2/MAPK1 (fig. 3) have been proposed to be responsible for congenital heart defects [Guris et al., 2001; Newbern et al., 2008]. In particular, ERK2/MAPK1 is included in the 22q11.2 distal deletion interval and its inactivation in the mouse leads to cardiac outflow tract abnormalities similar to those observed in the human patients. Nevertheless, a position effect on TBX1 expression cannot be excluded. Genes possibly involved in the growth retardation phenotype are UBE2L3 whose mutation has been associated with retarded pre-natal growth in the mouse [Harbers et al., 1996], and MIR130B showing cell growth promoting activity via silencing TP53INP1 [Ma et al., 2010].
The deleted interval demonstrated in our case overlaps both the 3-Mb proximal deletion in 22q11.2 and the distal deletion intervals and includes both the CRKL and the ERK2/MAPK1 genes (fig. 3). A similar deletion has been described in another patient, who, like our case, presents preterm delivery, growth retardation, head circumference less than the 3rd centile and congenital heart disease, albeit different from our case, atrial septal defect and ventricular septal defect [Ogilvie et al., 2009]. Moreover, the latter presented imperforate anus, a condition which has never hitherto been reported in the literature.
In conclusion, it is our opinion that pediatric geneticists should take into account this condition when they suspect a SRS and when they have the occasion to examine children with truncus arteriosus.
Acknowledgements
We are thankful to the parents for their cooperation and to Marco Bonazzi for photography work. This work was supported by grants from Telethon-Italia N. GGP07086, GGP11122 (A.R.), MIUR PRIN 2009 (A.R.), Associazione Italiana Ricerca sul Cancro (A.R.) and Ministry of Italian Health grant RC1101GM05 (M.C.).
References
- Baldini A. DiGeorge syndrome: the use of model organisms to dissect complex genetics. Hum Mol Genet. 2002;11:2363–2369. doi: 10.1093/hmg/11.20.2363. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce S, Hannula-Jouppi K, Puoskari M, Fransson I, Simola KO, et al. Submicroscopic genomic alterations in Silver-Russel syndrome and Silver-Russel-like patients. J Med Genet. 2010;47:816–822. doi: 10.1136/jmg.2009.069427. [DOI] [PubMed] [Google Scholar]
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, et al. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo CS, Jehee FS, Koiffmann CP. An inherited atypical 1 Mb 22q11.2 deletion within the DGS/VCFS 3 Mb region in a child with obesity and aggressive behavior. Am J Med Genet A. 2007;143A:1928–1932. doi: 10.1002/ajmg.a.31787. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Nevado J, Santos F, Heine-Suner D, Martinez-Glez V, et al. A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review. BMC Med Genet. 2009;10:48. doi: 10.1186/1471-2350-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Minaur S, Fantes J, Murray RS, Porteous ME, Strain L, et al. A novel atypical 22q11.2 distal deletion in father and son. J Med Genet. 2002;39:e62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- Harbers K, Müller U, Grams A, Li E, Jaenisch R, Franz T. Provirus integration into a gene encoding a ubiquitin-conjugating enzyme results in a placental defect and embryonic lethality. Proc Natl Acad Sci USA. 1996;93:12412–12417. doi: 10.1073/pnas.93.22.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, et al. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet. 2007;122:117–127. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Nakayama T, Osugi Y, Tsuda E, Masuno M, et al. Deletion mapping of 22q11 in CATCH22 syndrome: identification of a second critical region. Am J Hum Genet. 1996;58:1377–1381. [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Tsuda E, Kohama R, Nakayama T, Masuno M, et al. Another critical region for deletion of 22q11: a study of 100 patients. Am J Med Genet. 1997;72:180–185. doi: 10.1002/(sici)1096-8628(19971017)72:2<180::aid-ajmg10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Lafay-Cousin L, Payne E, Strother D, Chernos J, Chan M, Bernier FP. Goldenhar phenotype in a child with distal 22q11.2 deletion and intracranial atypical teratoid rhabdoid tumor. Am J Med Genet A. 2009;149A:2855–2859. doi: 10.1002/ajmg.a.33119. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, et al. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE. Genomic disorders on 22q11. Am J Hum Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail FM, Descartes M, Piotrowski A, Andersson R, Diaz de Stahl T, et al. A previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 encompassing the BRC gene. Am J Med Genet A. 2007;143A:2178–2184. doi: 10.1002/ajmg.a.31882. [DOI] [PubMed] [Google Scholar]
- Newbern J, Zhong J, Wickramasinghe RS, Li X, Wu Y, et al. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci USA. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie CM, Ahn JW, Mann K, Roberts RG, Flinter F. A novel deletion in proximal 22q associated with cardiac septal defects and microcephaly: a case report. Mol Cytogenet. 2009;2:9. doi: 10.1186/1755-8166-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Pfeiffer RA, Leipold G, Singer H, Tigges M, Hofbeck M. A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet. 1999;64:659–667. doi: 10.1086/302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Zink S, Zweier C, Thiel CT, Koch A, et al. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodningen OK, Prescott T, Eriksson AS, Rosby O. 1.4 Mb recurrent 22q11.2 distal deletion syndrome, two new cases expand the phenotype. Eur J Med Genet. 2008;51:646–650. doi: 10.1016/j.ejmg.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Saitta SC, McGrath JM, Mensch H, Shaikh TH, Zackai EH, Emanuel BS. A 22q11.2 deletion that excludes UFD1L and CDC45L in a patient with conotruncal and craniofacial defects. Am J Hum Genet. 1999;65:562–566. doi: 10.1086/302514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Sparago A, Russo S, Cerrato F, Ferraiuolo S, Castorina P, et al. Mechanisms causing imprinting defects in familial Beckwith-Wiedemann syndrome with Wilms' tumour. Hum Mol Genet. 2007;16:254–264. doi: 10.1093/hmg/ddl448. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Xu J, Fan YS, Siu VM. A child with features of Goldenhar syndrome and a novel 1.12 Mb deletion in 22q11.2 by cytogenetics and oligonucleotide array CGH: is this a candidate region for the syndrome? Am J Med Genet A. 2008;146A:1886–1889. doi: 10.1002/ajmg.a.32359. [DOI] [PubMed] [Google Scholar]