Abstract
We describe a patient with a rare interstitial deletion of chromosome 7p21.1–p14.3 detected by array-CGH. The deletion encompassed 74 genes and caused haploinsufficiency (or loss of allele) of 6 genes known to be implicated in different autosomal dominant genetic disorders: TWIST, DFNA5, CYCS, HOXA11, HOXA13, and GARS. The patient had several morphological abnormalities similar to Saethre-Chotzen syndrome (caused by TWIST mutations) including craniosynostosis of the coronal suture and anomalies similar to those seen in hand-foot-uterus syndrome (caused by HOXA13 mutations) including hypospadias. The combined phenotype of Saethre-Chotzen syndrome and hand-foot-uterus syndrome of our patient closely resembles a previously reported case with a cytogenetically visible small deletion spanning 7p21–p14.3. We therefore conclude that microdeletions of 7p spanning the TWIST gene and HOXA gene cluster lead to a clinically recognizable ‘haploinsufficiency syndrome’.
Key Words: array-CGH, Contiguous gene syndrome, Hand-foot-uterus syndrome, HOXA, Saethre-Chotzen syndrome, TWIST
Introduction
The short arm of chromosome 7 harbors different developmental regulator genes, including the TWIST gene on 7p21 and the HOXA gene cluster on 7p15. Haploinsufficiency of these genes either by deletion or mutation leads to Saethre-Chotzen syndrome (SCS) (OMIM#141400) and hand-foot-uterus syndrome (HFU) (OMIM#140000), respectively.
SCS is characterized by craniosynostosis, maxillary hypoplasia, prominent ear crura, and cutaneous syndactyly [Howard et al., 1997; Johnson et al., 1998; Zackai and Stolle, 1998]. Mutations of TWIST are also found in cases of isolated single-suture craniosynostosis (coronal or sagittal) [Seto et al., 2007].
In the past, Robinow-Sorauf syndrome was used to describe patients that in fact had SCS with hallucal duplication and valgus deformity. Similarly, blepharophimosis ptosis epicanthus syndrome (BPES) was used to describe patients who really had SCS with ptosis of the eyelids [Robinow and Sorauf, 1975; Maw et al., 1996; Dollfus et al., 2001].
HFU (or hand-foot-genital syndrome) is characterized by small hands with hypoplastic proximally placed thumbs and feet with small halluces. Postaxial polydactyly of the hands and short or uniphalangeal second toes with absent nails have also been reported. Males have hypospadias, while females have duplication of the uterus and sometimes the cervix, and they may have a septate vagina. Ureteral malformations are common in both sexes. HFU is caused by mutations of the HOXA13 gene on 7p15.2 [Devriendt et al., 1999; Goodman et al., 2000; Goodman, 2002; Frisen et al., 2003]. Guttmacher syndrome is allelic to HFU syndrome with phenotypic overlap (with the exception of postaxial polydactyly of the hands and short or uniphalangeal second toes with absent nails) [Innis et al., 2002]. Larger deletions of the HOXA gene cluster (HOXA1–HOXA13) result in HFU in combination with other malformations such as velopharyngeal insufficiency and a persistent patent ductus botali [Devriendt et al., 1999].
We report a patient with clinical features of both SCS and HFU due to a microdeletion on chromosome 7p21.1–p14.3 detected by array-CGH and encompassing the TWIST and HOXA gene clusters. This suggests that microdeletions spanning TWIST and HOXA represent a recognizable ‘haploinsufficiency syndrome’.
Patient and Methods
Case Report
A Greek boy was born at 38 weeks’ gestation by caesarian section due to a previous one. The patient was the second child of unrelated parents, and there was no relevant family history. The pregnancy was uncomplicated and prenatal tests (serologic, ultrasonographic) were normal. Birth weight was 2,225 g (2nd centile), length 45 cm (3rd centile), and head circumference 34.5 cm (50th centile). At delivery the neonate was cyanotic, hypotonic, and bradycardic, requiring resuscitation. APGAR scores were 1 and 5 at 1 and 9 min, respectively, and the child was admitted to the intensive care unit where he stayed for 2 months. Brain magnetic resonance imaging on the sixth day was within normal limits.
The infant was initially bottle fed, but feeding by nasogastric tube became necessary because of choking and cyanotic episodes. He had laryngomalacia and severe gastroesophageal reflux and failed to thrive.
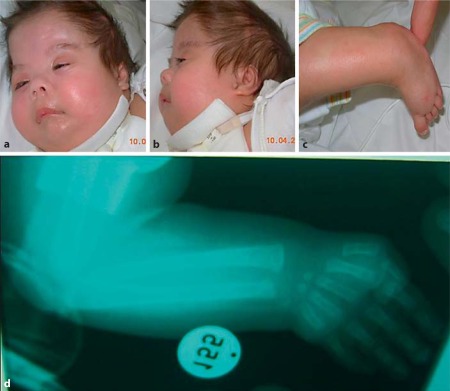
The patient had many dysmorphic features such as asymmetry of the face, overlapping cranial sutures, hypertrichosis of the forehead, and a low nasal bridge with anteverted nostrils. There were small palpebral fissures, hypertelorism with epicanthic folds and ptosis of the eyelids (fig. 1a, b). The ears were low-set with underdevelopment of the helix, hyperplastic anti-helix and anti-tragus, and a prominent intertragic notch. The philtrum was long and smooth. The patient had a high palate and clefting of the soft palate. Additional features included a short neck and widely spaced nipples. There were several limb anomalies: short upper limbs (particularly the forearms and hands), bilateral hypoplastic fifth fingers with clinodactyly, digital webbing between the second, third, and fourth fingers, and abnormal hand creases. Toes II–IV were short, whereas the halluces were relatively long, broad and medially deviated (fig. 1c). There was no radioulnar synostosis (fig. 1d). The scrotum was hypoplastic with left cryptorchidism and first-degree hypospadias.
Fig. 1.
Photographs of the patient. A Asymmetry of the face (particularly on the left side), hypertelorism, epicanthic folds, low nasal bridge, small nose with upturned nostrils, fish-shaped mouth, long and simple philtrum. B Low-set ears, interrupted ear tragus, hyperplastic anti-helix and anti-tragus and tracheostomy. C Brachydactyly of toes II–IV with a long and broad hallux. D X-ray of the lower arm and hand showed brachyphalangy, cone-shaped epiphyses, hypoplastic middle-phalanx of the 5th finger with clinodactyly. No radioulnar synostosis was noted.
Cardiac ultrasound showed an open foramen ovale and a mild aortic insufficiency. He had multiple episodes of prolonged apnea, and the sleep EEG was abnormal with predominance of delta waves on the left side. The hematological and biochemical tests were normal, and he never developed thrombocytopenia.
At 5 months, surgery was performed for severe unilateral craniostenosis of the left coronal suture (left metopic plagiocephaly). The post-surgical period was complicated by cardiorespiratory arrest, and neurological damage was suspected. He also developed moderate renal insufficiency. He required a tracheostomy and g-tube placement. At 18 months, he had severe neurological deficits with spastic quadriplegia and persistent opisthotonus responding only to noxious stimuli; it was therefore difficult to correctly estimate his hearing. His height, weight and head circumference were below the third centile. At the age of 2, the patient died after developing a severe respiratory infection. The parents refused an autopsy.
Molecular Analysis
Array comparative genomic hybridization analysis (array-CGH) consisted of hybridization of the patient and control DNA to the Agilent Human Genome CGH 4×44k microarrays (G4426B, AMADID#014950) (Agilent Technologies, Santa Clara, Calif., USA) according to the manufacturer's protocol with slight modifications. In brief, 150 ng of genomic DNA was labeled with Cy5 (patient) or Cy3 (control). After clean up, labeled fragments were pooled and 5 μl Cot-1 DNA (1 mg/ml), 10× blocking agent, and 2× hybridization buffer were added. This mixture was hybridized on the microarrays for 24 h at 65°C. After washing, the slides were scanned using an Agilent Microarray Scanner, quantified with Feature Extraction software 9.1 (Agilent Technologies), and analyzed with array-CGH base (http://medgen.ugent.be/arraycghbase/) [Menten et al., 2005].
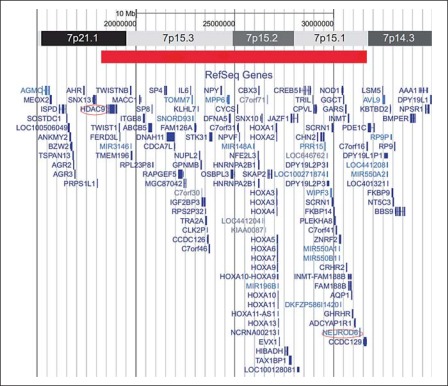
Array-CGH analysis (Agilent 44 K, resolution of ∼400 kb) revealed a microdeletion of ∼13 Mb on chromosome 7p21.1–p14.3 (fig. 2). Within the deleted region 74 genes are located, with 6 of them responsible for a variety of autosomal dominant diseases: TWIST, DFNA5, CYCS, HOXA11, HOXA13, and GARS. Array-CGH analysis of the parents and siblings of the patient were normal, indicating that the microdeletion was a de novo event in this patient and most likely caused his abnormalities. Because of the size of the microdeletion, confirmation by FISH analysis was unnecessary.
Fig. 2.
The results of the array-CGH, revealing a deletion of 13 Mb on chromosome 7p21.1–p14.3 (red bar) and showing the 2 breakpoints (circled) and the genes located within the deleted region.
Discussion
We describe a patient with a ∼13 Mb deletion on chromosome 7p21.1–p14.3 that was detected by array-CGH. Loss-of-function mutations in the TWIST gene, which encodes a basic helix-loop-helix transcription factor, are responsible for the SCS [Howard et al., 1997]. Our patient had the typical phenotypic features of SCS (craniostenosis, clefting of the soft palate, and limb anomalies), and his ears resembled those seen in SCS. He had an interrupted ear tragi and hyperplastic antihelices and antitragi but no prominent helical crura. He also had ptosis of the eyelids and short palpebral fissures (compatible with BPES syndrome) and the additional features of broad halluces medialy deviated which meet the clinical criteria for Robinow-Sorauf syndrome [Robinow and Sorauf, 1975; Chotai et al., 1994; Maw et al., 1996; Dollfus et al., 2001; Cox et al., 2002].
There are 4 HOX gene clusters (HOXA, HOXB, HOXC, and HOXD) which are the basic developmental regulators that define positional information for the embryo along the anterior-posterior axis of the body [Davis et al., 1995; Kmita et al., 2005; Zakany et al., 2007]. HOXA13 (OMIM #142959) mutations lead to the dominantly inherited hand-foot-uterus (OMIM #140000) and Guttmacher syndromes (OMIM #176305) [Davis et al., 1995; Goodman et al., 2000; Thompson et al., 2001; Kosaki et al., 2005]. Typical limb defects in these disorders include shortening of the carpals and tarsals and fifth finger clinodactyly which were observed in our case. However, the first-digit hypoplasia (typical of HFU) was absent which might have been due to the concurrent TWIST gene haploinsufficiency that is typically associated with broadening of the first digits. Our male proband had the typical genital anomalies found in patients with a HOXA13 mutation [Goodman et al., 2000].
The DFNA5 gene (OMIM #608798) is implicated in one of the autosomal dominant types of non-syndromic deafness (OMIM #600994). All 4 identified mutations result in the skipping of exon 8 of the DFNA5 gene, raising the hypothesis that there is a specific gain-of-function effect [Van Laer et al., 1998, 2004; Cheng et al., 2007]. Two patients (cases 1 and 3) described by Duno et al. [2004] had deletions of the complete DFNA5 gene without hearing loss. Unfortunately, we could not make a positive evaluation of our patient.
A single HOXA11 (OMIM #142958) truncating mutation has been reported to cause a specific syndrome with skeletal defects (radioulnar synostosis) and amegakaryocytic thrombocytopenia (OMIM #605432) [Thompson and Nguyen, 2000; Thompson et al., 2001]. Double mice mutants for Hoxa11 and Hoxd11 have a very small radius and ulna and severe kidney defects [Davis et al., 1995]. Our patient did not have thrombocytopenia (220–550 × 103/μl) or radioulnar synostosis.
A single missense mutation in the CYCS gene (OMIM #123970) encoding cytochrome c has been shown to cause autosomal dominant thrombocytopenia type 4 (OMIM #612004) [Morison et al., 2008], a syndrome not yet linked with haploinsufficiency which suggests it is a gain-of-function mutation.
Mutations of the GARS gene (OMIM #600287) are associated with Charcot-Marie-Tooth neuropathy type 2D (OMIM #601472) and distal spinal muscular atrophy V (OMIM #600794). Until now, only GARS missense mutations have been found [Antonellis et al., 2003], and it is unknown whether GARS haploinsufficiency leads to neurological deficits. Our patient did not have any neurological signs of Charcot-Marie-Tooth neuropathy or distal spinal muscular atrophy.
Several patients with deletions of chromosome 7p have been described in the literature [Chotai et al., 1994; Johnson et al., 1998; Cai et al., 1999; Cox et al., 2002; Hoover-Fong et al., 2003]. The boundaries of the deletions were not delineated in great detail because of the limited resolution of conventional cytogenetics, but the deletion breakpoints appeared distinct. The distal breakpoint of the deletion in the HDAC9 gene of our patient was very close to the distal inversion breakpoint of a patient described by Duno et al. [2004] (case 2) and a translocation breakpoint reported by David et al. [2003]. Also, the distal breakpoint of a deletion described by Johnson et al. [1998] was located close to the HDAC9 gene, which suggests that this region is a hot spot for recombination.
Only a single microdeletion encompassing both the TWIST and HOXA clusters has been described in the literature. Kosaki et al. [2005] described a patient with left coronal craniosynostosis, maxillary hypoplasia, prominent ear crura, rectoperineal fistula, anal atresia, patent ductus arteriosus, hypoplastic fifth finger, and psychomotor delay. Although no array-CGH was performed, fluorescent in situ hybridization analysis showed a deletion of the TWIST gene close to the HOXA gene cluster. The facial anomalies could be mainly attributed to haploinsufficiency of the TWIST gene, whereas the other anomalies were more difficult to assign to specific gene deletions. Chotai et al. [1994] reported a panel of 6 7p-deletion cases, 3 with craniosynostosis. There were 5 de novo deletions and 1 resulting from the unbalanced product of a paternal balanced insertion. The putative proximal locus at 7p13–p14 does not appear to be allelic with Greig cephalopolysyndactyly syndrome [Chotai et al., 1994].
In conclusion, microdeletion of 7p21 spanning the TWIST and the HOXA clusters leads to combined phenotype of Saethre-Chotzen syndrome and hand-foot-uterus syndrome. The phenotype of the microdeletion 7p21.1–p14.3 could not be explained only by variable expressivity of haploinsufficiency of other genes, but incomplete penetrance may play a critical role. Therefore, array-CGH testing is necessary to exclude a 7p21–p14 microdeletion in patients with craniosynostosis, limb anomalies, and genital hypoplasia.
References
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Yu P, Tagle DA, Xia J. Duplication of 7p21.2→pter due to maternal 7p;21q translocation: implications for critical segment assignment in the 7p duplication syndrome. Am J Med Genet. 1999;86:305–311. [PubMed] [Google Scholar]
- Cheng J, Han DY, Dai P, Sun HJ, Tao R, et al. A novel DFNA5 mutation, IVS8+4 A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a Chinese family. Clin Genet. 2007;72:471–477. doi: 10.1111/j.1399-0004.2007.00889.x. [DOI] [PubMed] [Google Scholar]
- Chotai KA, Brueton LA, van Herwerden L, Garrett C, Hinkel GK, et al. Six cases of 7p deletion: clinical, cytogenetic, and molecular studies. Am J Med Genet. 1994;51:270–276. doi: 10.1002/ajmg.1320510320. [DOI] [PubMed] [Google Scholar]
- Cox H, Stewart H, Hall L, Donnai D. Phenotypic spectrum of interstitial 7p duplication in mosaic and non-mosaic forms. Am J Med Genet. 2002;109:306–310. doi: 10.1002/ajmg.10368. [DOI] [PubMed] [Google Scholar]
- David D, Cardoso J, Marques B, Marques R, Silva ED, et al. Molecular characterization of a familial translocation implicates disruption of HDAC9 and possible position effect on TGFbeta2 in the pathogenesis of Peters' anomaly. Genomics. 2003;81:489–503. doi: 10.1016/s0888-7543(03)00046-6. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Devriendt K, Jaeken J, Matthijs G, Van Esch H, Debeer P, et al. Haploinsufficiency of the HOXA gene cluster, in a patient with hand-foot-genital syndrome, velopharyngeal insufficiency, and persistent patent ductus botalli. Am J Hum Genet. 1999;65:249–251. doi: 10.1086/302452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollfus H, Kumaramanickavel G, Biswas P, Stoetzel C, Quillet R, et al. Identification of a new TWIST mutation (7p21) with variable eyelid manifestations supports locus homogeneity of BPES at 3q22. J Med Genet. 2001;38:470–472. doi: 10.1136/jmg.38.7.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duno M, Hove H, Kirchhoff M, Devriendt K, Schwartz M. Mapping genomic deletions down to the base: a quantitative copy number scanning approach used to characterize and clone the breakpoints of a recurrent 7p14.2p15.3 deletion. Hum Genet. 2004;115:459–467. doi: 10.1007/s00439-004-1174-y. [DOI] [PubMed] [Google Scholar]
- Frisen L, Lagerstedt K, Tapper-Persson M, Kockum I, Nordenskjold A. A novel duplication in the HOXA13 gene in a family with atypical hand-foot-genital syndrome. J Med Genet. 2003;40:e49. doi: 10.1136/jmg.40.4.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR. Limb malformations and the human HOX genes. Am J Med Genet. 2002;112:256–265. doi: 10.1002/ajmg.10776. [DOI] [PubMed] [Google Scholar]
- Goodman FR, Bacchelli C, Brady AF, Brueton LA, Fryns JP, et al. Novel HOXA13 mutations and the phenotypic spectrum of hand-foot-genital syndrome. Am J Hum Genet. 2000;67:197–202. doi: 10.1086/302961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover-Fong JE, Cai J, Cargile CB, Thomas GH, Patel A, et al. Facial dysgenesis: a novel facial syndrome with chromosome 7 deletion p15.1–21.1. Am J Med Genet A. 2003;117A:47–56. doi: 10.1002/ajmg.a.10046. [DOI] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Innis JW, Goodman FR, Bacchelli C, Williams TM, Mortlock DP, et al. A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Hum Mutat. 2002;19:573–574. doi: 10.1002/humu.9036. [DOI] [PubMed] [Google Scholar]
- Johnson D, Horsley SW, Moloney DM, Oldridge M, Twigg SR, et al. A comprehensive screen for TWIST mutations in patients with craniosynostosis identifies a new microdeletion syndrome of chromosome band 7p21.1. Am J Hum Genet. 1998;63:1282–1293. doi: 10.1086/302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zakany J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Kosaki R, Higuchi M, Mitsui N, Matsushima K, Ohashi H, Kosaki K. Deletion involving the TWIST locus and the HOXA cluster: a contiguous gene syndrome on 7p? Congenit Anom (Kyoto) 2005;45:35–38. doi: 10.1111/j.1741-4520.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Maw M, Kar B, Biswas J, Biswas P, Nancarrow D, et al. Linkage of blepharophimosis syndrome in a large Indian pedigree to chromosome 7p. Hum Mol Genet. 1996;5:2049–2054. doi: 10.1093/hmg/5.12.2049. [DOI] [PubMed] [Google Scholar]
- Menten B, Pattyn F, De Preter K, Robbrecht P, Michels E, et al. ArrayCGH base: an analysis platform for comparative genomic hybridization microarrays. BMC Bioinformatics. 2005;6:124. doi: 10.1186/1471-2105-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Cramer Borde EM, Cheesman EJ, Cheong PL, Holyoake AJ, et al. A mutation of human cytochrome c enhances the intrinsic apoptotic pathway but causes only thrombocytopenia. Nat Genet. 2008;40:387–389. doi: 10.1038/ng.103. [DOI] [PubMed] [Google Scholar]
- Robinow M, Sorauf TJ. Acrocephalopolysyndactyly, type Noack, in a large kindred. Birth Defects Orig Artic Ser. 1975;11:99–106. [PubMed] [Google Scholar]
- Seto ML, Hing AV, Chang J, Hu M, Kapp-Simon KA, et al. Isolated sagittal and coronal craniosynostosis associated with TWIST box mutations. Am J Med Genet A. 2007;143:678–686. doi: 10.1002/ajmg.a.31630. [DOI] [PubMed] [Google Scholar]
- Thompson AA, Nguyen LT. Amegakaryocytic thrombocytopenia and radio-ulnar synostosis are associated with HOXA11 mutation. Nat Genet. 2000;26:397–398. doi: 10.1038/82511. [DOI] [PubMed] [Google Scholar]
- Thompson AA, Woodruff K, Feig SA, Nguyen LT, Schanen NC. Congenital thrombocytopenia and radio-ulnar synostosis: a new familial syndrome. B J Haematol. 2001;113:866–870. doi: 10.1046/j.1365-2141.2001.02834.x. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Huizing EH, Verstreken M, van Zuijlen D, Wauters JG, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–197. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Vrijens K, Thys S, Van Tendeloo VF, Smith RJ, et al. DFNA5: hearing impairment exon instead of hearing impairment gene? J Med Genet. 2004;41:401–406. doi: 10.1136/jmg.2003.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackai EH, Stolle CA. A new twist: some patients with Saethre-Chotzen syndrome have a microdeletion syndrome. Am J Hum Genet. 1998;63:1277–1281. doi: 10.1086/302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]