Abstract
As part of our ongoing investigation of filamentous fungi for anticancer leads, an active fungal extract was identified from the Mycosynthetix library (MSX 55526; from the Order Sordariales). Bioactivity-directed fractionation yielded the known ergosterol peroxide (2) and 5α,8α-epidioxyergosta-6,9(11),22-trien-3β-ol(3), and a new benzoate trimer, termed thielavin B methyl ester (1). The structure elucidation of 1 was facilitated by the use of HRMS coupled to an APPI (atmospheric pressure photoionization) source. Compound 1 proved to be moderately active against a panel of three cancer cell lines.
Keywords: Fungi, Cytotoxicity, Benzoate, Thielavin, APPI
The pursuit of drug leads from fungi has yielded thousands of interesting compounds, many of which have become important drugs. Penicillin and lovastatin may be the most well-known examples from the 20th century,1 but even in the last 12 months, the U.S. FDA approved a new treatment for multiple sclerosis, fingolimod (trade name, Gilenya), which was optimized via medicinal chemistry from the fungal secondary metabolite, myriocin.2 Hence, fungi have, and continue to be, a reservoir for drug discovery.
Through the ongoing exploration of extracts of filamentous fungi for anticancer drug leads,3 the crude 1:1 CHCl3/MeOH extract of fungus MSX 555264 displayed promising cytotoxicity (i.e., >97% inhibition of the growth of H460 cells when tested at 20 µlg/mL). This extract was partitioned with 3:3:4 MeOH/CH3CN/hexanes, and the residue from the bottom layer (1.14 g) was then fractionated by flash silica-gel chromatography. The material eluting at ~2–3% MeOH in CHCl3 was active (>95% inhibition of H460 cell growth when tested at 2 µg/mL). Preparative scale RP-HPLC (gradient of CH3CN in H2O on C18) was utilized to isolate all compounds of interest in >95% purity, as evaluated by UPLC-ELSD.5 Compounds 2 and 3 were known compounds, ergosterol peroxide and 5α,8α-epidioxyergosta-6,9(11),22-trien-3β-ol, respectively; their NMR data were in good agreement with the literature.6,7 Compound 1 (1.1 mg), however, was a new benzoate trimer (Fig. 1), the structure elucidation of which was facilitated greatly by the use of high resolution mass spectrometery (HRMS) with an atmospheric pressure photoionization (APPI) source.8
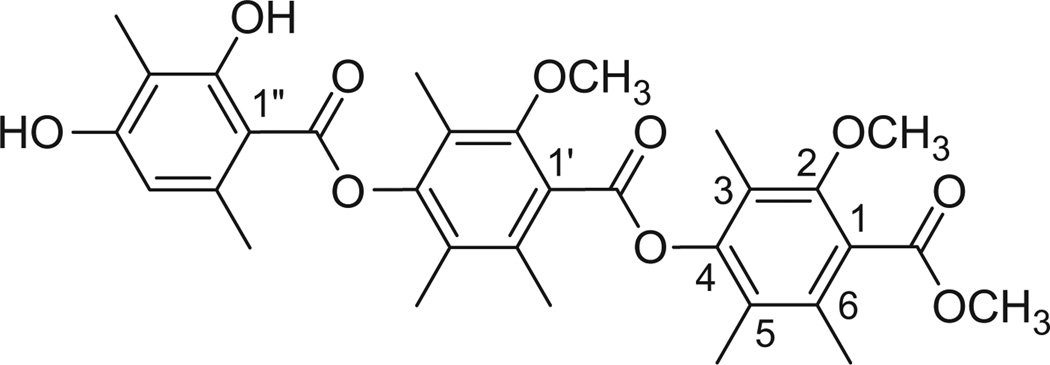
Figure 1.
Structure of Thielavin B methyl ester (1).
The HRMS of compound 1 showed [M+H]+ at 581.2378, corresponding to a molecular formula of C32H36O10 (calcd 581.2387 for [M+H]+), corresponding to an index of hydrogen deficiency of 15 and indicative of a high degree of unsaturation. The 1H NMR spectrum of 1 was somewhat atypical (see Supplementary data), particularly for a natural product of m.w. >500 amu, as it showed all singlets; eight singlets were consistent with aromatic methyl groups, three singlets were consistent with methoxy groups, a one-proton singlet that was consistent with an aromatic proton, and a one-proton singlet that was consistent with an intramolecular hydrogen-bonded phenol.
A search of the formula in the Dictionary of Natural Products9 returned ten hits, seven of which were compounds of fungal origin. Two of the hits, thielavin C10 and Antibiotic PS-990,11 had spectroscopic data that were similar, but not entirely consistent, with compound 1. Specifically, both thielavin C and PS-990 had nine aromatic methyl groups, two methoxy groups, and no aromatic proton. The UV spectrum of 1 from the diode-array detector of the HPLC showed a maximum at 276 nm and a shoulder at approximately 310 nm, and of the two known compounds, these data more closely matched those of thielavin C.10
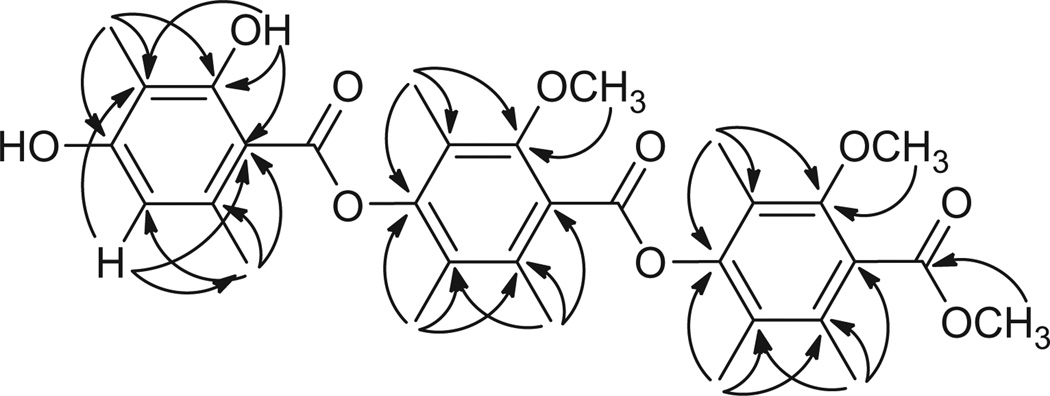
From the HMBC spectrum (Fig. 2), it was apparent that the compound was indeed closely related to thielavin C, containing three benzoates linked by two p-phenoxy groups. The key differences between thielavin C and 1 were that one of the rings in 1 was not fully substituted, and compound 1 terminated at the carboxyl end with a methyl ester instead of a carboxylic acid. HMBC data further revealed that two of the benzoate rings had identical substitution patterns (2-methoxy-4-phenoxy-3,5,6-trimethyl), while the substitution pattern for the third benzoate was 2,4-diphenoxy-3,6-dimethyl. The question that could not be answered by HMBC was the sequence of the three benzoates.
Figure 2.
HMBC correlations of Thielavin B methyl ester (1).
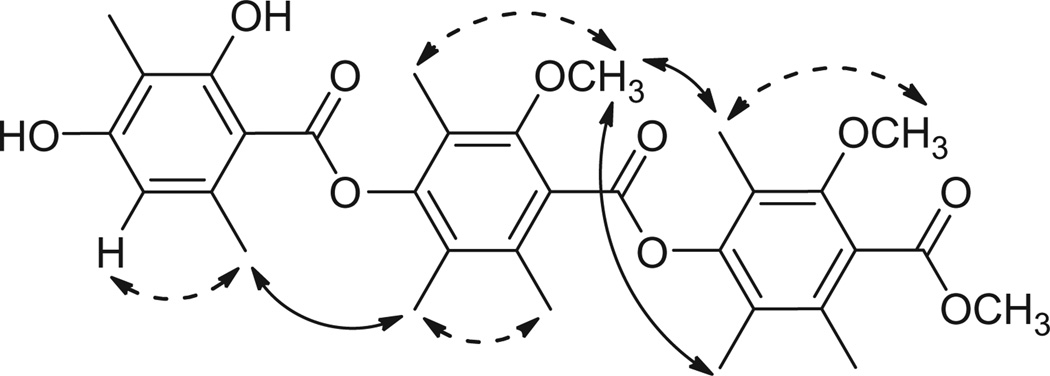
Two methods were used to elucidate the sequence of the benzoate rings. The first was NOESY spectroscopy (see Supplementary data for spectrum). The NOESY spectrum showed both intra-ring correlations (Fig. 3, dashed lines), and inter-ring correlations (Fig. 3, solid lines), which proved that the methyl ester was part of one of the hexa-substituted rings, while the sequence terminated at the phenolic end with the penta-substituted ring.
Figure 3.
NOESY correlations of Thielavin B methyl ester (1).
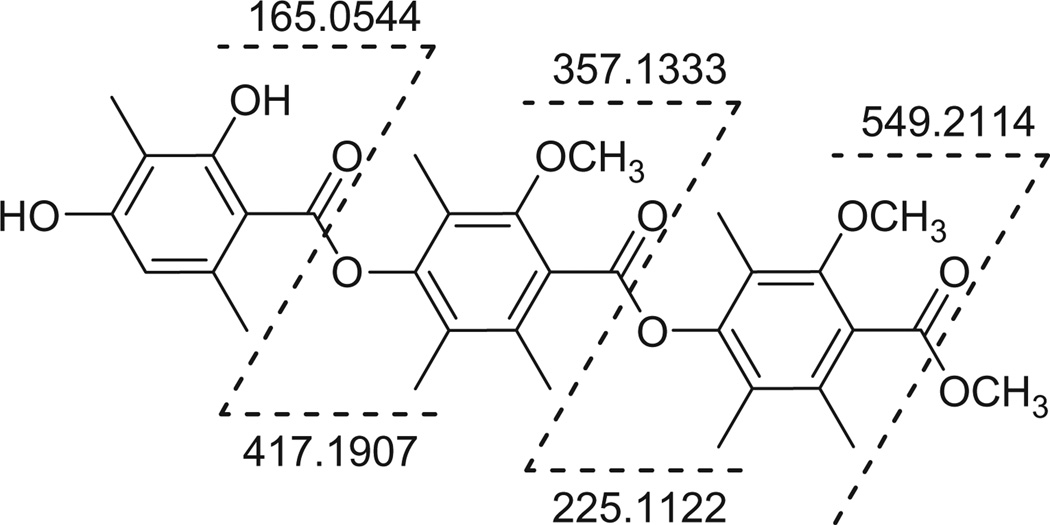
The second method, which confirmed the ring sequence, was HRMS coupled to an APPI source (Fig. 4, see Supplementary data for spectrum). APPI is an ionization technique that utilizes UV photons to generate the quasi-molecular ion (i.e., [M+H]+) through charge transfer reactions between the solvent and the analyte of interest.12 APPI has been demonstrated to ionize compounds with a high degree of unsaturation preferentially, as was observed with 1.13 The use of APPI is routine in the analysis of crude oil,14–18 whereas, in the analysis of natural products, APPI has been used largely for the measurement or identification of targeted compounds, such as aflatoxins,19 pesticide residues,20 and naphthalenes.21 Thus, to the best of our knowledge, this represents the first example of its utility in the de novo structure elucidation of bioactive secondary metabolites, particularly when coupled to HRMS. In positive mode, in-source fragmentation occurred at the ester bonds (Fig. 4), revealing all of the possible fragments and confirming the sequence as deduced by NOESY. These data indicated compound 1 was the methyl ester of thielavin B, and hence was ascribed the trivial name thielavin B methyl ester.
Figure 4.
APPI-HRMS in-source fragmentation of 1.
Compound 1 was assayed against the MCF-7 (human breast carcinoma), H460 (human non-small cell lung carcinoma), and SF268 (human astrocytoma) cell lines exactly as described previously.22 The IC50 values for 1 were 7.3, 6.6, and 8.1 µM, respectively. In our hands, activity of this order of magnitude was considered ‘moderate’, since positive controls (i.e., camptothecin) have IC50 values in the sub-µM range.22
In conclusion, a new congener (1) in the thielavin series was isolated via bioactivity-directed purification. Compound 1 was shown by a combination of 2D NMR and APPI-HRMS to be the methyl ester of thielavin B. APPI-HRMS was demonstrated to be an effective method for elucidating the sequence of benzoate rings for this class of compounds, and the MS conclusions were confirmed by 2D NOESY spectroscopy. Thielavin B methyl ester (1) exhibited moderate cytotoxicity. Other members of this class of tribenzoates have shown inhibition of prostaglandin biosynthesis10,23 and inhibition of phospholipase A2,24 both implicated in inflammation. The thielavins have also shown inhibitory activity against glucose-6-phosphatase, a possible target for anti-diabetic drug leads.25 Antibiotic PS-990 was shown to be a cyclic nucleotide phosphodiesterase inhibitor, a potential target for treatment of neurodegenerative conditions.11
Supplementary Material
Acknowledgments
This research was supported by P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. The Golden LEAF Foundation (Rocky Mount, NC) provided partial support to D.J.K. Mycology technical support was provided by Blaise Darveaux and Maurica Lawrence.
Footnotes
Supplementary data
Supplementary data (1D and 2D NMR spectra and data, APPIHRMS spectrum) associated with this article can be found, in the online version, at doi:10.1016/j.tetlet.2011.08.125.
References and notes
- 1.Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. In: Bioactive Compounds from Natural Sources. 2nd ed. Tringali C, editor. London: Natural Products as Load Compounds in Drog Discovery, Taylor & Francis; 2011. pp. 37–63. [Google Scholar]
- 2.Strader CR, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:900–907. doi: 10.1021/np2000528. [DOI] [PubMed] [Google Scholar]
- 3.Kinghorn AD, Carache de Blanco EJ, Chai HB, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mycosynthetix fungal strain 55526 was isolated in May, 1991 by Dr. Barry Katz of MYCOsearch from leaf litter collected in the North Carolina Smokey Mountains. DNA analysis was performed by MIDI Labs, Inc. (Newark, DE), and the D2 variable region of the Large Subunit (LSU) rRNA was sequenced and compared to their database; the closest match could only determine that this fungus was of the Order Sordariales; these data were deposited in Genbank (accession No. JN257135). The culture was stored on a malt extract slant and was transferred periodically. A fresh culture was grown on a similar slant, and a piece was transferred to a medium containing 2% soy peptone, 2% dextrose, and 1% yeast extract (YESD media). Following incubation (7 days) at 22 °C with agitation, the culture was used to inoculate 50 mL of a rice medium, prepared using rice to which was added a vitamin solution and twice the volume of rice with H2O, in a 250 mL Erlenmeyer .ask. This was incubated at 22 °C until the culture showed good growth (approximately 14 days)
- 5.The 1:1 CHCl3/MeOH extract of MSX 55526 was partitioned in a 3:3:4 mixture of MeOH/CH3CN/hexanes. After drying, the residue from the bottom layer (1.14 g) was then eluted at 40 mL/min on a RediSep Rf silica gel column (40 g) using a Teledyne ISCO CombiFlash Rf. The solvent gradient was 100% hexanes to 100% CHCl3 over 10 column volumes (CV), 100% CHCl3 for 7 CV, then from 100:0 to 80:20 CHCl3/MeOH over 24 CV. The material eluting from ~3–4% MeOH was active in the cytotoxicity assay and pooled (122.6 mg). This fraction was subjected to preparative HPLC (Phenomenex Gemini C18, 250 × 21.2 mm,5µm, 16 mL/min, 20–100% CH3CN in H2O over 20 min, hold 100% CH3CN for 15min). Compound 1 eluted at 21.4 min (2.55 mg), 3 eluted at 28.8 min (2.54 mg), and 2 eluted at 31.3 min. (11.93 mg). Compound 1 was re-purified by semi-preparative HPLC (YMC-Pack ODS-A, 250 × 10 mm, 5 µm, 4 mL/min, 30–100% CH3CN in H2O over 30 min). Compound 1 eluted at 28.7 min and was isolated at >95% purity by UPLC-ELSD (1.08 mg)
- 6.Gunatilaka AAL, Gopichand Y, Schmitz FJ, Djerassi CJ. Org. Chem. 1981;46:3860–3866. [Google Scholar]
- 7.Della Greca M, Mangoni L, Mollinaro A, Monaco P, Previtera L. Gazz. Chim. Ital. 1990;120:391–392. [Google Scholar]
- 8.High resolution mass analysis was conducted on a LTQ Orbitrap XL mass spectrometer (ThermoFisher, Breman, Germany) equipped with an IonMax atmospheric pressure photoionization source. Samples were directly injected into the mass spectrometer via syringe pump operated at a .ow rate of 15 µL/ min. Source conditions were set at 250 ଌ for the nebulizer heater, 9.0 V for the capillary voltage, the capillary temperature was 250 ଌC, and the tube lens voltage was 130.0V. Nitrogen was utilized for both the sheath and auxiliary gases and set at 15 and 5 arb, respectively
- 9.Anonymous Dictionary of Natural Products. http://www.chemnetbase.com.
- 10.Kitahara N, Haruyama H, Hata T, Takahashi SJ. Antibiot. 1983;36:599–600. doi: 10.7164/antibiotics.36.599. [DOI] [PubMed] [Google Scholar]
- 11.Toki S, Ando K, Yoshida M, Matsuda YJ. Antibiot. 1994;47:1175–1181. doi: 10.7164/antibiotics.47.1175. [DOI] [PubMed] [Google Scholar]
- 12.Robb DB, Covey TR, Bruins AP. Anal. Chem. 2000;72:3653–3659. doi: 10.1021/ac0001636. [DOI] [PubMed] [Google Scholar]
- 13.Kauppila TJ, Kuuranne T, Meurer EC, Eberlin MN, Kotiaho T, Kostiainen R. Anal. Chem. 2002;74:5470–5479. doi: 10.1021/ac025659x. [DOI] [PubMed] [Google Scholar]
- 14.Marshall AG, Rodgers RP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18090–18095. doi: 10.1073/pnas.0805069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell JM, Hendrickson CL, Rodgers RP, Marshall AG. Anal. Chem. 2006;78:5906–5912. doi: 10.1021/ac060754h. [DOI] [PubMed] [Google Scholar]
- 16.Purcell JM, Hendrickson CL, Rodgers RP, Marshall AG. J. Am. Soc. Mass. Spectrom. 2007;18:1682–1689. doi: 10.1016/j.jasms.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Purcell JM, Juyal P, Kim DG, Rodgers RP, Hendrickson CL, Marshall AG. Energy Fuels. 2007;21:2869–2874. [Google Scholar]
- 18.Purcell JM, Rodgers RP, Hendrickson CL, Marshall AG. J.Am. Soc. Mass. Spectrom. 2007;18:1265–1273. doi: 10.1016/j.jasms.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Takino M, Tanaka T, Yamaguchi K, Nakahara T. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2004;21:76–84. doi: 10.1080/02652030310001632538. [DOI] [PubMed] [Google Scholar]
- 20.Takino M, Yamaguchi K, Nakahara TJ. Agric. Food Chem. 2004;52:727–735. doi: 10.1021/jf0343377. [DOI] [PubMed] [Google Scholar]
- 21.Rauha JP, Vuorela H, Kostiainen RJ. Mass Spectrom. 2001;36:1269–1280. doi: 10.1002/jms.231. [DOI] [PubMed] [Google Scholar]
- 22.Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitahara N, Endo A, Furuya K, Takahashi SJ. Antibiot. 1981;34:1562–1568. doi: 10.7164/antibiotics.34.1562. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto K, Tanaka K, Matsutani S, Sakazaki R, Hinoo H, Uotani N, Tanimoto T, Kawamura Y, Nakamoto S, Yoshida TJ. Antibiot. 1995;48:106–112. doi: 10.7164/antibiotics.48.106. [DOI] [PubMed] [Google Scholar]
- 25.Sakemi S, Hirai H, Ichiba T, Inagaki T, Kato Y, Kojima N, Nishida H, Parker JC, Saito T, Tonai-Kachi H, VanVolkenburg MA, Yoshikawa N, Kojima YJ. Antibiot. 2002;55:941–951. doi: 10.7164/antibiotics.55.941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.