Abstract
Dystrobrevin is a component of the dystrophin-associated protein complex and has been shown to interact directly with dystrophin, α1-syntrophin, and the sarcoglycan complex. The precise role of α-dystrobrevin in skeletal muscle has not yet been determined. To study α-dystrobrevin's function in skeletal muscle, we used the yeast two-hybrid approach to look for interacting proteins. Three overlapping clones were identified that encoded an intermediate filament protein we subsequently named desmuslin (DMN). Sequence analysis revealed that DMN has a short N-terminal domain, a conserved rod domain, and a long C-terminal domain, all common features of type 6 intermediate filament proteins. A positive interaction between DMN and α-dystrobrevin was confirmed with an in vitro coimmunoprecipitation assay. By Northern blot analysis, we find that DMN is expressed mainly in heart and skeletal muscle, although there is some expression in brain. Western blotting detected a 160-kDa protein in heart and skeletal muscle. Immunofluorescent microscopy localizes DMN in a stripe-like pattern in longitudinal sections and in a mosaic pattern in cross sections of skeletal muscle. Electron microscopic analysis shows DMN colocalized with desmin at the Z-lines. Subsequent coimmunoprecipitation experiments confirmed an interaction with desmin. Our findings suggest that DMN may serve as a direct linkage between the extracellular matrix and the Z-discs (through plectin) and may play an important role in maintaining muscle cell integrity.
The severe muscle wasting disorder, Duchenne muscular dystrophy, is caused by abnormalities in the dystrophin gene (1). The dystrophin protein is expressed in heart and skeletal muscle, where it is part of the dystrophin-associated protein complex. Dystrophin's N-terminal domain binds to actin, whereas the WW domain and the total cysteine-rich domain bind to β-dystroglycan (2), a component of the dystroglycan subcomplex. This subcomplex links to laminin, a major component of the basal membrane, thereby forming the linkage between an intracellular protein, actin, and the extracellular matrix.
A second subcomplex of the dystrophin-associated protein complex includes four transmembrane proteins (α-, β-, γ-, and δ-sarcoglycan) (3). Each has been shown to be involved in different forms of limb-girdle muscular dystrophy (LGMD 2D, 2E, 2C, and 2F) (4–8). α-Sarcoglycan is a type 1 transmembrane protein and is expressed in heart and skeletal muscle. β-, γ-, and δ-sarcoglycans are type 2 transmembrane proteins containing a cluster of cysteine residues in their extracellular domains. These four proteins form the sarcoglycan complex, which is thought to be involved in some type of signaling pathway (9).
A third subcomplex of the dystrophin-associated protein complex involves α-dystrobrevin (10–12) and the syntrophins (α1, β1, and β2) (13–15). These intracellular proteins directly bind to dystrophin (16, 17). In addition, the N-terminal region of α-dystrobrevin associates with the sarcoglycan complex (18). There are at least five different forms of α-dystrobrevin generated by alternative splicing. The largest splice variant α-dystrobrevin 1 shows sequence homology to the cysteine-rich and C-terminal domains of dystrophin, the Torpedo 87-kDa protein (19), the rabbit 94-kDa protein (A0) (20), and β-dystrobrevin (21). α-Dystrobrevin 1 has a unique C-terminal region with multiple sites for tyrosine phosphorylation and is highly expressed in muscle and brain. This protein has two predicted α-helical coiled-coil motifs and has been shown to interact directly with α1-syntrophin (16, 17) and dystrophin (12). The α-dystrobrevin 2 splice form is slightly different in that it lacks the unique C-terminal region and thus would not be phosphorylated. α-Dystrobrevin 3 has an alternatively spliced 3′ end that is more truncated than that of α-dystrobrevin 2. α-Dystrobrevin 4 and 5 have a different 5′ start site relative to variants 1–3.
To better understand the role of α-dystrobrevin in skeletal muscle, we looked for interacting proteins by using the yeast two-hybrid technique. We isolated three overlapping clones and confirmed their interaction with dystrobrevin with in vitro coimmunoprecipitation (CoIP). A full-length cDNA clone of the interacting protein was isolated. This protein, desmuslin (DMN), colocalizes to the Z-lines with desmin. Our results suggest that DMN forms a linkage between desmin and extracellular matrix and therefore provides an important structural support in muscle.
Materials and Methods
Yeast Two-Hybrid Library Screening.
α-Dystrobrevin cDNA (nucleotides 7–1617) was inserted downstream of the Gal-4 DNA-binding domain in the pGBT9 bait vector. A yeast two-hybrid cDNA library derived from human skeletal muscle was screened for interacting proteins with the use of the Matchmaker two-hybrid system as described by the distributor (CLONTECH).
In brief, transformation mixtures were spread on synthetic dropout/−His/−Leu/−Trp/+3-amino-1,2,4-triazole plates and incubated at 30°C until colonies appeared (6 days). Colonies able to grow on minimal plates were screened for β-galactosidase activity with the use of a filter-lift assay as described by the manufacturer. Yeast DNA isolated from colonies positive for β-galactosidase activity was used to electroporate Escherichia coli to recover the interacting cDNA. The sequence of the interacting cDNA was analyzed on an ABI 373 or 377 automated sequencer with the use of fluorescent dye terminator chemistry (Applied Biosystems).
Phage cDNA Library Screening.
A λgt 11 human skeletal muscle library (CLONTECH) was screened with a 152-bp hybridization probe homologous to DMN's 5′ region (Fig. 2A, nucleotides −23 to 129). The probe was labeled with the use of a Random Primer DNA Labeling System (GIBCO/BRL) in the presence of 5 μCi of [α-32P]dCTP. Filters were prehybridized in hybridization buffer [5× SSC (0.75 M NaCl/0.075 M sodium citrate)/50 mM sodium phosphate (pH 7.4)/1× Denhardt's solution (0.02% Ficoll [type 400]/0.02% polyvinylpyrrolidone/0.02% BSA)] for 5.5 h at 65°C. The denatured probe (8.9 × 105 cpm/ml) was added to the hybridization buffer, and the filters were hybridized for 18 h at 65°C. Filters were washed in 2× SSC and 0.1% SDS for 30 min at 55°C with several changes of washing solution. Filters were wrapped in Saran wrap and exposed to x-ray film at −80°C for 18 h. DNA from positive clones was purified with a Qiagen (Chatsworth, CA) Lambda Midi Kit. The identity of the cDNA as overlapping with the original clone was verified by sequencing.
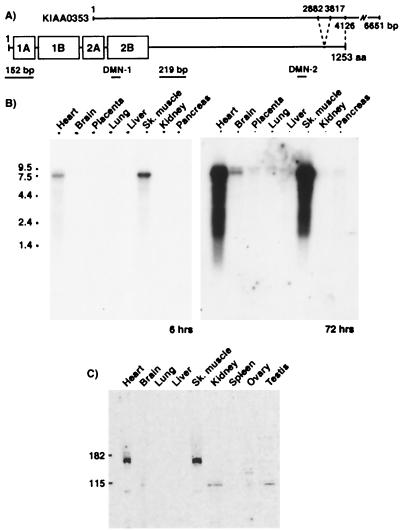
Figure 2.
Summary of DMN, probes, and antibodies (A) and primary expression of DMN in heart and skeletal muscle (B and C). (A) DMN has a central α-helical coiled-coil rod domain (1A to 2B), which is highly conserved; a nonhelical N-terminal head domain (10 aa); and a C-terminal tail domain (931 aa). The rod domain contains nonhelical spacer elements. DMN structure and KIAA0353 sequence are schematically aligned. KIAA0353 starts 573 bp downstream of DMN but includes a region from 2882 to 3817 that was not present on any muscle cDNA clones. Locations of epitopes for anti-DMN antibodies (DMN-1 and DMN-2 indicate those used in Western blot and immunohistochemical analyses, respectively) and probes (152 bp and 219 bp indicate those used for hybridization to Northern blots) are shown. (B) Northern blot analysis with the 219-bp hybridization probe (A) detects a single 7.5-kb relatively abundant transcript in heart and skeletal muscle after a 6-h exposure. A longer exposure of the same Northern blot shows a faint doublet in brain (72 h). (C) Western blot analysis with an anti-DMN-1 antibody (A) detects an ≈160-kDa protein in heart and skeletal muscle but not in brain.
Northern Blot Analysis.
A 219-bp hybridization probe against DMN's 3′ region (Fig. 2A, nucleotides 1268–1486) was prepared by PCR amplification. This probe was then labeled with the use of a Random Primer DNA Labeling System (GIBCO/BRL) and used to hybridize to a human multiple-tissue RNA blot (CLONTECH) according to the manufacturer's protocol. DMN mRNA was visualized by autoradiography. A second hybridization probe, the 152-bp DMN probe (Fig. 2A, nucleotides −23 to 129) used for screening the phage cDNA library, was prepared similarly and used to probe the blot.
Constructs.
Truncated forms of α-dystrobrevin were prepared by PCR amplification: (i) exons 1–16 (nucleotides 16–1617), (ii) exons 1–14 (nucleotides 16–1386), (iii) exons 8–16 (long) (nucleotides 904-1617), (iv) exons 8–16 (short) (nucleotides 973-1617), and (v) exons 10–16 (nucleotides 1033–1617). The PCR products were subcloned into the pMGT1 T7 expression vector.
DMN cDNA from clones 4C and 5D was excised from the pGAD10 library vector with the use of restriction enzymes and was subcloned into pFHR2. The 5D sequence was cloned in two pieces by cleaving the clone with EcoRI [see position 434 in KIAA0353 (22)] (Fig. 1A). The pFHR2 vector coexpresses the FLAG epitope on the N-terminal of the subcloned protein. Because of its location downstream of the FLAG sequence and with the absence of a stop codon, the short 5′ untranslated region of the subcloned cDNAs also was expressed from the pFHR2 vector along with the rest of the protein. Proteins expressed from these vectors were termed FLAG 4C, FLAG 5D-1, or FLAG 5D-2 (Fig. 1B).
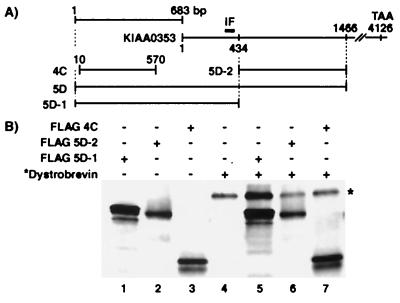
Figure 1.
CoIP analysis of DMN clones with dystrobrevin. (A) Alignment of KIAA0353 (22) and DMN clones 4C, 5D, 5D-1, and 5D-2. IF designates a region with an intermediate filament signature. (B) When simultaneously expressed, FLAG 5D-1 coimmunoprecipitates dystrobrevin (compare lane 5 to lane 4). FLAG 5D-2 and FLAG 4C do not coimmunoprecipitate dystrobrevin (lanes 6 and 7). Our controls show that the FLAG antibody specifically precipitates proteins with the FLAG epitope (lanes 1–3) and precipitates background levels of dystrobrevin (lane 4). * indicates 35S-labeled dystrobrevin. +/− indicates the presence or absence of a particular construct.
We also made several truncated versions of DMN to identify the subregion that interacted with α-dystrobrevin. Constructs were prepared by PCR amplification, using full-length DMN as the template. The DMN PCR product covered the sequence from −3 to 1011 and was subcloned into pFHR2 to express FLAG 1A-1B-2A-2B (amino acids −1 to 337). Constructs 1A-1B-2A (amino acids −1 to 225), 2A-2B (amino acids 161–337), 1B-2A (amino acids 62–225), C-terminal-1 (amino acids 336–933), and C-terminal-2 (amino acids 932-1253) were cloned in a similar fashion. Clone 4C inserted into pFHR2 was used to express FLAG 1A-1B (amino acids −34 to 153).
Desmin was cloned by PCR amplification from total cellular RNA isolated from human skeletal muscle (CLONTECH). The PCR product was subcloned into pMGT1 and sequenced.
Antibodies.
Synthetic peptides corresponding to DMN-1 amino acids 232–250 (C-QEAEALRREALGLEQLRAR) and DMN-2 amino acids 1038–1053 (C-SLSRQRSPAPGSPDEE) (Fig. 2A) were generated and used to inject New Zealand White rabbits (Research Genetics, Huntsville, AL). The amino-terminal cysteine is not part of the DMN sequence, but was added for subsequent affinity purification with the use of a SulfoLink coupling gel (Pierce). Anti-FLAG mAb was purchased from Sigma.
In Vitro Transcription/Translation and CoIP.
Proteins encoding the various constructs were labeled with [35S]methionine with a TNT Quick Coupled Transcription/Translation System (Promega). Depending on the experiment, the proteins were expressed either individually or simultaneously. Five microliters of protein lysate was added to 25 μl of CoIP buffer [150 mM NaCl/50 mM Tris⋅HCl (pH 7.4)/1% Nonidet P-40/Protease Inhibitor Mixture Tablet (Roche)/50 ml] and mixed for 2 h at 4°C. Subsequently, 2 μl of anti-FLAG mAb (Sigma) and 18 μl of the CoIP buffer were added, and the mixture was then reincubated for 3 h at 4°C. After this step, 50 μl of suspended protein G-Sepharose (Sigma) was added and shaken at 4°C overnight. The next day, the mixture was centrifuged at 1,000 × g for 1 min, and the pellet was washed three times with CoIP buffer and then resuspended in 2× Tris-glycine sample buffer (Novex) with 50 mM DTT. The samples were heated to 85°C for 2 min and separated by electrophoresis on 10% Tris-glycine acrylamide gels (Novex, San Diego). Proteins were visualized by exposing the gels to a phosphor plate and scanning with a PhosphorImager (Molecular Dynamics).
Immunoblot Analysis.
Human protein medleys (CLONTECH) were separated by electrophoresis on 4–20% acrylamide gels, and the proteins were transferred to a nitrocellulose membrane in a transfer buffer (48 mM Tris/39 mM glycine/13 mM SDS/20% methanol) at 15 V for 20 min with a Trans-Blot SemiDry apparatus (Bio-Rad). The membrane was blocked with blocking buffer (1× PBS/0.1% Tween 20/5% nonfat milk) overnight at 4°C. The membrane was incubated with anti-DMN-1 antibody diluted in blocking buffer for 2 h at room temperature, washed with 1× PBS/0.1% Tween 20, and then incubated for 1 h with horseradish peroxidase-conjugated donkey anti-rabbit IgG (H + L) secondary antibody (Jackson ImmunoResearch). The membrane was washed in 1× PBS/0.1% Tween 20, and the horseradish peroxidase-conjugated protein was detected by chemiluminescence.
Immunofluorescent Analysis.
Human skeletal muscle was obtained from biopsies of patients without neuromuscular disorders. Muscle sections were fixed in cold methanol for 3 min, blocked in 10% FCS in 1× PBS for 1 h at 4°C, and stained with anti-DMN-2 antibody (Fig. 2A) overnight at 4°C. The slides were washed in 1× PBS for 1 h and incubated with a Cy3-conjugated Affinipure donkey anti-rabbit IgG (H + L) secondary antibody (Jackson ImmunoResearch) for 1 h at room temperature. The slides were washed in 1× PBS for 1 h and mounted with a ProLong antifade kit (Molecular Probes). Analysis was done with a Zeiss Axioplan 2 microscope.
Immunoelectron Microscopic Analysis.
Human skeletal muscle was obtained from biopsies of patients without neuromuscular disorders. After fixation in 2% paraformaldehyde/0.01% glutaraldehyde, the biopsies were cut into small (1-mm cubes) before immersion in 2.3 M sucrose/0.1 M PBS. The samples subsequently were mounted on cutting stubs, shock-frozen, and stored in liquid nitrogen. Thin sections (70–100 nm) were cut with a Reichert Ultracut S Ultramicrotome with a FC4S cryo-attachment. Sections were lifted in a small drop of sucrose, mounted on Formvar-coated carbon grids, and washed three times in PBS containing 0.5% BSA and 0.15% glycine (pH 7.4) (buffer A). This procedure was followed by a 30-min incubation with purified goat IgG (50 mg/ml) at 25°C and three additional washes with buffer A. All of the preceding steps were designed to ensure minimal nonspecific reactions to the antibodies used. Sections then were incubated for 60 min with primary antibody (either a rabbit polyclonal antibody directed to DMN-2 or to desmin), followed by three washes in buffer A and a 60-min incubation in gold-labeled second antibody (1–2 mg/ml). The sections were then washed six times (5 min per wash) in buffer A and then rewashed thoroughly in buffer A (five changes) and in PBS (three changes), followed by a brief fixation step in 2.5% glutaraldehyde in PBS to ensure that the antibodies did not dissociate. Subsequent steps included three further washes in PBS, five washes in water, and counterstaining with uranyl acetate and embedding in 1.25% methylcellulose. Observation was done with a Jeol 1210 electron microscope.
Results
Identification of an α-Dystrobrevin-Interacting Protein.
To identify proteins that interact with α-dystrobrevin, we screened a human skeletal muscle cDNA library with the yeast two-hybrid approach. The 5′ region of α-dystrobrevin (nucleotides 7–1617) was subcloned into the pGBT9 bait vector as described in Materials and Methods. Of the 21 positive clones, three represented either the same (3C and 4C) or overlapping (4C and 5D) sequences (Fig. 1A) from a yet uncharacterized protein. Database searching with clone 5D revealed that the protein contains sequence identity to 1,466 bp of KIAA0353 (22). The 5D clone also contained an additional 683 bp upstream from the KIAA0353 sequence. KIAA0353 is a partial cDNA sequence, isolated from a brain cDNA library, which contains the intermediate filament (IF) signature (VATYRALLE). Clone 4C is identical to the 5D sequence from 10 to 570. The exact relationship between 4C, 5D, and KIAA0353 is given schematically in Fig. 1A.
CoIP of the Novel Protein and α-Dystrobrevin.
To verify the results of the two-hybrid screen, in vitro CoIP experiments were performed with different tagged portions of the protein (FLAG 5D-1, FLAG 5D-2, FLAG 4C) (Fig. 1B) and untagged dystrobrevin exons 1–16. As a control, these proteins were also individually precipitated. Proteins were expressed and 35S-labeled with the TNT quick coupled transcription/translation system (Promega). After incubation with protein G-Sepharose, each 35S-labeled translated protein was immunoprecipitated with the anti-FLAG antibody and then analyzed by SDS/PAGE gel (Fig. 1B). The anti-FLAG antibody is able to specifically immunoprecipitate each protein individually (Fig. 1B, lanes 1–4). When dystrobrevin was simultaneously expressed with either FLAG 5D-2 (lane 6) or FLAG 4C (lane 7), the anti-FLAG antibody did not coimmunoprecipitate dystrobrevin to a greater degree than that seen in lane 4, where there were no FLAG epitopes. However, when dystrobrevin and FLAG 5D-1 were simultaneously expressed (lane 5), dystrobrevin was coimmunoprecipitated, as indicated by the increase in the dystrobrevin band over background (compare lanes 4 and 5). These results support an interaction between α-dystrobrevin and the clones isolated from the yeast two-hybrid screen.
Characterizing DMN.
With the use of a 152-bp probe (Fig. 2A), a phage human skeletal muscle cDNA library was screened by hybridization to obtain the full-length DMN cDNA. DMN's approximate transcription start site was identified by comparing the 5′ sequence of the 16 DMN phage clones with that of the clones isolated from the yeast two-hybrid screening (clones 4C and 5D) and that obtained by two experiments with 5′ rapid amplification of cDNA ends (data not shown). Most of the clones fell within 10–20 bases 5′ of one another. Comparing the DMN cDNA sequence with that of genomic DNA (Homo sapiens, clone RP11–28O17), a stop codon was found 39 bases upstream of the predicted transcription start site and 150 bases upstream of the first ATG codon, suggesting that this ATG likely serves as the translation start site. In addition, the sequence GGC AAG ATG CTG contains an adequate Kozak consensus sequence (23).
blast analysis with DMN cDNA revealed that it is an IF family member protein (24). DMN has a unique 10-aa N-terminal domain, followed by the hallmark rod domain, which is broken into regions 1A, 1B, 2A, and 2B and a long 931-aa C-terminal domain. The ORF (3,762 bp) encodes a 1,253-aa protein. Compared with the KIAA0353 sequence, which was isolated from neuronal cells, muscle DMN cDNA contains an additional sequence at the 5′ end and lacks KIAA0353 region 2882–3817 (Fig. 2A).
DMN Is Expressed Predominantly in Heart and Skeletal Muscle.
To determine DMN's expression profile, multitissue Northern blot analysis was performed with a 219-bp hybridization probe (Fig. 2A). A 7.5-kb transcript was observed in heart and skeletal muscle after a 6-h exposure (Fig. 2B). Interestingly, a faint doublet also was detected in brain after long exposures (72 h) (Fig. 2B). With the use of the 152-bp DMN probe, the same probe that was used for screening the phage cDNA library, DMN mRNA was again detected in both heart and skeletal muscle. No doublet was detected in brain with this probe, even after long exposures (data not shown). The doublet in brain detected with the 219-bp probe suggests that there may be tissue-specific DMN isoforms.
Sequence analysis of DMN's cDNA predicts that the encoded protein would have a molecular mass of ≈140 kDa. Western blot analysis with a DMN-1-specific antibody (Fig. 2A) detected a protein calculated to be 160 kDa, expressed solely in heart and skeletal muscle (Fig. 3C), thereby supporting the results of the Northern blot.
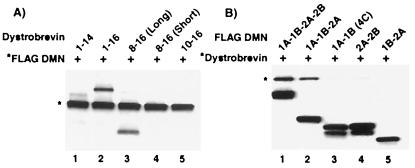
Figure 3.
Portions of the DMN rod domain specifically interact with dystrobrevin. (A) CoIP experiments using the anti-FLAG antibody show that FLAG 1A-1B-2A-2B DMN specifically interacts with dystrobrevin exons 1–16 (lane 2) and 8–16 (long) (lane 3). Other regions of α-dystrobrevin do not coimmunoprecipitate in the presence of DMN (lanes 1, 4, and 5). (B) By varying the length of the DMN rod domain, CoIP experiments show that dystrobrevin exons 1–16 interacts with 1A-1B-2A-2B (lane 1) and 1A-1B-2A (lane 2), but not with the shorter versions of the DMN rod domain (lanes 3–5). Proteins are identified by an *.
DMN Rod Domain Interacts with Exon 8–16 of α-Dystrobrevin.
By both yeast two-hybrid and CoIP analyses, FLAG 5D-1 (amino acids −37 to 337) interacts with dystrobrevin exons 1–16 (Fig. 1). As a consequence of cloning 5D-1 with its 5′ untranslated region downstream of the N-terminal FLAG, this construct contains an additional 37 aa before the initiation methionine. To eliminate the possibility that this small amino acid segment affected our results, we removed the 5′ untranslated region from the cDNA of clone 5D-1 and recloned it back into pFHR2. Coincidentally, 5D-1 encodes the whole rod domain for DMN such that its expression from pFHR2 would encode the protein FLAG 1A-1B-2A-2B (amino acids −1 to 337).
To refine the interacting domain between DMN and α-dystrobrevin, we performed CoIP experiments with subfragments of the two proteins. The following fragments of α-dystrobrevin were amplified by PCR and subcloned into the non-FLAG pMGT1 expression vector: (i) exons 1–14, (ii) exons 8–16 (long), (iii) exons 8–16 (short), and (iv) exons 10–16. The PCR primers used for subcloning were internal to the first and last listed exon; consequently, these exons are not complete. The exons 8–16 (long) are 23 aa longer at the N terminus than the exons 8–16 (short). In each lane, the proteins were labeled with [35S]methionine and expressed simultaneously with the TNT quick coupled transcription/translation system. CoIP experiments were performed by coexpressing FLAG 1A-1B-2A-2B DMN with different dystrobrevin fragments and immunoprecipitating with the anti-FLAG antibody (Fig. 3A). Our results show that FLAG 1A-1B-2A-2B interacts specifically with dystrobrevin exons 1–16 (lane 2) and dystrobrevin exons 8–16 (long) (lane 3) but not exons 1–14 (lane 1), exons 8–16 (short) (lane 4), and exons 10–16 (lane 5). These results imply that dystrobrevin exons 8–16 are important for interactions with DMN.
We also wanted to determine which portions of the DMN rod domain interact with dystrobrevin. To perform this experiment, we subcloned various portions of the DMN rod domain into pFHR2 so that they would be expressed with the N-terminal FLAG epitope. CoIP experiments were done by simultaneous translation of dystrobrevin exons 1–16 with truncated versions of the DMN rod domain (Fig. 3B). These experiments show that binding requires the DMN domain 1A-1B-2A, although the inclusion of 2B adds some stability to the interaction (compare lanes 1 and 2). When DMN's entire 2A-2B, 1A-1B, or flanking 1A and 2B regions were deleted, all interactions were either abolished or greatly diminished (lanes 3–5).
DMN Is Present at the Z-lines.
To get a better idea of DMN's subcellular localization within normal muscle, the anti-DMN-2 antibody was used in immunofluorescent studies. As shown in Fig. 4E, DMN was detected as a mosaic in cross sections with minimal sarcolemmal labeling. The expression of DMN was stronger for type 2 fast fibers (data not shown). In longitudinal sections, the fibers stain in a stripe-like pattern (Fig. 4F). These results are consistent with DMN's being an IF protein.
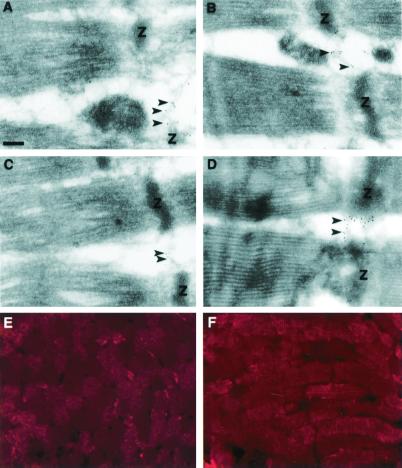
Figure 4.
DMN is located in the Z-lines. When muscle was stained with anti-DMN-2 antibody, a mosaic staining pattern was detected in cross sections (E), and a stripe-like staining pattern was detected in longitudinal sections (F). Three different muscle tissues were stained for DMN (A–C), thereby localizing DMN to the filamentous structures running between the Z-lines (Z) (arrowheads). These tissues are the regions that also stain for desmin (D) (arrowheads). (Bar = 200 nm.)
Immunoelectron microscopy was used to validate the putative Z-line staining seen by immunofluorescence (Fig. 4). Fig. 4 A–C shows typical staining for DMN in skeletal muscles from three different patients without neuromuscular diseases. DMN appears to be localized between adjacent myofibrils at the level of the Z-lines. To confirm this, an equivalent experiment was performed on the same muscle biopsies with the use of antibodies to desmin (Fig. 4D), which envelops all myofibrils at the Z-lines. Interestingly, we find that DMN colocalizes with desmin.
DMN Rod Domain Interacts with Desmin.
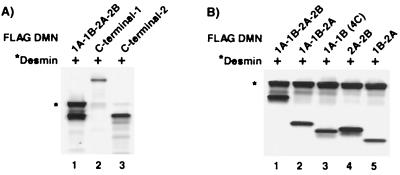
Because DMN and desmin colocalize, we checked for an interaction between DMN and desmin with CoIP experiments. These experiments were done by coexpressing DMN (FLAG 1A-1B-2A-2B, FLAG C-terminal-1, or FLAG C-terminal-2) with untagged desmin and then immunoprecipitating the 35S-labeled proteins with the use of the anti-FLAG antibody (Fig. 5A). Our results show that desmin coimmunoprecipitates with FLAG 1A-1B-2A-2B (lane 1) but not with either FLAG C-terminal-1 (lane 2) or FLAG C-terminal-2 (lane 3). To further define which portion of the DMN rod domain interacts with desmin, we simultaneously expressed desmin and truncated versions of the FLAG DMN rod domain (Fig. 5B). This CoIP experiment shows that desmin interacts with all of the rod domain constructs we generated (lanes 1–5), suggesting that any portion of this domain is sufficient for interaction.
Figure 5.
Interaction of DMN with desmin. (A) CoIP analysis shows that the DMN FLAG 1A-1B-2A-2B subfragment (lane 1) interacts with desmin, whereas the DMN FLAG C-terminal subfragments do not (lanes 2 and 3). (B) CoIP experiments with full-length human desmin show that desmin interacts with all five truncated FLAG DMN constructs (lanes 1–5). Desmin (identified with an *) was 35S-labeled.
Comparison of DMN with Other IF Proteins.
DMN and various other IF proteins are grouped in Table 1, along with three IF-associated proteins. Amino acid prediction analysis was performed with macvector software (Oxford Molecular Group). When the amino acid numbers of N-terminal, rod, and C-terminal domains were compared, human DMN most closely resembled rat nestin (25), frog tanabin (26), chicken paranemin (27), and chicken synemin (28), all of which have a short N-terminal and long C-terminal domains. These proteins, together with nestin, should be classified as a type 6 IF protein or even a type 7 IF protein. Comparing DMN's rod domain sequence with other IF proteins, human DMN has 60% homology with chicken synemin, indicating that DMN is more similar to chicken synemin than it is to rat nestin, frog tanabin, and chicken paranemin. DMN is not likely to be the human ortholog of chicken synemin because other IF proteins cloned in both species, for example vimentin, are greater than 80% homologous to one another.
Table 1.
Comparison of DMN with other major IF and IF-associated proteins
| Type | Protein | No. of
residues
|
Homology with DMN, % | ||
|---|---|---|---|---|---|
| N terminus | Rod | C terminus | |||
| 1 | Human keratin 14 | 114 | 312 | 46 | 41 |
| 2 | Human keratin 5 | 167 | 314 | 109 | 43 |
| 3 | Human desmin | 107 | 309 | 54 | 49 |
| 4 | Human neurofilament M | 100 | 312 | 504 | 47 |
| 5 | Human lamin C | 30 | 357 | 185 | 40 |
| 6 | Rat nestin | 7 | 307 | 1,491 | 49 |
| Frog tanabin | 12 | 308 | 1,424 | 50 | |
| Chicken paranemin | 15 | 308 | 1,283 | 51 | |
| Chicken synemin | 10 | 304 | 1,290 | 60 | |
| Human DMN | 10 | 312 | 931 | 100 | |
Representative proteins from each of the different IF classes are listed for comparison with human DMN. The sum of identical and similar amino acids is given for only the conserved rod domain of DMN and the other IF proteins. IF-associated but unclassified proteins are blank for IF protein type.
Discussion
α-Dystrobrevin 1 is the largest of the five α-dystrobrevin splice variants. We used the first 16 of the 21 α-dystrobrevin exons as the bait in a yeast two-hybrid screen to clone desmuslin. Because these exons are also shared with α-dystrobrevin 2, it is likely that DMN can also interact with both of these α-dystrobrevin isoforms.
Sequence analysis suggests that DMN contains an IF signature (24), suggesting that DMN may be a structural protein. Of the various IF protein family members, DMN is most similar to synemin (28), paranemin (27), tanabin (26), and nestin (25) in terms of domain structure. All have short N-terminal, conserved rod, and long C-terminal domains (Table 1). Currently, nestin is the only one classified as a type 6 IF protein. The others are either not yet classified or are called IF-associated proteins, although Steinert et al. (29) have proposed that they all be grouped as type 6 IF proteins. By domain structure, we believe that DMN should also be grouped as a type 6 IF protein.
Like DMN, chicken synemin also shares homology with parts of KIAA0353. Although several parts of synemin's C-terminal domain and the extra C-terminal end (50 aa) are almost identical to KIAA0353 (28), the remainder is not homologous. On the other hand, DMN shares 100% homology with the entire KIAA0353 cDNA, except at the 5′ terminus, where DMN has an additional 572 bases, and at the 3′ terminus, where DMN lacks region 2882–3817. However, unlike chicken synemin, our in vitro CoIP experiments show that the C-terminal domain of DMN does not interact with α-actinin (data not shown). Because of differences in binding preference and sequence homology, we do not believe that DMN is the human homolog of chicken synemin.
Recently, syncoilin, an IF protein that interacts with α-dystrobrevin, was reported to be concentrated at the neuromuscular junction of normal skeletal muscle (30). In dystrophic muscle, syncoilin's expression was increased such that the protein was found throughout the sarcolemma. This finding is in contrast to DMN, which is expressed similarly in control and DMD muscle (data not shown). This difference in their expression profile suggests that DMN and syncoilin have different functions. Moreover, syncoilin shares the greatest homology with type 3 and 4 IF proteins rather than a type 6 IF protein like DMN. These results suggest that DMN is a yet uncharacterized protein present in muscle.
Two observations led us to identify proteins other than α-dystrobrevin that are capable of interacting with DMN. First, electron microscopic analysis colocalized DMN with desmin, another IF group family member. Second, desmin had been shown to be capable of interacting with synemin (28), a protein that shares some homology with DMN. As such, we attempted to determine whether DMN could interact with desmin with the use of CoIP experiments. As expected, DMN can interact with desmin, and the site of interaction was localized to DMN's rod domain (Fig. 5A). α-Dystrobrevin, a membrane protein, also interacts with DMN at the rod domain. Therefore, we speculate that there are at least two distinct DMN subpopulations, one in which DMN interacts exclusively with desmin within the Z-lines, and another in which DMN interacts with both dystrobrevin and desmin at the costamere. It is possible that posttranslational modifications or other interacting proteins could modulate DMN's intracellular location.
IF proteins like DMN and desmin are thought to help maintain the structural integrity of tissues by mechanically reinforcing protein connections. Desmin encircles Z-lines and then makes longitudinal filamentous bridges between neighboring Z-lines. These bridges are supported by plectin (31) and αB-crystallin (32). Plectin links desmin to the Z-discs and desmin to itself (31). Mutations in the desmin gene can cause desmin myopathies characterized by muscle weakness, cardiac impairment, and intracytoplasmic accumulation of desmin deposits (33–36). The accumulation of desmin may actually be a secondary feature, as other proteins also accumulate in deposits. Although we have not yet investigated this possibility, it is possible that DMN deposits would be found in muscle from a desmin myopathy, and/or desmin deposits may be found in muscle with mutations in the DMN gene.
In summary, we have cloned and subsequently characterized an α-dystrobrevin-interacting protein, which we have termed desmuslin (DMN). Our results support a model in which DMN forms a linkage connecting the extracellular matrix to the Z-discs. The DMN linkage we have identified is a linkage between a component of dystrophin-associated protein complex (α-dystrobrevin) and a component of the Z-lines (desmin). This linkage can be extended to the extracellular matrix by recognizing that the extracellular matrix protein laminin interacts with α-dystroglycan (2). Disruption of any of the proteins in this linkage could potentially damage muscle cell integrity.
Acknowledgments
S.C.W. is supported by a grant from the Muscular Dystrophy Association. L.M.K. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- CoIP
coimmunoprecipitation
- DMN
desmuslin
- IF
intermediate filament
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF359284).
References
- 1.Koenig M, Monaco A P, Kunkel L M. Cell. 1988;53:219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 2.Ibraghimov-Beskrovnaya O, Ervasti J M, Leveille C J, Slaughter C A, Sernett S W, Campbell K P. Nature (London) 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 3.Ozawa E, Noguchi S, Mizuno Y, Hagiwara Y, Yoshida M. Muscle Nerve. 1998;21:421–438. doi: 10.1002/(sici)1097-4598(199804)21:4<421::aid-mus1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Roberds S L, Leturcq F, Allamand V, Piccolo F, Jeanpierre M, Anderson R D, Lim L E, Lee J C, Tome F M S, Romero N B, et al. Cell. 1994;78:625–633. doi: 10.1016/0092-8674(94)90527-4. [DOI] [PubMed] [Google Scholar]
- 5.Bonnemann C G, Modi R, Noguchi S, Mizuno Y, Yoshida M, Gussoni E, McNally E M, Duggan D J, Angelini C, Hoffman E P, et al. Nat Genet. 1995;11:266–273. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- 6.Lim L E, Duclos F, Broux O, Bourg N, Sunada Y, Allamand V, Meyer J, Richard I, Moomaw C, Slaughter C, et al. Nat Genet. 1995;11:257–265. doi: 10.1038/ng1195-257. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi S, McNally E M, Othmane K B, Hagiwara Y, Mizuno Y, Yoshida M, Yamamoto H, Bonnemann C G, Gussoni E, Denton P H, et al. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 8.Nigro V, Piluso G, Belsito A, Politano L, Puca A A, Papparella S, Rossi E, Viglietto G, Esposito M G, Abbondanza C, et al. Hum Mol Genet. 1996;5:1179–1186. doi: 10.1093/hmg/5.8.1179. [DOI] [PubMed] [Google Scholar]
- 9.Thompson T G, Chan Y, Hack A A, Brosius M, Rajala M, Lidov H G W, McNally E M, Watikins S C, Kunkel L M. J Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadoulet-Puccio H M, Khurana T S, Cohen J B, Kunkel L M. Hum Mol Genet. 1996;5:489–496. doi: 10.1093/hmg/5.4.489. [DOI] [PubMed] [Google Scholar]
- 11.Sadoulet-Puccio H M, Feener C A, Schaid D J, Thibodeau S N, Michels V V, Kunkel L M. Neurogenetics. 1997;1:37–42. doi: 10.1007/s100480050006. [DOI] [PubMed] [Google Scholar]
- 12.Sadoulet-Puccio H M, Rajala M, Kunkel L M. Proc Natl Acad Sci USA. 1997;94:12413–12418. doi: 10.1073/pnas.94.23.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams M E, Butler M H, Dwyer T M, Peters M F, Murnane A A, Froehner S C. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- 14.Ahn A H, Yoshida M, Anderson M S, Feener C A, Selig S, Hagiwara Y, Ozawa E, Kunkel L M. Proc Natl Acad Sci USA. 1994;91:4446–4450. doi: 10.1073/pnas.91.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang B, Ibraghimov-Beskrovnaya O, Moomaw C R, Slaughter C A, Campbell K P. J Biol Chem. 1994;269:6040–6044. [PubMed] [Google Scholar]
- 16.Suzuki A, Yoshida M, Ozawa E. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn A H, Kunkel L M. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida M, Hama H, Ishikawa-Sakurai M, Imamura M, Mizuno Y, Araishi K, Wakabayashi-Takai E, Noguchi S, Sasaoka T, Ozawa E. Hum Mol Genet. 2000;9:1033–1040. doi: 10.1093/hmg/9.7.1033. [DOI] [PubMed] [Google Scholar]
- 19.Wagner K R, Cohen J B, Huganir R L. Neuron. 1993;10:511–512. doi: 10.1016/0896-6273(93)90338-r. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida M, Yamamoto H, Noguchi S, Mizuno Y, Hagiwara Y, Ozawa E. FEBS Lett. 1995;367:311–314. doi: 10.1016/0014-5793(95)00574-s. [DOI] [PubMed] [Google Scholar]
- 21.Puca A A, Nigro V, Piluso G, Belsito A, Sampaolo S, Quaderi N, Rossi E, Iorio G D, Ballabio A, Franco B. FEBS Lett. 1998;425:7–13. doi: 10.1016/s0014-5793(98)00097-0. [DOI] [PubMed] [Google Scholar]
- 22.Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 23.Kozac M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 24.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. New York: Scientific American Books; 1995. pp. 1106–1116. [Google Scholar]
- 25.Lendahl U, Zimmerman L B, McKay R D G. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 26.Hemmati-Brivanlou A, Mann R W, Harland R M. Neuron. 1992;9:417–428. doi: 10.1016/0896-6273(92)90180-l. [DOI] [PubMed] [Google Scholar]
- 27.Hemken P M, Bellin R M, Sernett S W, Becker B, Huiatt T W, Robson R M. J Biol Chem. 1997;272:32489–32499. doi: 10.1074/jbc.272.51.32489. [DOI] [PubMed] [Google Scholar]
- 28.Bellin R M, Sernett S W, Becker B, Ip W, Huiatt T W, Robson R M. J Biol Chem. 1999;274:29493–29499. doi: 10.1074/jbc.274.41.29493. [DOI] [PubMed] [Google Scholar]
- 29.Steinert P M, Chou Y-H, Prahlad V, Parry D A D, Marekov L N, Wu K C, Jang S-I, Goldman R D. J Biol Chem. 1999;274:9881–9890. doi: 10.1074/jbc.274.14.9881. [DOI] [PubMed] [Google Scholar]
- 30.Newey S E, Howman E V, Ponting C P, Benson M A, Nawrotzki R, Loh N Y, Davies K E, Blake D J. J Biol Chem. 2001;276:6645–6655. doi: 10.1074/jbc.M008305200. [DOI] [PubMed] [Google Scholar]
- 31.Hijikata T, Murakami T, Imamura M, Fujimaki N, Ishikawa H. J Cell Sci. 1999;112:867–876. doi: 10.1242/jcs.112.6.867. [DOI] [PubMed] [Google Scholar]
- 32.Vicart P, Caron A, Guicheney P, Li Z, Prevost M C, Faure A, Chateau D, Chapon F, Tome F, Dupret J M, et al. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 33.Goldfarb L G, Park K, Cervenakova L, Gorokhova S, Lee H, Vasconcelos O, Nagle J W, Semino-Mora C, Sivakumar K, Dalakas M C. Nat Genet. 1998;19:402–403. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]
- 34.Munoz-Marmol A M, Strasser G, Isamat M, Coulombe P A, Yang Y, Roca X, Vela E, Mate J L, Coll J, Fernandez-Figueras M T, et al. Proc Natl Acad Sci USA. 1998;95:11312–11317. doi: 10.1073/pnas.95.19.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalakas M C, Park K Y, Semino-Mora C, Lee H S, Sivakumar K, Goldfarb L G. N Engl J Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 36.Sjoberg G, Saavedra-Matiz C A, Rosen D R, Wijsman E M, Borg K, Horowitz S H, Sejersen T. Hum Mol Genet. 1999;12:2191–2198. doi: 10.1093/hmg/8.12.2191. [DOI] [PubMed] [Google Scholar]