Abstract
Recently developed techniques for reconstruction of high-resolution 3D images from fetal MR scans allows us to study the morphometry of developing brain tissues in utero. However, existing adult brain analysis methods cannot be directly applied as the anatomy of the fetal brain is significantly different in terms of geometry and tissue morphology. We describe an approach to atlas-based segmentation of the fetal brain with particular focus on the delineation of the germinal matrix, a transient structure related to brain growth. We segment 3D images reconstructed from in utero clinical MR scans and measure volumes of different brain tissue classes for a group of fetal subjects at gestational age 20.5–22.5 weeks. We also include a partial validation of the approach using manual tracing of the germinal matrix at different gestational ages.
1 Introduction
Imaging of the human fetus using magnetic resonance (MR) imaging is emerging as an important clinical tool in the early detection of brain abnormalities [1, 2]. The ability to evaluate morphometric measures of brain development that can be reliably derived from clinical imaging promises a range of new quantitative biomarkers that can be used in clinical evaluation of pregnancy and to form a better understanding of brain development [3, 4]. Typical clinical MR image (MRI) acquired in utero is severely corrupted by motion of the fetus. Recently developed methods of image reconstruction using registration to correct for fetal motion [5–7] have permitted the formation of true 3D MR images of the fetal brain and have opened up the possibility of using methodology developed for adult brain image analysis to study the developing brain in utero. However, the underlying anatomy of the developing fetal brain is significantly different both in terms of geometry as well as underlying tissue morphology [8].
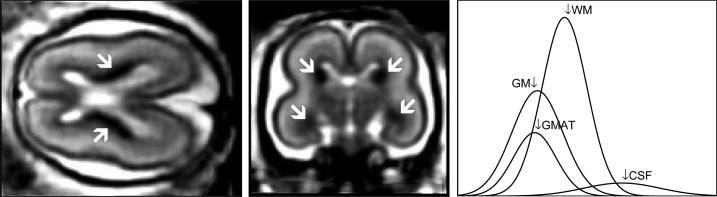
The fetal brain consists of a mixture of developed white matter and grey matter regions and other transient structures related to brain growth. One of the most important of these is the germinal matrix, a deep brain region of developing cells adjacent to ventricules that is present in the fetal brain between 8 and 28 weeks gestational age (GA) [9, 10]. During embryology and early fetal life, the germinal matrix is the site of production of both neurons and glial cells which then migrate out to their final location. Due to its high cell-packing density, the germinal matrix appears hypointense on T2-weighted (T2w) MR images as shown in Fig. 1. The volume of the germinal matrix increases exponentially reaching its peak at about 23–26 weeks GA and decreases subsequently [9, 10]. There is little published material about the volume or the shape of the germinal matrix in normal fetuses and most of the work is derived from pathology [10]. The recent development of 3D MRI reconstruction techniques allows us to study the morphology and morphometry of developing tissues in utero. In this work we describe an approach to atlas-based tissue segmentation which is aimed at extracting key tissue regions from reconstructed 3D fetal MRI data, with particular focus on the delineation of the germinal matrix from developed white and grey matter regions.
Fig. 1.
Axial and coronal views of an MR T2w image with clearly visible hypointense regions of the germinal matrix. Distribution of voxel intensities for grey matter (GM), the germinal matrix (GMAT), white matter (WM), and cerebrospinal fluid (CSF).
2 Method
2.1 Problem overview
Current clinical practice at our institution makes use of fast T2-weighted MR imaging to provide contrast for main tissue types in the fetal brain. Figure 1 illustrates the structures visible in this type of data. Segmentation of developing anatomy, even in neonates [11] and young children [12], is challenging due to evolving states of tissue and how this is reflected in MR images. In fetal imaging, there is a number of basic tissue states that are of interest including developed grey matter (GM) and white matter (WM), the germinal matrix (GMAT) and the coritcal plate (CP). As the germinal matrix generally appears with a similar intensity to the grey matter as shown in Fig. 1, its interpretation is dependent on the location around the ventricles. There are key areas where ventricle, GMAT and GM cortex are adjacent, particularly in regions of the parietal and occipital lobes. As some form of spatial context will be required to achieve meaningful segmentation, we approach the problem using an atlas-based tissue segmentation methodology.
2.2 MR imaging procedure
MR imaging was performed on a 1.5T scanner according to an IRB-approved protocol without sedation or contrast agent administration. A quick low-resolution localizer sequence was obtained during maternal free breathing to determine the location of the fetal head. Then, single-shot fast spin-echo (SSFSE) T2-weighted images were acquired using an eight channel torso phased array coil during normal maternal breathing. Based on the initial localizer, sets of contiguous slices were obtained in the approximately axial, coronal and sagittal planes with respect to the fetal brain. All images were acquired in an interleaved manner to reduce saturation of spins in adjacent slices. The following MR parameters were used: relaxation time TR ranging from 4000 ms to 8000 ms, effective echo time TE = 91 ms, flip angle 130°, in plane resolution 0.469mm × 0.469mm, slice thickness ≈ 3mm. These MR parameters were originally designed for clinical scans and have not been adjusted for tissue segmentation in this study or any other type of image analysis.
The following study was performed using clinical MR scans of 5 women at 20.5–22.5 weeks of pregnancy. The patients were referred for fetal MRI due to questionable abnormalities on prenatal ultrasound or a prior abnormal pregnancy. All women had normal fetal MRI and all newborns have had normal postnatal neurodevelopment.
2.3 Atlas construction
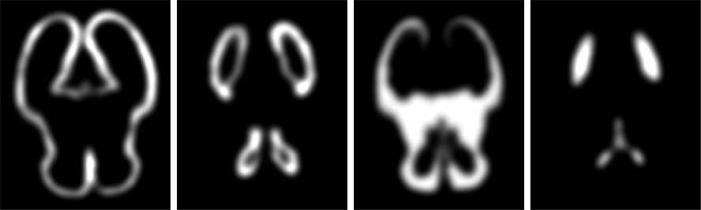
To create a reference anatomy, a set of 3D high-resolution isotropic volumes (0.469mm × 0.469mm × 0.469mm) was formed from subject MR scans using the registration-based reconstruction technique [5]. An average shape and intensity image was constructed from these subject images by spatial normalization of the scans using a sequence of global linear registrations driven by normalized mutual information [13], followed by multiple elastic deformations driven by mutual information [14, 15]. These were used to iteratively form an averagely shaped anatomy in an approach similar to [16]. The intensity of each scan was normalized and the spatially normalized images were averaged to form a high quality reference image. The reference average image was manually segmented into regions of white matter, grey matter, CSF, ventricular CSF and germinal matrix by expert observers. For this preliminary work, we have not delineated other subregions of white matter such as the cortical plate. Tissue label maps were extracted from the manual segmentation, convolved with a Gaussian kernel (σ = 3.0 × voxelsize) and normalized to simulate a probabilistic atlas shown in Fig. 2.
Fig. 2.
Probabilistic atlas for grey matter, germinal matrix, white matter and cerebrospinal fluid (left to right, respectively) derived from the segmentation of the average MRI anatomy model.
2.4 Volume segmentation
General EM framework
Expectation-Maximization (EM) is a general technique for finding maximum likelihood estimates of model parameters in problems with missing data. The EM algorithm maximizes the likelihood of the observed data by interleaving the expectation step (E-step) which performs statistical classification of the observed data into K classes and the maximization step (M-step) which updates the current parameter estimation. In the context of brain MRI segmentation [17, 18], the observed data are the voxel intensities y = {y1, y2, ..., yN}, the missing data are the voxel labels c = {c1, c2, ..., cN} (image segmentation), and the model parameters are the class-conditional intensity distribution parameters θ = {θ1, θ2, ..., θK}.
Assuming that each voxel value is selected at random from one of the K classes and each class k is modeled by a Gaussian distribution Gσk(yi – μk) [17] with mean μk and variance , the probability density that class k generated the voxel value yi at position xi is p(yi|k) = Gσk(yi – μk) and the corresponding class posterior probability p(k|yi) computed in the E-step is
| (1) |
where P(k) is the prior probability for tissue class k. The estimation of current class distribution parameters in the M-step is performed according to
| (2) |
and the intermediate segmentation of the MR image is given by voxel labels c(xi) assigned using the maximum posterior probability rule.
| (3) |
Bias estimation
Intensity inhomogeneity or bias field can be major problems for automated MR image segmentation. Although it may not be visible for a human reader, such a bias may cause tissue mislabeling in intensity-based segmentation. This is particularly an issue for our fetal MR scans where we have an eight channel torso phased array coil.
Assuming a standard multiplicative bias model, intensity inhomogeneity is approximated by a linear combination of spatially smooth basis functions (low order polynomials). The bias field parameters are calculated in each iteration of the EM algorithm as the weighted least-squares fit to the difference between measured intensities yi and predicted intensities ỹi calculated from intermediate estimates of class distribution parameters [19].
Prior probabilities
The independent segmentation model from Eqns. 1 and 2 performs labeling of MRI voxels based solely on their intensities yi and assumes that different classes are well separated in the intensity space. This, however, is not the case for fetal brain segmentation where the overlap of intensities between tissue classes is substantial, especially for grey matter and germinal matrix, as shown in Fig. 1. Moreover, the resulting brain segmentation may be noisy or not anatomically feasible. To address these issues, we use the probabilistic atlas described in Section 2.3 to provide spatially varying class prior probabilities Pa(k|xi) at every voxel xi.
The voxel labeling process is be further constrained by introduction of neighborhood dependencies where the probability that a voxel belongs to tissue k depends on the tissue types of its neighbors. We use an additional neighborhood-based prior Pn(k|xi) calculated from voxels xj located not farther than dk from xi (|xi – xj| ≤ dk) and currently assigned to class k (c(xj) = k).
| (4) |
As a result, the prior probabilities P(k) in Eqn. 1 become spatially varying as they are calculated from both the atlas and the neighborhood of the voxel.
| (5) |
3 Results
High-resolution 3D volumes reconstructed from the subject scans were non-rigidly registered to the average anatomy model described in Section 2.3. The number of classes, K = 6, was selected to cover four types of brain tissue (GM, GMAT, WM, CSF) and two types of non-brain voxels (the fluid around the brain and the skull). Class probabilities p(k|yi) for brain tissues were initialized with values from the atlas and used to calculate initial estimates of intensity distribution parameters {μk, σk} according to Eqn. 2. The values of μk and σk for the two non-brain classes were copied from estimates for GM and CSF, respectively.
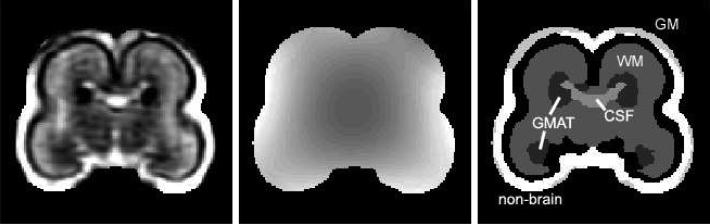
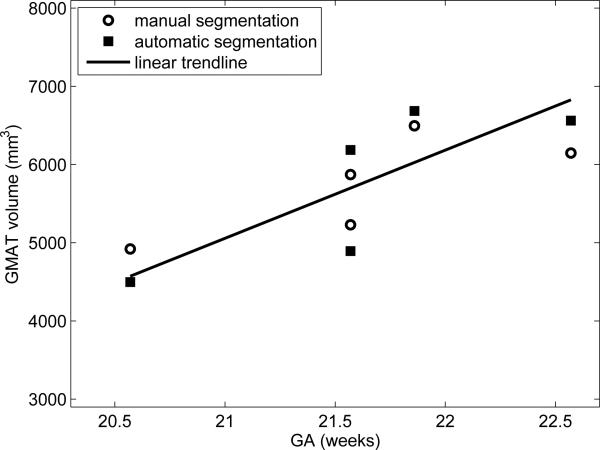
The segmentation was confined to regions where the atlas indicated the probability of any brain tissue to be larger than 0.01 as shown in Fig. 3. For each volume, 20 steps of the EM-based segmentation algorithm were performed as no significant changes were observed afterwards. Volumes of brain tissue were originally measured in the reference/atlas space and then corrected for the non-rigid transformation to achieve actual volumes in the original space of each subject. Additionally, for two subjects of different gestational age (20.57 and 22.57 weeks), the germinal matrix was manually traced by a field expert. The resulting tissue volumes and the relative difference between GMAT volumes obtained from automatic and manual segmentation are reported in Table 1.
Fig. 3.
A coronal view of the original MRI (left), bias field estimation (center) and final segmentation (right) for subject C.
Table 1.
Volumes of brain tissues (in mm3) for fetal subjects of different gestational age (GA, in weeks) obtained from automatic and manual segmentation.
| Fetus | GA | automatic segmentation | man. seg. | rel. diff. | |||
|---|---|---|---|---|---|---|---|
| CSF | GM | WM | GMAT | GMAT | GMAT | ||
| A | 20.57 | 3155 | 13436 | 19287 | 4497 | 4920 | 8.6% |
| B | 21.57 | 2831 | 13985 | 19555 | 4892 | 5229 | 6.4% |
| C | 21.57 | 3588 | 18543 | 26549 | 6186 | 5872 | 5.3% |
| D | 21.86 | 3223 | 20523 | 26215 | 6685 | 6495 | 2.9% |
| E | 22.57 | 3894 | 18834 | 25339 | 6561 | 6146 | 6.7% |
4 Conclusions
In this paper we have described the development of the first in utero 3D MRI atlas of the human fetus which incorporates both developing and developed tissue classes. The approach builds on earlier work on 3D fetal image reconstruction techniques from clinical MRI data, making use of standard clinical protocols. This initial work has focussed on a narrow age range capturing the early brain development visible using MRI, with particular focus on extracting the germinal matrix, which is a key area of cell formation in the developing brain.
We have created an unbiased intensity and shape average template from a set of example MRI studies and formed an ideal segmentation of tissue classes in this reference anatomy. This is then used as a target for spatial normalization of individual scans to provide spatial priors for an EM-based segmentation scheme. We have automatically delineated 3D structures in the developing brain from clinically acquired MR images and have calculated volumes for both developed and transient tissue types.
The presented study will be further extended by analysis of other WM classes such as the cortical plate, incorporating manual tracing of multiple subjects in the probabilistic atlas and forming multiple atlases for different developmental stages.
Fig. 4.
The volume of germinal matrix (GMAT) for different gestational age (GA).
References
- 1.Coakley FV, Glenn OA, Qayyum A, Barkovich AJ, Goldstein R, Filly RA. Fetal MRI: a developing technique for the developing patient. Am. J. Roentgenol. 2004;182(1):243–252. doi: 10.2214/ajr.182.1.1820243. [DOI] [PubMed] [Google Scholar]
- 2.Prayer D, Kasprian G, Krampl E, Ulm B, Witzani L, Prayer L, Brugger PC. MRI of normal fetal brain development. Eur. J. Radiol. 2006;57(2):199–216. doi: 10.1016/j.ejrad.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Huppi PS, Warfield SK, Kikinis R, Barnes PD, Zientara GP, Jolesz FA, Tsuji MK, Volpe JJ. Quantitative magnetic resonance imaging of brain development in premature and mature newborns. Ann. Neurol. 1998;43(2):224–235. doi: 10.1002/ana.410430213. [DOI] [PubMed] [Google Scholar]
- 4.Grossman R, Hoffman C, Mardor Y, Biegon A. Quantitative MRI measurements of human fetal brain development in utero. Neuroimage. 2006;33(2):463–470. doi: 10.1016/j.neuroimage.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich AJ, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad. Radiol. 2006;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans. Med. Imaging. 2007;26(7):967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- 7.Kim K, Hansen MF, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Proc. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. Paris, France: 2008. Intersection-based registration of slice stacks to form 3d images of the human fetal brain. pp. 1167–1170. [Google Scholar]
- 8.Kostovic I, Judas M, Rados M, Hrabac P. Laminar organization of the human fetal cerebrum revealed by histochemical markers and magnetic resonance imaging. Cereb. Cortex. 2002;12(5):536–544. doi: 10.1093/cercor/12.5.536. [DOI] [PubMed] [Google Scholar]
- 9.Battin MR, Maalouf EF, Counsell SJ, Herlihy AH, Rutherford MA, Azzopardi D, Edwards AD. Magnetic resonance imaging of the brain in very preterm infants: visualization of the germinal matrix, early myelination, and cortical folding. Pediatrics. 1998;101(6):957–962. doi: 10.1542/peds.101.6.957. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita Y, Okudera T, Tsuru E, Yokota A. Volumetric analysis of the germinal matrix and lateral ventricles performed using MR images of postmortem fetuses. Am. J. Neuroradiol. 2001;22(2):382–388. [PMC free article] [PubMed] [Google Scholar]
- 11.Prastawa M, Gilmore JH, Lin W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9(5):457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Murgasova M, Dyet L, Edwards D, Rutherford M, Hajnal J, Rueckert D. Segmentation of brain MRI in young children. Acad. Radiol. 2007;14(11):1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognit. 1999;32(1):71–86. [Google Scholar]
- 14.Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Proc. Information Processing in Medical Imaging. Brest, France: 1995. Automated multi-modality image registration based on information theory. pp. 263–274. [Google Scholar]
- 15.Viola P, Wells WM. Alignment by maximization of mutual information. Int. J. Comput. Vis. 1997;24(2):137–154. [Google Scholar]
- 16.Guimond A, Meunier J, Thirion JP. Average brain models: a convergence study. Comput. Vis. Image Underst. 2000;77(2):192–210. [Google Scholar]
- 17.Wells WM, Grimson WEL, Kikinis R, Jolesz FA. Adaptive segmentation of MRI data. IEEE Trans. Med. Imaging. 1996;15(4):429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- 18.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Trans. Med. Imaging. 1999;18(10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 19.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imaging. 1999;18(10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]