Abstract
Automatic segmentation using multi-atlas label fusion has been widely applied in medical image analysis. To simplify the label fusion problem, most methods implicitly make a strong assumption that the segmentation errors produced by different atlases are uncorrelated. We show that violating this assumption significantly reduces the efficiency of multi-atlas segmentation. To address this problem, we propose a regression-based approach for label fusion. Our experiments on segmenting the hippocampus in magnetic resonance images (MRI) show significant improvement over previous label fusion techniques.
1. Introduction
Atlas-based segmentation methods label an unknown image by referring to a labeled image through deformable registration. Due to its wide applicability, i.e., one can apply it to segment any anatomical structures by registering it to an atlas, and the wide availability of registration tools, atlas-based segmentation has been one of the most popular techniques used in medical image analysis. As an extension, multi-atlas based segmentation makes use of more than one reference atlas to compensate potential errors imposed by using any single atlas. As extensive empirical studies have verified in the recent literature, e.g. [9, 2], it is more accurate than single atlas based segmentation.
Errors in atlas-based segmentation can be attributed to dissimilarity in anatomy and/or appearance between the atlas and the target image. Recent research has been focusing on addressing this problem. For instance, research has been done on optimally constructing a single atlas from training data that is the most representative of a population [11]. Constructing multiple representative atlases from training data has been considered as well, and usually works better than single-atlas based approaches. Multi-atlas construction can be done either by constructing one representative atlas for each mode obtained from clustering training images [1] or by simply selecting the most relevant atlases for the unknown image on the fly [18]. Either way, one needs to combine the segmentation results obtained by referring to different atlases to produce the final solution.
To combine segmentations, most label fusion techniques implicitly assume that different atlases produce uncorrelated errors with respect to the target segmentation [13]. Under this assumption, according to the law of large numbers, errors produced by using any single atlas can be efficiently reduced when multiple atlases are used. Furthermore, when different atlases produce different segmentation accuracies, more efficiency can be achieved by assigning higher non-negative weights to atlases that are expected to produce more accurate results. In this regard, image similarity-based local weighting has been shown to be the most accurate label fusion strategy [2, 21]. Although the un-correlated error assumption significantly simplifies the label fusion problem, it is often invalid in practice [9]. To address this problem, our main contribution is to propose a regression technique for label fusion.
We apply our method to segment the hippocampus from MRI and show significant improvements over similarity-based local weighting label fusion.
2. Label fusion based multi-atlas segmentation
Atlas-based segmentation is motivated by the observation that segmentation strongly correlates with image appearance. A target image can be segmented by referring to labeled images that have similar image appearance. Accordingly, an atlas contains a reference image and its segmentation. After registering an atlas’s reference image to the target image via deformable registration, one can directly transfer labels from the atlas to the target image.
Segmentation errors produced by this method are mainly due to registration errors, i.e. that the registration associates wrong regions from an atlas to the target image. Under the assumption that the segmentation errors produced by using different atlases are uncorrelated, errors obtained from any single atlas can be efficiently corrected when multiple atlases are used. For example, the majority voting method [7, 12] simply counts the vote for each label from each registered atlas and chooses the label receiving the most votes.
Let TF be a target image and be n registered atlases. and denote the ith warped atlas image and the corresponding warped manual segmentation. The majority voting algorithm produces the final segmentation T̂S by:
| (1) |
where l indexes through labels and L is the number of labels. x indexes through image pixels. I(·) is an indicator function that outputs 1 if the input is true and 0 otherwise.
Majority voting makes a strong assumption that different atlases produce equal quality registrations for the target image. To address this problem, recent work focuses on developing more accurate registration quality estimations. In a broader context, most of these label fusion techniques can be modeled under the maximum a posterior (MAP) inference framework [21]. The posterior label probability given the target image can be estimated by:
| (2) |
p(l|Ai, x) is the label posterior probability defined by atlas Ai. Typically, for deterministic atlases that have one unique label for every image location, p(l|Ai, x) is 1 if and 0 otherwise. Continuous label posterior probability can be used as well especially when probabilistic atlases are involved. Even for deterministic atlases, continuous label posterior probability still can be derived, see [21] for some examples. p(Ai|TF, x) is the probability that atlas Ai has the correct label for TF at location x, which can be interpreted as a weight assigned to the atlas. As mentioned above, this term mainly reflects the quality of registration. One way to estimate this probability is based on local image similarity. When summed square distance (SSD) and a Gaussian model is used [21], it can be estimated by:
| (3) |
where
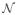 (x) defines a neighborhood centered around x. In our experiment, we use a cubic neighborhood definition, specified by a radius r. The radius specifies the Manhattan distance from the center of the cubic region, i.e. the voxel being considered, to the neighborhood boundary. Hence, an image patch contains (2r + 1)3 voxels. Note that the registration quality is estimated locally, which accommodates nonuniform registration qualities over the entire image. The inverse distance weighting has been applied to estimate this term as well [2]:
(x) defines a neighborhood centered around x. In our experiment, we use a cubic neighborhood definition, specified by a radius r. The radius specifies the Manhattan distance from the center of the cubic region, i.e. the voxel being considered, to the neighborhood boundary. Hence, an image patch contains (2r + 1)3 voxels. Note that the registration quality is estimated locally, which accommodates nonuniform registration qualities over the entire image. The inverse distance weighting has been applied to estimate this term as well [2]:
| (4) |
where σ and β are model parameters.
To reduce the noise effect, one can spatially smooth the weights for each atlas. In our experiment, we use mean filter smoothing with the smoothing window
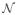 , the same neighborhood used for computing the similarity-based weights. After smoothing, the weights are re-normalized s.t. for any x,
.
, the same neighborhood used for computing the similarity-based weights. After smoothing, the weights are re-normalized s.t. for any x,
.
3. Problems raised by violating the assumption of uncorrelated errors
Given a certain registration accuracy at location x, the label posterior probability produced by any single warped atlas can be modeled as obtained from the true label posterior probability plus some error [24], i.e.:
| (5) |
where
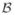 (Ai, x) is the bias and ε(Ai, x) is a zero-mean random error. Averaging over all registrations that produce the same error distribution, the error produced by Ai at x can be quantified by:
(Ai, x) is the bias and ε(Ai, x) is a zero-mean random error. Averaging over all registrations that produce the same error distribution, the error produced by Ai at x can be quantified by:
| (6) |
where V is the variance of the random error. After combining multiple atlases, the combined error is:
| (7) |
The second term measures the error correlation between atlases. Under the assumption that the errors produced by different atlases are uncorrelated, the correlation terms disappear. By properly assigning larger weights to better registered atlases, one can significantly reduce uncorrelated errors through label fusion. For instance, if all atlases produce equal quality label probabilities with the same error e, applying uniform weights, , produces the combined error e.
In reality, segmentation errors produced by different atlases are often correlated. When the errors produced by two atlases are negatively correlated, i.e. they tend to make opposite mistakes, their correlation term is negative. Combining them is even more effective in reducing errors. The problem arises when the errors are positively correlated, i.e. different atlases tend to make similar errors. Intuitively, when several atlases produce the same wrong label, it is easier for the wrong label to accumulate higher posteriors than the correct label, therefore causing errors. Theoretically, when different atlases tend to make similar errors, their error correlation terms are positive, which increases the combined error (see (7)). In the worst case, when all error correlation terms are positive and are greater than the error produced by the best single atlas, for the best label fusion result, zero weights have to be applied to all atlases except the best one. The multi-atlas method reduces to a single-atlas approach.
4. Regression-based label fusion
When errors are positively correlated, simply combining atlases by assigning higher weights to better registered atlases becomes inefficient. To address this problem, we propose to estimate the posterior label probability through regression.
Given the image appearance of a target image at a local patch
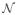 (x), our goal is to estimate the label posterior distribution at x, i.e. p(l|TF(
(x), our goal is to estimate the label posterior distribution at x, i.e. p(l|TF(
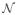 (x))). This is a high dimensional function of local appearance. Each registered atlas provides one observed value for this function
at one data point
. One common way to estimate a function’s values at unobserved data points is interpolation or regression analysis. Note that the image similarity-based weighting methods can be interpreted as applying the nearest neighbor interpolation [19]. However, as we show above, nearest neighbor interpolation does not properly handle correlated errors.
(x))). This is a high dimensional function of local appearance. Each registered atlas provides one observed value for this function
at one data point
. One common way to estimate a function’s values at unobserved data points is interpolation or regression analysis. Note that the image similarity-based weighting methods can be interpreted as applying the nearest neighbor interpolation [19]. However, as we show above, nearest neighbor interpolation does not properly handle correlated errors.
Since we work on high dimensional regression problems, over-fitting is a key issue to be addressed. Hence, we study low order polynomial regression for label fusion in this paper. To this end, we model the label posterior probability as a second order polynomial function of image intensities from the local patch:
| (8) |
and are first order and second order weights, respectively. Since the regression model is anatomy dependent, the regression weights may be different for different locations. For a simple notation, we rewrite the equation:
| (9) |
and T = [TF(y1); …; TF(ym);TF(y1)2; …; TF(ym)2], where yi ∈
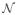 (x) for i = 1, … m and m is the size of the local image patch
(x) for i = 1, … m and m is the size of the local image patch
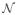 (x). t stands for matrix transpose.
(x). t stands for matrix transpose.
Using the observations obtained from the registered atlases, one can estimate β by solving the following linear equations:
| (10) |
A = [AF; (AF)2] and is the matrix of the local appearances from the registered atlases. is a vector of size m × 1 that stores the local image intensity patch from . Pl = [p(l|A1, x); …; p(l|An, x)] is the vector of the corresponding label posterior probabilities from all atlases. When enough atlases are used, we can solve β via linear least square fitting:
| (11) |
Note that the matrix AAt has dimension 2m × 2m. In practice, the number of atlases used are usually limited. For instance, for small image patches with dimension 3 × 3× 3 or 5 × 5 ×5, the patch size is usually greater than the number of atlases used for label fusion. In such cases, the matrix does not have a full rank and the matrix inverse is not well defined. To address this problem, we propose the following approximation to implement the regression analysis.
| (12) |
| (13) |
| (14) |
(14) is obtained by substituting (10). When n = 2m, A(AtA)−1At is an identity matrix. (14) gives the accurate solution. When n < 2m, (14) approximates the label posterior probability via least square fitting. With this approximation, we transfer the problem of solving inverse of a large matrix [AAt]2m×2m into solving inverse of a smaller matrix [AtA]n×n. If this smaller matrix is still near singular, one can add an identity matrix weighted by a small positive number λ to it. Adding a weighted identity matrix can be interpreted as penalizing large weights that exploit correlations beyond some level of precision in the data sampling process [20].
In (14), each registered atlas has one single weight computed by wn×1 = (AtA)−1AtT, i.e.:
| (15) |
Hence, as previous label fusion techniques, our regression-based method applies weighted-voting as well. One main difference is that the weights computed by our method can be either positive or negative, while the weights used by other similarity-based weighting approaches are nonnegative. When the errors produced by different atlases are positively correlated, applying negative weights to some of the atlases allows to cancel out the positively correlated errors between these negatively weighted atlases and other positively weighted atlases, which results in smaller combined error (see equation (7)). Furthermore, most similarity based weighting methods rely on some pre-selected weighting models, e.g. (3). The optimal model parameter, e.g. the standard deviation in the Gaussian model, is usually determined empirically. By contrast, our method automatically determines the optimal weights.
To see that our approximation applies linear least square fitting, note that w is the linear least square fitting solution of the following linear equations when n < 2m:
| (16) |
We compute weights such that both the original target image and its squared image can be linearly interpolated by the warped atlas reference images and their squared images, respectively. In other words, the weighted local appearance of the warped atlases center around the local appearance of the target image. Since atlas-based segmentation relies on the assumption that segmentations are strongly correlated with appearances, putting the local target image at the center of the weighted warped local atlas images can be interpreted as an effort of putting the local segmentation of the target image at the center of the weighted warped local atlas segmentations, therefore the weighted segmentation errors are uncorrelated.
4.1. Error analysis
Since we attempt to model the relationship between image appearance and segmentation labels, the errors produced by our method can be categorized into two classes: 1) the appearance-label model selection error, i.e., the errors caused by employing the polynomial function to model the appearance-label relationship and 2) the model fitting error, i.e., the target image can not be perfectly represented as a linear combination of the atlas reference images.
The model fitting errors usually can be alleviated by employing enough atlases for label fusion. The actual number of atlases required for accurate model fitting depends on the complexity of the appearance-label model. Typically, more complex appearance-label models require more atlases. Hence, when limited atlases are used, to avoid significant model fitting errors one should use simple appearance-label models, like ours. However, segmentation labels usually correlate to image appearances in a more ambiguous fashion. Due to the factors such as the limitations in the imaging process and employing local image appearance for label fusion, most local appearance patterns may have more than one plausible segmentation and vice versa. Under these circumstances, the performance of our method also depends on how well the polynomial model captures the relationship between appearances and segmentation labels.
4.2. A toy example
Fig. 1 demonstrates applying our regression approach to a toy example in 2D. In this binary segmentation problem, we have five atlases. After registration, suppose that the foreground label patterns of the target image and the warped atlases on a local patch with size 3 × 3 have the structures shown in Fig. 1. For simplicity, we consider deterministic atlases. The label posterior produced by each atlas is either 1 or 0. The registration quality is assumed to be constant within the patch. Furthermore, suppose that the image appearances have the same patterns as the foreground labels such that image appearances indeed linearly correlate to segmentation labels.
Figure 1.
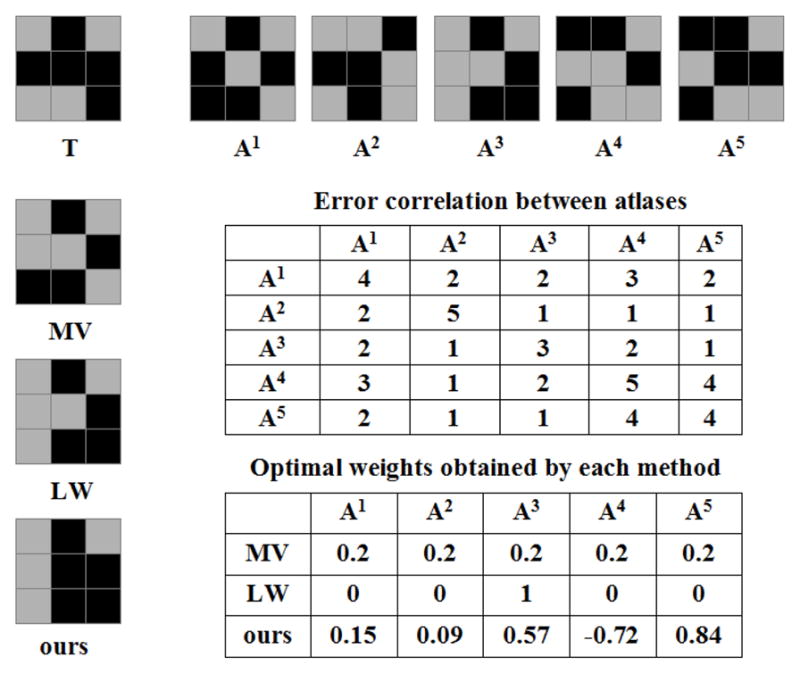
Illustration of label fusion on a toy example. The target segmentation, T, is shown on the top left corner, followed by five registered atlases, A1 to A5. For simplicity, we use binary label posteriors and assume that the images have the same appearance patterns as the segmentations. Error correlations are all positive, indicating a strong positive correlation between the label errors produced by the atlases. As a result, majority voting (MV) produces a result biased towards atlas A1 and A4. Similarity-based Gaussian weighting label fusion (LW) reduces to single-atlas segmentation, i.e. only the most similar atlas, A3, has a non-zero weight. To compensate the overall bias towards A4, when linear regression is applied, only A4 receives a negative weight to cancel out the consistent errors among all atlases. For LW and our method, the atlas weights computed for the center pixel are also used for non-center pixels in this example.
The pairwise correlations of label posterior errors between the atlases are all positive, indicating a consistent bias in the atlases. Also note that A1 and A4 have the largest combined inter-atlas error correlations, indicating that they contain the most common segmentation errors produced by all atlases. As a result, the segmentation obtained from majority voting has a structure more similar to A1 and A4, with five mislabeled pixels. Due to the strong error correlation, the optimal image similarity based weighting label fusion approach reduces to the single-atlas based segmentation, i.e. applying zero weights to all atlases except the most similar atlas A3. Applying our regression technique, only atlas A4 receives a negative weight. This allows to cancel some of the consistent errors among all the atlases. Correspondingly, our method gives the best solution on this local patch with two mislabeled pixels. Since the polynomial model accurately captures the appearance-label relationship for this example, the errors are purely due to the model fitting error.
4.3. Improving the robustness of model fitting
Since we face a high dimensional regression problem, the computed weights may be sensitive to outliers/noises caused by registration errors. In this section, we propose two techniques to reduce the model fitting error.
4.3.1 Similarity-rankig based regularization
Note that our regression approach represents the target image as a linear combination of the registered atlas images. It is reasonable to expect that the atlases whose reference images are more similar to the target image will contribute differently from those whose reference images are less similar to the target image. Such similarity-ranking based weighting strategy has been explored in the classifier combining literature, [10, 16]. To alleviate the over-fitting problem, we propose to accommodate this prior knowledge to regularize our regression weights. To this end, we rewrite equation (16) as follows:
| (17) |
where π(1), …, π(n) is a permutation of 1, …, n.
is the appearance vector of the ith most similar atlas to the target image T = [TF(
 (x)); TF(
(x)); TF(
 (x))2], measured by SSD. Now, the regression weights assigned to atlases are associated with their similarity rankings.
(x))2], measured by SSD. Now, the regression weights assigned to atlases are associated with their similarity rankings.
To estimate these similarity-ranking based weights, again we use the atlases. Since all atlases are registered to the same target image, these registrations also establish the pairwise correspondence between the atlases via the target image. Hence, the image patches TF(
 (x)) and
for i = 1, …, n all represent the same anatomy structure. Following the logic that one can estimate the label posterior probability for the target image using the atlases, given the label posterior probability of the target image, we should be able to regress the label posterior probability for any atlas using the target image and the remaining atlases as well. For this purpose, we still need represent the warped atlas reference image as a linear combination of the target image and the remaining warped atlas reference images. For a simpler notation, let the appearance vectors of the target image and the warped atlases be represented by F1, …, Fn+1. Similar to equation (17), any image Fj can be represented as a similarity-ranking based linear combination of the remaining images as follows:
(x)) and
for i = 1, …, n all represent the same anatomy structure. Following the logic that one can estimate the label posterior probability for the target image using the atlases, given the label posterior probability of the target image, we should be able to regress the label posterior probability for any atlas using the target image and the remaining atlases as well. For this purpose, we still need represent the warped atlas reference image as a linear combination of the target image and the remaining warped atlas reference images. For a simpler notation, let the appearance vectors of the target image and the warped atlases be represented by F1, …, Fn+1. Similar to equation (17), any image Fj can be represented as a similarity-ranking based linear combination of the remaining images as follows:
| (18) |
where Fπj(i) is the ith most similar image to Fj from the remaining images and πj(1), …, πj(n) is a permutation of 1, …, j − 1, j + 1, …, n + 1.
Under this formulation, each image provides a set of linear constraints for solving the weights w. Simultaneously solving all these linear equations produces the desired weights. The main advantage of employing similarity-ranking based weights is that it significantly increases the training data through a cross validation process, therefore the results are less sensitive to noises.
4.3.2 Reducing extrapolation via local searching
When the warped atlas images are scattered around the target image, the target image is located within the reference range defined by the atlases. The posterior label distribution can be reliably estimated via interpolating label distributions obtained from the atlases. When the warped atlases are strongly biased, most warped atlases deviate from the target segmentation in a consistent way and the target image may be completely outside the reference range. Our method is more akin to extrapolation. Extrapolation is based on a strong assumption that the fitted regression model is still valid outside the reference range defined by the training samples. Hence, extrapolation is usually more prone to errors than interpolation. To alleviate such unreliability caused by extrapolation, we propose to use the most similar image patches from each atlas for label fusion.
Note that the goal of image-based registration is to correspond the most similar image patches between the registered images. However, the correspondence obtained from registration may not give the maximal similarity between all corresponding regions. For instance, deformable image registration usually needs to balance the image matching constraint and the regularization prior on deformation fields. A global regularization constraint on the deformation fields is necessary to clarify the ambiguous appearance-label relationship arising from employing small image patches for matching. However, enforcing a global regularization constraint on the deformation fields may compromise the local image matching constraint. In such cases, the correspondence that maximizes the appearance similarity between the warped atlas and the target image may be within a small neighborhood of the registered correspondence.
Motivated by this observation, instead of using the original registered correspondence, we propose to remedy the risk of extrapolation by searching for the correspondence, that gives the most similar appearance matching, i.e. minimal SSD distance, within a small neighborhood centered around the registered correspondence in each atlas. The locally searched optimal correspondence is:
| (19) |
xi is the location from ith warped atlas with the best image matching for location x in the target image within the local area
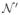 (x). Again, we use a cubic neighborhood definition, specified by a radius rs.
(x). Again, we use a cubic neighborhood definition, specified by a radius rs.
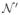 and
and
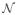 may represent different neighborhoods and they are the only free parameters in our method. Instead of the registered corresponding patch Ai(
may represent different neighborhoods and they are the only free parameters in our method. Instead of the registered corresponding patch Ai(
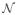 (x)), we apply the searched patch Ai(
(x)), we apply the searched patch Ai(
 (xi)) to produce the fused label at x for the target image, i.e. (2) becomes
. Note that a similar local searching technique was recently proposed by [5] to reduce noise effects for similarity-based local weighting label fusion.
(xi)) to produce the fused label at x for the target image, i.e. (2) becomes
. Note that a similar local searching technique was recently proposed by [5] to reduce noise effects for similarity-based local weighting label fusion.
With more similar data for regression, the local searching can significantly reduce the risk of extrapolation. In this regard, larger searching neighborhoods are more desirable. However, using larger local searching windows also compromises the regularization prior on the deformation fields. This drawback makes the appearance-label relationship more ambiguous on local patches, resulting greater model selection errors. Hence, avoiding model-fitting errors and avoiding model-selection errors can not be satisfied simultaneously. It is reasonable to expect an optimal searching range that balances these two factors.
5. Experiments
In this section, we apply our method to segment the hippocampus using T1-weighted MRI. The hippocampus plays an important role in memory function. Macroscopic changes in brain anatomy, detected and quantified by magnetic resonance imaging (MRI), consistently have been shown to be predictive of Alzheimer’s disease (AD) pathology and sensitive to AD progression [22]. Accordingly, automatic hippocampus segmentation from MR images has been widely studied.
We use the data in the Alzheimer’s Disease Neuroimaging Initiative (ADNI, www.loni.ucla.edu/ADNI). Our study is conducted using 3 T MRI and only includes data from mild cognitive impairment (MCI) patients and controls. Overall, the data set contains 139 images (57 controls and 82 MCI patients). The images are acquired sagitally, with 1 × 1 mm in-plane resolution and 1.2 mm slice thickness. To obtain manual segmentation, we first apply a landmark-guided atlas-based segmentation method [17] to produce the initial segmentation for each image. Each fully-labeled hippocampus is manually edited by one of the authors following a previously validated protocol [8].
For cross-validation evaluation, we randomly select 20 images to be the atlases and another 20 images for testing. Image guided registration is performed by the Symmetric Normalization (SyN) algorithm implemented by ANTS [3], which was a top performer in a recent evaluation study [14], between each pair of the atlas reference image and the testing image. The cross-validation experiment is repeated 10 times. In each cross-validation experiment, a different set of atlases and testing images are randomly selected from the ADNI dataset.
We focus on comparing with similarity-based local weighting methods, which are shown to be the most accurate label fusion methods in recent experimental studies, e.g. [2, 21]. We use majority voting (MV) and the STAPLE algorithm [23] to define the baseline performance. For each method, we use binary label posteriors obtained from the deterministic atlases. For similarity-based label fusion, we apply Gaussian weighting (3) (LWGau) and inverse distance weighting (4) (LWInv).
Our method has two parameters, r for the local appearance window used in regression analysis, rs for the local searching window used in reducing the risk of extrapolation. For each cross-validation experiment, the parameters are optimized by evaluating a range of values (r ∈ {1, 2, 3, 4}; rs ∈ {0, 1, 2, 3}) using the atlases in a leave-one-out cross-validation strategy. We measure the average overlap between the automatic segmentation of each atlas obtained via the remaining atlases and the reference segmentation of that atlas, and find the optimal parameters that maximize this average overlap. Similarly, The optimal local searching window and local appearance window are determined for LWGau and LWInv as well. In addition, the optimal model parameters are also determined for LWGau and LWInv, with the searching range σ ∈ [0.005, 0.1, …, 1] and β ∈ [0.5, 1, …, 10], respectively.
For robust image matching, instead of using the raw image intensities, we normalize the intensity vector obtained from each local image intensity patch such that the normalized vector has zero mean and unit variance. To reduce the noise effect, we spatially smooth the weights computed by each method for each atlas. We use mean filter smoothing with the smoothing window
 , the same neighborhood used for local appearance patches.
, the same neighborhood used for local appearance patches.
Fig. 2 shows some parameter selection experiments for LWGau in the first cross-validation experiment. The results are quantified in terms of Dice overlaps [6] between automatic and reference segmentations of the atlases. The Dice overlap between two regions, A and B, measures the volume consistency as . For this cross-validation experiment, the optimal parameters for LWGau are σ = 0.05, r=2, rs = 2. Note that local searching only slightly improves the performance for LWGau. Similar results are observed for LWInv as well.
Figure 2.
Visualizing some of the parameter selection experiments for LWGau using leave-one-out on the atlases for the first cross-validation experiment. The figures show the performance of LW-Gau with respect to the Gaussian weighting function (left), local appearance window (middle) and local searching window (right), respectively when the other two parameters are fixed (the fixed parameters are shown in the figure’s title).
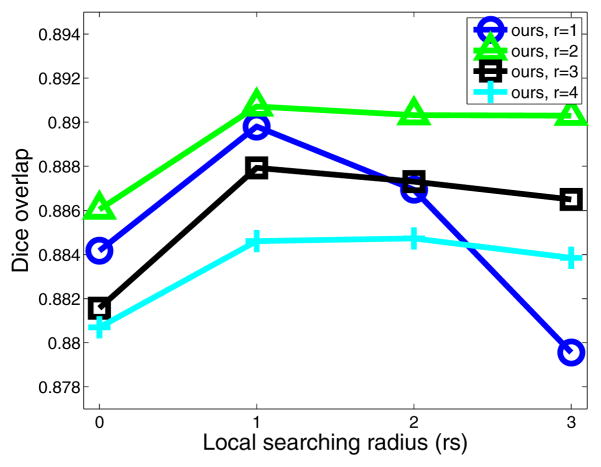
For our regression-based label fusion, we apply a conditioning identity matrix with weight λ=0.01. Fig. 3 shows the performance of our method when applied on the atlases in a leave-one-out fashion in the first cross-validation experiment. Comparing to LWGau, local searching yields more improvement for our method. Most improvement is obtained by applying searching windows with rs = 1. This result indicates that by using more similar patches for regression, local searching significantly reduces the risk of extrapolation. Overall, our method produces ~ 1% Dice improvement over LWGau and LWInv on the atlases in this cross-validation experiment.
Figure 3.
Leave-one-out performance by our method on the atlases for the first cross-validation experiment when different appearance and searching windows are used.
As discussed above, although using larger local searching neighborhoods reduces the chance of extrapolation, it also increases the ambiguity in the appearance-label relationship. For small image patches with r = 1, the appearance-label relationship is the most ambiguous. Good appearance matchings using small patches do not necessarily indicate good label matchings. Applying larger local searching windows makes the appearance-label relationship even more ambiguous. Hence, using large local searching windows with rs > 1 significantly reduces the performance. Using larger image patches reduces ambiguity in the appearance-label relationship, i.e., good appearance matchings using large patches are more likely to indicate good label matchings. Hence, using larger patches we can afford larger local searching windows. Note that applying larger appearance windows yields smoother local appearance similarity variations, therefore results in smoother local weights for label fusion. Over-smoothing the local weights for label fusion by using larger local appearance windows reduces the label fusion accuracy.
Table 1 shows the results produced by each method. Overall, LWGau and LWInv produce similar results, both significantly outperform majority voting and the STAPLE algorithm. Our method outperforms similarity based local weighting approaches by 0.7% of Dice overlap, which is substantial because when segmenting from scratch, our manual rater produces average intra-rater segmentation overlap of 0.90. Our improvement is statistically significant as well, with p < 0.0001 on the paired Student’s t-test for each cross-validation experiment. Fig. 4 shows some segmentations produced by LWGau and our method.
Table 1.
Results in terms of Dice overlap produced by each method.
| method | left | right |
|---|---|---|
| MV | 0.836±0.084 | 0.829±0.069 |
| STAPLE | 0.846±0.086 | 0.841±0.086 |
| LWGau | 0.886±0.027 | 0.875±0.030 |
| LWInv | 0.885±0.027 | 0.873±0.030 |
| LWReg | 0.892±0.025 | 0.882±0.028 |
Figure 4.
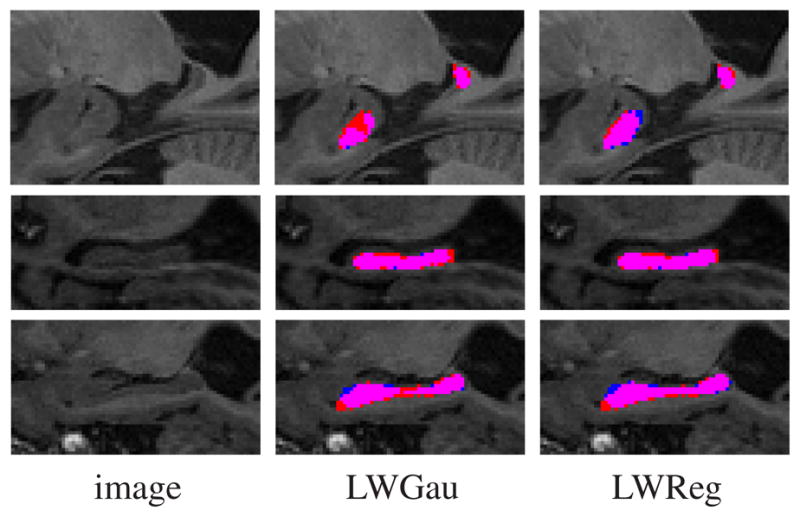
Sagittal views of hippocampus segmentations produced by LWGau and our method. Red: manual; Blue: automatic; Pink: overlap between manual and automatic segmentation.
Comparing to the state of the art [4, 5, 15] present the highest published hippocampus segmentation results. All these methods are based on similarity-based local weighting label fusion. The experiments in [4, 5] are conducted in a leave-one-out strategy on data set containing 80 control subjects. They report average Dice overlaps of 0.887 and 0.884 respectively. For controls, we produce Dice overlap of 0.896 ± 0.021. [15] uses a template library of 55 atlases. However, for each test image, both the original image and its flipped mirror image are used. Hence, [15] effectively uses 110 atlases for label fusion. [15] reports results in Jaccard index ( ) for the left side hippocampus of 10 controls, 0.80 ± 0.03, and 10 MCI patients, 0.81 ± 0.04. Our results for the left hippocampus are0.820 ± 0.033 for controls and 0.795 ± 0.042 for MCI patients. Overall, using significantly fewer atlases, we produce results compare favorably to the state-of-the-art.
6. Conclusions
We proposed a regression-based label fusion technique. Unlike previous image similarity based local weighting techniques, our method does not assume that the segmentation errors produced by different atlases are uncorrelated. To ensure robust regression in high dimensional space, we proposed a similarity-ranking based regularization technique and a local searching technique. To validate our method, we conducted segmentation experiments on a hippocampus segmentation problem. In our experiment, our method significantly outperformed the state of the art label fusion technique, the similarity-based local weighting label fusion method. Using significantly fewer atlases, our hippocampus segmentation results still compare favorably to the state of the art in published work.
References
- 1.Allassonniere S, Amit Y, Trouve A. Towards a coherent statistical framework for dense deformable template estimation. Journal of the Royal Statistical Society: Series B. 2007;69(1):3–29. [Google Scholar]
- 2.Artaechevarria X, Munoz-Barrutia A, de Solorzano CO. Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Tran Medical Imaging. 2009;28(8):1266–1277. doi: 10.1109/TMI.2009.2014372. [DOI] [PubMed] [Google Scholar]
- 3.Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins D, Pruessner J. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuro Image. 2010;52(4):1355–1366. doi: 10.1016/j.neuroimage.2010.04.193. [DOI] [PubMed] [Google Scholar]
- 5.Coupe P, Manjon J, Fonov V, Pruessner J, Robles N, Collins D. Nonlocal patch-based label fusion for hippocampus segmentation. MICCAI. 2010 doi: 10.1007/978-3-642-15711-0_17. [DOI] [PubMed] [Google Scholar]
- 6.Dice L. Measure of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 7.Hansen LK, Salamon P. Neural network ensembles. IEEE Trans on PAMI. 1990;12(10):993–1001. [Google Scholar]
- 8.Hasboun D, Chantome M, Zouaoui A, Sahel M, Deladoeuille M, Sourour N, Duymes M, Baulac M, Marsault C, Dormont D. MR determination of hippocampal volume: Comparison of three methods. Am J Neuroradiol. 1996;17:1091–1098. [PMC free article] [PubMed] [Google Scholar]
- 9.Heckemann R, Hajnal J, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuro Image. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 10.Ho TK, Hull J, Srihari S. Decision combination in multiple classifier systems. IEEE Trans on Pattern Analysis and Machine Intelligence. 1994;16(1):66–75. [Google Scholar]
- 11.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphism atlas construction for computational anatomy. Neuro Image. 2004;23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 12.Kittler J. Combining classifiers: A theoretical framework. Pattern Analysis and Application. 1998;1:18–27. [Google Scholar]
- 13.Kittler J, Alkoot F. Sum versus vote fusion in multiple classifier systems. IEEE Trans on PAMI. 2003;25(1):110–115. [Google Scholar]
- 14.Klein A. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuro Image. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung K, Barnes J, Ridgway G, Bartlett J, Clarkson M, Macdonald K, Schuff N, Fox N, Ourselin S. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s Disease. Neuro Image. 2010;51:1345–1359. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melnik O, Vardi Y, Zhang C-H. Mixed group ranks: Preference and confidence in classifier combination. IEEE Trans on PAMI. 2004;26(8):973–981. doi: 10.1109/TPAMI.2004.48. [DOI] [PubMed] [Google Scholar]
- 17.Pluta J, Avants B, Glynn S, Awate S, Gee J, Detre J. Appearance and incomplete label matching for diffeomorphic template based hippocampus segmentation. Hippocampus. 2009;19:565–571. doi: 10.1002/hipo.20619. [DOI] [PubMed] [Google Scholar]
- 18.Rohlfing T, Brandt R, Menzel R, Maurer C. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuro Image. 2004;21(4):1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Rohlfing T, Brandt R, Menzel R, Russakoff DB, CRM Quo vadis, atlas-based segmentation? The Handbook of Medical Image AnalysisVolume III: Registration Models. 2005:435–486. [Google Scholar]
- 20.Roweis S, Saul L. Nonlinear dimensionality reduction bu locally linear embedding. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 21.Sabuncu M, Yeo B, Leemput KV, Fischl B, Golland P. A generative model for image segmentation based on label fusion. IEEE Trans on Medical Imaging. 2010;29(10):1714–1720. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scahill R, Schott J, Stevens J, Fox MRN. Mapping the evolution of regional atrophy in Alzheimer’s Disease: unbiased analysis of fluidregistered serial MRI. Proc Natl Acad Sci U S A. 2002;99(7):4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warfield S, Zou K, Wells W. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans on Medical Imaging. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warfield S, Zou K, Wells W. Validation of image segmentation by estimating rater bias and variance. Philosophical Transactions of the Royal Society. 2008;366(1874):2361–2375. doi: 10.1098/rsta.2008.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]