Abstract
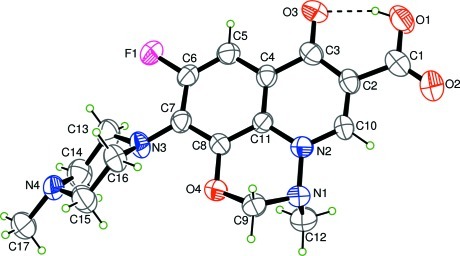
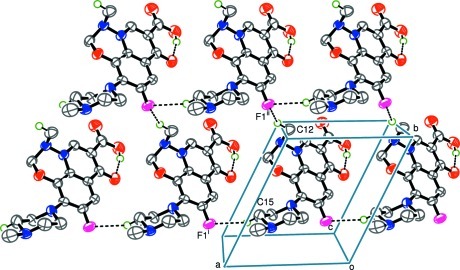
In the title compound, [systematic name: 9-fluoro-2,3-dihydro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-7H-pyrido[1,2,3-ij][1,2,4]benzoxadiazine-6-carboxylic acid], C17H19FN4O4, the carbonyl and carboxyl groups are coplanar with the quinoline ring, making a dihedral angle of 2.39 (2)°. The piperazine ring adopts a chair conformation and the oxadiazinane ring displays an envelope conformation with the CH2 group at the flap displaced by 0.650 (2) Å from the plane through the other five atoms. The molecular structure exhibits an S(6) ring motif, owing to an intramolecular O—H⋯O hydrogen bond. In the crystal, weak C—H⋯F hydrogen bonds link molecules into layers parallel to the ab plane.
Related literature
Marbofloxacin is a third-generation fluoroquinolone for veterinary use, the antimicrobial activity of which depends upon its inhibition of DNA-gyrase and topoisomerase IV (Paradis et al., 2001 ▶; Thomas et al., 2001 ▶; Voermans et al., 2006 ▶). With a broad spectrum bactericidal activity and good efficacy, marbofloxacin is indicated for dermatological, respiratory and urinary tract infections resulting from both Gram-positive and Gram-negative bacteria (Lefebvre et al., 1998 ▶) and Mycoplasma (Spreng et al., 1995 ▶; Dossin et al., 1998 ▶; Carlotti et al., 1999 ▶; Ishak et al., 2008 ▶).
Experimental
Crystal data
C17H19FN4O4
M r = 362.36
Triclinic,
a = 8.0145 (5) Å
b = 8.9218 (6) Å
c = 13.0874 (8) Å
α = 91.65 (3)°
β = 99.65 (3)°
γ = 115.091 (10)°
V = 830.26 (16) Å3
Z = 2
Mo Kα radiation
μ = 0.11 mm−1
T = 296 K
0.31 × 0.13 × 0.03 mm
Data collection
Rigaku RAXIS-RAPID/ZJUG diffractometer
Absorption correction: multi-scan (Higashi, 1995 ▶) T min = 0.956, T max = 0.997
6601 measured reflections
2925 independent reflections
1428 reflections with I > 2σ(I)
R int = 0.052
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.203
S = 1.00
2925 reflections
241 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.29 e Å−3
Δρmin = −0.34 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 2006 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku, 2007 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812009312/nr2019sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812009312/nr2019Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812009312/nr2019Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O3 | 0.83 (3) | 1.77 (2) | 2.560 (4) | 159 (5) |
| C12—H12B⋯F1i | 0.96 | 2.62 | 3.422 (3) | 140 (4) |
| C15—H15B⋯F1ii | 0.97 | 2.54 | 3.446 (3) | 155 (5) |
Symmetry codes: (i) 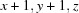 ; (ii)
; (ii) 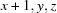 .
.
Acknowledgments
The project was supported by the Zhejiang Provincial Natural Science Foundation of China (J200801).
supplementary crystallographic information
Comment
Marbofloxacin is a third-generation fluoroquinolone for veterinary use, the antimicrobial of which depends upon its inhibition of DNA-gyrase and topoisomerase IV (Paradis et al., 2001; Thomas et al., 2001; Voermans et al., 2006). With a broad spectrum bactericidal activity and good efficacy, marbofloxacin is indicated for dermatological, respiratory and urinary tract infections due to both Gram-positive and Gram-negative bacteria (Lefebvre et al., 1998) and Mycoplasma (Spreng et al., 1995; Dossin et al., 1998; Carlotti et al., 1999; Ishak et al., 2008). But up till now, no single-crystal structure of marbofloxacin has been reported. In the prestent study, we report the crystal structure of marbofloxacin, recrystallized from methanol.
In the crystal structure of marbofloxacin (Fig.1), the carbonyl and carboxyl group are coplanar with the quinoline ring system. The least-squares plane through atoms O1, C1, C2, C3 and O3 is rotated by 2.39 (2)° with respect to the least-scqures plane of quinolinemoiety. The quionline moiety is planar, with the maximum displacement from the least-squares plane being observed for atom C2 [0.020 Å].
The piperazine ring adopts a chair conformation, with the distance of 0.663 (7) Å, -0.662 (7) Å for N4 and N3 to the plane of C13, C14, C15, C16, respectively. The oxadiazinane ring diaplays an envelop conformation. The methyl substituent on N1 is perpendicular to the quinoline moiety, with a C12—N1—N2—C10 torsion angle of -88.8 (4)°.
The carboxyl atom O1 and carbonyl atom O3 is connected by intramolecular hydrogen bond O1—H1···O3 and formed a six-membered ring. Weak intermolecular C15—H15B···F1i[Symmetric code:(i)1 + x,y,z] interaction link molecules into chains along a axis, which is stacked along b axis through another weak intermolecular interaction C12—H12B···F1ii[Symmetric code:(ii)1 + x,1 + y,z].
Experimental
The crude product is supplied by Zhejiang Excel Pharmaceutical Co.,Ltd. It was recrystallized from methanol solution, giving yellow crystal of marbofloxacin suitable for X-ray diffraction.
Refinement
Atom H1 was placed from the difference fourier density and refined free with restraints to the OH bond of O1—H1=0.82 (1) Å. All other H atoms were placed in calculated positions with C—H = 0.93–0.97 Å and included in the refinement in riding model, with Uiso(H) = 1.2Ueq or 1.5Ueq(carrier atom).
Figures
Fig. 1.
Molecular structure of marbofloxacin showing atom-labelling scheme and displacement ellipsoids at 40% probability level. H atoms are shown as small circles of arbitary radii.
Fig. 2.
Part of the crystal packing of Marbofloxacin. Weak Hydrogen bonds are shown as dashed lines. H atoms not involved in hydrogen bonding have been omitted for clarity. Symmetric code: (i)1 + x,y,z; (ii)1 + x,1 + y,z.
Crystal data
| C17H19FN4O4 | Z = 2 |
| Mr = 362.36 | F(000) = 380 |
| Triclinic, P1 | Dx = 1.449 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.0145 (5) Å | Cell parameters from 4017 reflections |
| b = 8.9218 (6) Å | θ = 3.1–27.4° |
| c = 13.0874 (8) Å | µ = 0.11 mm−1 |
| α = 91.65 (3)° | T = 296 K |
| β = 99.65 (3)° | Plates, yellow |
| γ = 115.091 (10)° | 0.31 × 0.13 × 0.03 mm |
| V = 830.26 (16) Å3 |
Data collection
| Rigaku RAXIS-RAPID/ZJUG diffractometer | 2925 independent reflections |
| Radiation source: rolling anode | 1428 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.052 |
| Detector resolution: 10.00 pixels mm-1 | θmax = 25.0°, θmin = 3.1° |
| ω scans | h = −9→9 |
| Absorption correction: multi-scan (Higashi, 1995) | k = −10→10 |
| Tmin = 0.956, Tmax = 0.997 | l = −15→15 |
| 6601 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.203 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.072P)2 + 0.8525P] where P = (Fo2 + 2Fc2)/3 |
| 2925 reflections | (Δ/σ)max < 0.001 |
| 241 parameters | Δρmax = 0.29 e Å−3 |
| 1 restraint | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.2105 (3) | 0.1945 (3) | 0.64667 (19) | 0.0642 (7) | |
| O1 | 0.2871 (5) | 0.7762 (4) | 1.0967 (2) | 0.0680 (9) | |
| H1 | 0.230 (6) | 0.693 (4) | 1.053 (3) | 0.09 (2)* | |
| O2 | 0.5831 (5) | 0.9686 (4) | 1.1352 (2) | 0.0732 (10) | |
| O3 | 0.1921 (4) | 0.5352 (4) | 0.9550 (2) | 0.0594 (8) | |
| O4 | 0.8358 (3) | 0.6152 (3) | 0.7475 (2) | 0.0539 (8) | |
| N1 | 0.9081 (4) | 0.8423 (4) | 0.8761 (3) | 0.0499 (9) | |
| N2 | 0.7233 (4) | 0.7572 (4) | 0.8983 (2) | 0.0455 (8) | |
| N3 | 0.5854 (4) | 0.3404 (4) | 0.6136 (2) | 0.0494 (9) | |
| N4 | 0.7031 (5) | 0.1924 (4) | 0.4634 (3) | 0.0551 (9) | |
| C1 | 0.4644 (7) | 0.8439 (6) | 1.0818 (3) | 0.0576 (11) | |
| C2 | 0.5010 (5) | 0.7549 (5) | 0.9964 (3) | 0.0451 (10) | |
| C3 | 0.3568 (6) | 0.6070 (5) | 0.9367 (3) | 0.0467 (10) | |
| C4 | 0.4097 (5) | 0.5369 (4) | 0.8514 (3) | 0.0414 (9) | |
| C5 | 0.2802 (5) | 0.3970 (5) | 0.7853 (3) | 0.0475 (10) | |
| H5 | 0.1554 | 0.3464 | 0.7926 | 0.057* | |
| C6 | 0.3386 (5) | 0.3344 (5) | 0.7091 (3) | 0.0484 (10) | |
| C7 | 0.5238 (5) | 0.4014 (4) | 0.6918 (3) | 0.0436 (9) | |
| C8 | 0.6516 (5) | 0.5420 (5) | 0.7594 (3) | 0.0441 (9) | |
| C9 | 0.9600 (5) | 0.7160 (5) | 0.8421 (3) | 0.0557 (11) | |
| H9A | 0.9574 | 0.6446 | 0.8969 | 0.067* | |
| H9B | 1.0876 | 0.7688 | 0.8302 | 0.067* | |
| C10 | 0.6778 (5) | 0.8265 (5) | 0.9749 (3) | 0.0463 (10) | |
| H10 | 0.7678 | 0.9247 | 1.0140 | 0.056* | |
| C11 | 0.5951 (5) | 0.6118 (4) | 0.8364 (3) | 0.0400 (9) | |
| C12 | 0.9089 (7) | 0.9603 (5) | 0.7983 (4) | 0.0665 (13) | |
| H12A | 0.8252 | 0.8996 | 0.7343 | 0.100* | |
| H12B | 1.0339 | 1.0196 | 0.7856 | 0.100* | |
| H12C | 0.8681 | 1.0380 | 0.8246 | 0.100* | |
| C13 | 0.4605 (6) | 0.2587 (6) | 0.5132 (3) | 0.0628 (12) | |
| H13A | 0.3799 | 0.3129 | 0.4920 | 0.075* | |
| H13B | 0.3817 | 0.1428 | 0.5190 | 0.075* | |
| C14 | 0.5809 (7) | 0.2709 (6) | 0.4340 (3) | 0.0684 (13) | |
| H14A | 0.5007 | 0.2181 | 0.3667 | 0.082* | |
| H14B | 0.6560 | 0.3872 | 0.4272 | 0.082* | |
| C15 | 0.8245 (6) | 0.2699 (6) | 0.5640 (3) | 0.0651 (13) | |
| H15A | 0.9056 | 0.3855 | 0.5587 | 0.078* | |
| H15B | 0.9034 | 0.2138 | 0.5841 | 0.078* | |
| C16 | 0.7106 (6) | 0.2613 (5) | 0.6461 (3) | 0.0546 (11) | |
| H16A | 0.6367 | 0.1459 | 0.6557 | 0.065* | |
| H16B | 0.7939 | 0.3180 | 0.7120 | 0.065* | |
| C17 | 0.8116 (7) | 0.1965 (6) | 0.3831 (4) | 0.0840 (17) | |
| H17A | 0.7272 | 0.1399 | 0.3185 | 0.126* | |
| H17B | 0.8906 | 0.1422 | 0.4042 | 0.126* | |
| H17C | 0.8881 | 0.3101 | 0.3740 | 0.126* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0504 (14) | 0.0478 (13) | 0.0771 (17) | 0.0064 (11) | 0.0123 (12) | −0.0136 (12) |
| O1 | 0.077 (2) | 0.081 (2) | 0.0573 (19) | 0.0394 (19) | 0.0278 (17) | 0.0012 (18) |
| O2 | 0.084 (2) | 0.072 (2) | 0.0611 (19) | 0.0311 (18) | 0.0186 (17) | −0.0087 (17) |
| O3 | 0.0514 (17) | 0.0659 (18) | 0.0658 (19) | 0.0245 (15) | 0.0267 (15) | 0.0086 (15) |
| O4 | 0.0438 (15) | 0.0543 (16) | 0.0548 (17) | 0.0115 (13) | 0.0164 (13) | −0.0057 (13) |
| N1 | 0.0444 (19) | 0.0450 (18) | 0.056 (2) | 0.0129 (15) | 0.0172 (16) | 0.0015 (16) |
| N2 | 0.0415 (18) | 0.0460 (18) | 0.0441 (18) | 0.0137 (15) | 0.0112 (14) | 0.0023 (15) |
| N3 | 0.053 (2) | 0.060 (2) | 0.0391 (17) | 0.0302 (17) | 0.0041 (15) | −0.0071 (15) |
| N4 | 0.067 (2) | 0.0451 (19) | 0.052 (2) | 0.0196 (17) | 0.0221 (17) | −0.0016 (16) |
| C1 | 0.066 (3) | 0.068 (3) | 0.051 (3) | 0.038 (2) | 0.020 (2) | 0.010 (2) |
| C2 | 0.059 (3) | 0.048 (2) | 0.036 (2) | 0.029 (2) | 0.0121 (18) | 0.0061 (17) |
| C3 | 0.051 (2) | 0.053 (2) | 0.044 (2) | 0.0278 (19) | 0.0146 (19) | 0.0114 (19) |
| C4 | 0.049 (2) | 0.0373 (19) | 0.040 (2) | 0.0200 (17) | 0.0092 (17) | 0.0053 (16) |
| C5 | 0.042 (2) | 0.045 (2) | 0.055 (2) | 0.0169 (18) | 0.0115 (18) | 0.0051 (19) |
| C6 | 0.042 (2) | 0.038 (2) | 0.056 (2) | 0.0102 (17) | 0.0063 (18) | −0.0031 (18) |
| C7 | 0.051 (2) | 0.0363 (19) | 0.046 (2) | 0.0205 (17) | 0.0110 (18) | 0.0056 (17) |
| C8 | 0.042 (2) | 0.046 (2) | 0.044 (2) | 0.0186 (17) | 0.0121 (17) | 0.0003 (18) |
| C9 | 0.042 (2) | 0.057 (2) | 0.059 (3) | 0.0149 (19) | 0.0090 (19) | −0.007 (2) |
| C10 | 0.054 (2) | 0.048 (2) | 0.038 (2) | 0.0236 (19) | 0.0104 (18) | −0.0031 (17) |
| C11 | 0.041 (2) | 0.0352 (18) | 0.0396 (19) | 0.0131 (16) | 0.0082 (16) | 0.0025 (16) |
| C12 | 0.069 (3) | 0.058 (3) | 0.068 (3) | 0.018 (2) | 0.025 (2) | 0.010 (2) |
| C13 | 0.066 (3) | 0.068 (3) | 0.052 (3) | 0.031 (2) | 0.001 (2) | −0.010 (2) |
| C14 | 0.083 (3) | 0.070 (3) | 0.049 (3) | 0.032 (3) | 0.010 (2) | −0.001 (2) |
| C15 | 0.063 (3) | 0.079 (3) | 0.057 (3) | 0.034 (2) | 0.015 (2) | −0.005 (2) |
| C16 | 0.063 (3) | 0.061 (3) | 0.053 (2) | 0.038 (2) | 0.015 (2) | 0.005 (2) |
| C17 | 0.098 (4) | 0.073 (3) | 0.074 (3) | 0.021 (3) | 0.048 (3) | −0.007 (3) |
Geometric parameters (Å, º)
| F1—C6 | 1.361 (4) | C5—C6 | 1.368 (5) |
| O1—C1 | 1.339 (5) | C5—H5 | 0.9300 |
| O1—H1 | 0.83 (3) | C6—C7 | 1.407 (5) |
| O2—C1 | 1.210 (5) | C7—C8 | 1.395 (5) |
| O3—C3 | 1.267 (4) | C8—C11 | 1.401 (5) |
| O4—C8 | 1.377 (4) | C9—H9A | 0.9700 |
| O4—C9 | 1.448 (4) | C9—H9B | 0.9700 |
| N1—N2 | 1.434 (4) | C10—H10 | 0.9300 |
| N1—C9 | 1.439 (5) | C12—H12A | 0.9600 |
| N1—C12 | 1.484 (6) | C12—H12B | 0.9600 |
| N2—C10 | 1.341 (4) | C12—H12C | 0.9600 |
| N2—C11 | 1.387 (4) | C13—C14 | 1.507 (6) |
| N3—C7 | 1.397 (4) | C13—H13A | 0.9700 |
| N3—C13 | 1.465 (5) | C13—H13B | 0.9700 |
| N3—C16 | 1.470 (5) | C14—H14A | 0.9700 |
| N4—C14 | 1.438 (6) | C14—H14B | 0.9700 |
| N4—C15 | 1.451 (5) | C15—C16 | 1.506 (5) |
| N4—C17 | 1.464 (5) | C15—H15A | 0.9700 |
| C1—C2 | 1.491 (5) | C15—H15B | 0.9700 |
| C2—C10 | 1.368 (5) | C16—H16A | 0.9700 |
| C2—C3 | 1.425 (5) | C16—H16B | 0.9700 |
| C3—C4 | 1.470 (5) | C17—H17A | 0.9600 |
| C4—C5 | 1.387 (5) | C17—H17B | 0.9600 |
| C4—C11 | 1.399 (5) | C17—H17C | 0.9600 |
| C1—O1—H1 | 106 (4) | H9A—C9—H9B | 107.9 |
| C8—O4—C9 | 111.3 (3) | N2—C10—C2 | 121.0 (3) |
| N2—N1—C9 | 106.7 (3) | N2—C10—H10 | 119.5 |
| N2—N1—C12 | 109.9 (3) | C2—C10—H10 | 119.5 |
| C9—N1—C12 | 113.8 (3) | N2—C11—C4 | 119.3 (3) |
| C10—N2—C11 | 122.3 (3) | N2—C11—C8 | 120.0 (3) |
| C10—N2—N1 | 118.4 (3) | C4—C11—C8 | 120.7 (3) |
| C11—N2—N1 | 119.1 (3) | N1—C12—H12A | 109.5 |
| C7—N3—C13 | 121.3 (3) | N1—C12—H12B | 109.5 |
| C7—N3—C16 | 117.4 (3) | H12A—C12—H12B | 109.5 |
| C13—N3—C16 | 110.8 (3) | N1—C12—H12C | 109.5 |
| C14—N4—C15 | 110.1 (3) | H12A—C12—H12C | 109.5 |
| C14—N4—C17 | 111.2 (4) | H12B—C12—H12C | 109.5 |
| C15—N4—C17 | 111.6 (4) | N3—C13—C14 | 108.0 (4) |
| O2—C1—O1 | 120.9 (4) | N3—C13—H13A | 110.1 |
| O2—C1—C2 | 123.9 (4) | C14—C13—H13A | 110.1 |
| O1—C1—C2 | 115.2 (4) | N3—C13—H13B | 110.1 |
| C10—C2—C3 | 121.6 (3) | C14—C13—H13B | 110.1 |
| C10—C2—C1 | 116.6 (3) | H13A—C13—H13B | 108.4 |
| C3—C2—C1 | 121.7 (4) | N4—C14—C13 | 111.6 (4) |
| O3—C3—C2 | 123.6 (3) | N4—C14—H14A | 109.3 |
| O3—C3—C4 | 120.3 (3) | C13—C14—H14A | 109.3 |
| C2—C3—C4 | 116.0 (3) | N4—C14—H14B | 109.3 |
| C5—C4—C11 | 118.8 (3) | C13—C14—H14B | 109.3 |
| C5—C4—C3 | 121.6 (4) | H14A—C14—H14B | 108.0 |
| C11—C4—C3 | 119.6 (3) | N4—C15—C16 | 111.0 (4) |
| C6—C5—C4 | 119.0 (4) | N4—C15—H15A | 109.4 |
| C6—C5—H5 | 120.5 | C16—C15—H15A | 109.4 |
| C4—C5—H5 | 120.5 | N4—C15—H15B | 109.4 |
| F1—C6—C5 | 118.2 (3) | C16—C15—H15B | 109.4 |
| F1—C6—C7 | 117.0 (3) | H15A—C15—H15B | 108.0 |
| C5—C6—C7 | 124.8 (3) | N3—C16—C15 | 109.4 (4) |
| N3—C7—C8 | 119.3 (3) | N3—C16—H16A | 109.8 |
| N3—C7—C6 | 125.6 (3) | C15—C16—H16A | 109.8 |
| C8—C7—C6 | 115.1 (3) | N3—C16—H16B | 109.8 |
| O4—C8—C7 | 118.6 (3) | C15—C16—H16B | 109.8 |
| O4—C8—C11 | 119.9 (3) | H16A—C16—H16B | 108.2 |
| C7—C8—C11 | 121.5 (3) | N4—C17—H17A | 109.5 |
| N1—C9—O4 | 112.4 (3) | N4—C17—H17B | 109.5 |
| N1—C9—H9A | 109.1 | H17A—C17—H17B | 109.5 |
| O4—C9—H9A | 109.1 | N4—C17—H17C | 109.5 |
| N1—C9—H9B | 109.1 | H17A—C17—H17C | 109.5 |
| O4—C9—H9B | 109.1 | H17B—C17—H17C | 109.5 |
| C9—N1—N2—C10 | 147.4 (4) | N3—C7—C8—C11 | 177.4 (4) |
| C12—N1—N2—C10 | −88.8 (4) | C6—C7—C8—C11 | −1.6 (6) |
| C9—N1—N2—C11 | −36.1 (5) | N2—N1—C9—O4 | 62.1 (4) |
| C12—N1—N2—C11 | 87.7 (4) | C12—N1—C9—O4 | −59.3 (4) |
| O2—C1—C2—C10 | −3.8 (7) | C8—O4—C9—N1 | −56.7 (4) |
| O1—C1—C2—C10 | 176.1 (4) | C11—N2—C10—C2 | 0.7 (6) |
| O2—C1—C2—C3 | 179.1 (4) | N1—N2—C10—C2 | 177.0 (4) |
| O1—C1—C2—C3 | −1.0 (6) | C3—C2—C10—N2 | −1.8 (6) |
| C10—C2—C3—O3 | 178.9 (4) | C1—C2—C10—N2 | −178.9 (4) |
| C1—C2—C3—O3 | −4.2 (6) | C10—N2—C11—C4 | 2.2 (6) |
| C10—C2—C3—C4 | 0.0 (6) | N1—N2—C11—C4 | −174.1 (3) |
| C1—C2—C3—C4 | 177.0 (4) | C10—N2—C11—C8 | −178.1 (4) |
| O3—C3—C4—C5 | 3.6 (6) | N1—N2—C11—C8 | 5.6 (5) |
| C2—C3—C4—C5 | −177.5 (4) | C5—C4—C11—N2 | 176.4 (4) |
| O3—C3—C4—C11 | −176.1 (4) | C3—C4—C11—N2 | −3.9 (6) |
| C2—C3—C4—C11 | 2.8 (5) | C5—C4—C11—C8 | −3.3 (6) |
| C11—C4—C5—C6 | 1.5 (6) | C3—C4—C11—C8 | 176.4 (4) |
| C3—C4—C5—C6 | −178.2 (4) | O4—C8—C11—N2 | 2.0 (6) |
| C4—C5—C6—F1 | 178.3 (4) | C7—C8—C11—N2 | −176.2 (4) |
| C4—C5—C6—C7 | 0.2 (7) | O4—C8—C11—C4 | −178.3 (3) |
| C13—N3—C7—C8 | −148.1 (4) | C7—C8—C11—C4 | 3.4 (6) |
| C16—N3—C7—C8 | 70.4 (5) | C7—N3—C13—C14 | 157.3 (4) |
| C13—N3—C7—C6 | 30.9 (6) | C16—N3—C13—C14 | −58.9 (5) |
| C16—N3—C7—C6 | −110.7 (5) | C15—N4—C14—C13 | −59.0 (5) |
| F1—C6—C7—N3 | 2.7 (6) | C17—N4—C14—C13 | 176.7 (3) |
| C5—C6—C7—N3 | −179.2 (4) | N3—C13—C14—N4 | 59.4 (5) |
| F1—C6—C7—C8 | −178.3 (3) | C14—N4—C15—C16 | 57.3 (5) |
| C5—C6—C7—C8 | −0.2 (6) | C17—N4—C15—C16 | −178.7 (4) |
| C9—O4—C8—C7 | −159.0 (4) | C7—N3—C16—C15 | −156.2 (3) |
| C9—O4—C8—C11 | 22.7 (5) | C13—N3—C16—C15 | 58.4 (5) |
| N3—C7—C8—O4 | −0.8 (6) | N4—C15—C16—N3 | −56.9 (5) |
| C6—C7—C8—O4 | −179.9 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O3 | 0.83 (3) | 1.77 (2) | 2.560 (4) | 159 (5) |
| C12—H12B···F1i | 0.96 | 2.62 | 3.422 (3) | 140 (4) |
| C15—H15B···F1ii | 0.97 | 2.54 | 3.446 (3) | 155 (5) |
Symmetry codes: (i) x+1, y+1, z; (ii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NR2019).
References
- Carlotti, D. N., Guaguere, E., Pin, D., Jasmin, P., Thomas, E. & Guiral, V. (1999). J. Small Anim. Pract 40, 265–270. [DOI] [PubMed]
- Dossin, O., Gruet, P. & Thomas, E. (1998). J. Small Anim. Pract 39, 286–289. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Ishak, A. M., Dowers, K. L., Cavanaugh, M. T., Powell, C. C., Hawley, J. R., Radecki, S. V. & Lappin, M. R. (2008). J. Vet. Intern. Med. 22, 288–292. [DOI] [PubMed]
- Lefebvre, H. P., Schneider, M., Dupouy, V., Laroute, V., Costes, G., Delesalle, L. & Toutain, P. L. (1998). J. Vet. Pharmacol. Ther. 21, 453–461. [DOI] [PubMed]
- Paradis, M., Abbey, L., Baker, B., Coyne, M., Hannigan, M., Joffe, D., Pukay, B., Trettien, A., Waisglass, S. & Wellington, J. (2001). Vet. Dermatol. 12, 163–169. [DOI] [PubMed]
- Rigaku (2006). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku (2007). CrystalStructure Rigaku, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spreng, M., Deleforge, J., Thomas, V., Boisrame, B. & Drugeon, H. (1995). J. Vet. Pharmacol. Ther. 18, 284–289. [DOI] [PubMed]
- Thomas, E., Caldow, G. L., Borell, D. & Davot, J. L. (2001). J. Vet. Pharmacol. Ther. 24, 353–358. [DOI] [PubMed]
- Voermans, M., van Soest, J. M., van Duijkeren, E. & Ensink, J. M. (2006). J. Vet. Pharmacol. Ther. 29, 555–560. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812009312/nr2019sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812009312/nr2019Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812009312/nr2019Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report