Abstract
Neurotrophic factor deprivation causes apoptosis by a mechanism that requires macromolecular synthesis. This fact suggests that gene expression is necessary to achieve cell death. To identify mRNA that is expressed in apoptotic cells we used subtractive hybridization with cDNA prepared from neuronal pheochromocytoma cells. Monoamine oxidase (MAO) expression was increased in cells during nerve growth factor withdrawal-induced apoptosis. The increased apoptosis and induction of MAO was prevented by inhibition of the p38 mitogen-activated protein (MAP) kinase pathway. MAO may contribute to the apoptotic process because inhibition of MAO activity suppressed cell death. Together, these data indicate that MAO may be a target of pro-apoptotic signal transduction by the p38 MAP kinase pathway.
Apoptosis contributes to the normal development of the nervous system and to the progression of some neurodegenerative diseases. For example, neurons that fail to make a functional connection with survival factor-producing target cells during development are eliminated by apoptosis (1). Furthermore, subpopulations of neurons associated with pathological neurodegeneration, including Parkinson's, Huntington's, and Alzheimer's diseases, may die by apoptosis (2–6). The death of these neurons is thought to contribute to the symptoms of these neurodegenerative disorders. Thus, the degeneration of nigrostriatal dopaminergic neurons causes Parkinson's syndrome (7, 8). Although the degeneration of these cells has been attributed to apoptosis (7, 8), the mechanisms that contribute to the apoptotic process remain to be established.
Pheochromocytoma (PC12) cells can be considered to be a model for the study of both developmental and disease-associated apoptosis. When treated with nerve growth factor (NGF), PC12 cells differentiate into a postmitotic neuronal cell type (9). Because neuronal PC12 cells synthesize, store, and secrete large amounts of dopamine they most closely resemble dopaminergic neurons of the sympathetic nervous system (9, 10). Expression of mutated amyloid precursor protein, exposure to iron, or exposure to reactive oxygen species (e.g., H2O2 or peroxynitrites) causes neuronal PC12 cells to undergo apoptosis, mimicking the etiology of neurodegenerative disease (11–13). Depriving differentiated PC12 cells of NGF reproduces a developmental scenario whereby the failure of a neuron to connect to a source of trophic support causes that neuron to undergo programmed cell death (1). The wide variety of stimuli that can cause apoptosis suggests that several molecular pathways leading to death exist within these cells (14, 15). NGF withdrawal-induced apoptosis of primary neurons and PC12 cells is blocked by the addition of macromolecular synthesis inhibitors up to 14 h after the removal of NGF (16, 17). These data indicate that gene expression is required, either to enhance death signals or to suppress survival signals, to carry out the death program.
Previous studies designed to characterize genes expressed in apoptotic PC12 cells have led to the identification of rat orthologs of human TAFII70, Ring 3, and the Drosophila fsh gene (18, 19). In addition, the cell cycle gene cyclin D1 and the immediate-early response transcription factors c-Jun and c-Fos are induced in apoptotic PC12 cells (20, 21). An important role for c-Jun in neuronal apoptosis has been established in experiments using dominant-negative approaches and the analysis of mice with germline mutations in the c-Jun gene (22–24). Furthermore, c-Jun NH2-terminal kinase (JNK) is implicated in death mediated by this pathway (22, 25–27). Similarly, the p38 mitogen-activated protein kinase (MAPK) contributes to neuronal apoptosis (26, 28, 29) by mechanisms that include the MEF2-dependent induction of c-Jun expression (30). Importantly, both the JNK and p38 MAPKs are activated during neurotrophic factor withdrawal (25–27, 29). It is thought that these MAPK signal transduction pathways may mediate pro-apoptotic gene expression (31). Targets of the JNK pathway in neurons include increased expression of FasL (32), but little is known about potential targets of the p38 MAPK pathway.
The purpose of this study was to identify p38 MAPK-dependent gene expression in apoptotic neuronal PC12 cells. We report the identification of MAO as a gene that is regulated by the p38 MAPK pathway in apoptotic cells. Interestingly, MAO activity strongly potentiates neurotrophic factor withdrawal-induced apoptosis.
Materials and Methods
Cell Culture.
PC12 cells were maintained at 37°C with 10% CO2 in DMEM containing 10% heat-inactivated horse serum (GIBCO), 5% FBS (Omega Scientific, Tarzana, CA), penicillin/streptomycin, and 2 mM glutamine (Life Technologies, Rockville, MD). PC12 cells were differentiated on plates coated with 0.2 mg/ml rat-tail collagen (Biomedical Technologies, Stoughton, MA) in DMEM supplemented with 1% heat-inactivated horse serum (Life Technologies), penicillin, streptomycin, and glutamine as listed above and 75 ng/ml NGF (Austral Biological) for 10–12 days. The medium was changed every other day. NGF withdrawal was initiated by washing differentiated PC12 cells once with NGF-free medium and the addition of NGF-free medium containing a neutralizing antibody (Sigma) to the 2.5S subunit of NGF (13 μg/ml; 1:1000 dilution). Control cells were incubated in the NGF containing differentiation media after the wash in NGF-free medium. Clorgyline (0–10 μM; Research Biochemicals, Natick, MA), deprenyl (0–10 μM; Research Biochemicals), PD169316 (10 μM; Calbiochem), and PD98059 (25 μM; Calbiochem) were added to the culture medium. To treat cells with dopamine, the medium was replaced with differentiation media with 300 μM dopamine plus 75 ng/ml of NGF.
Gene Expression Analysis.
Poly(A)+ RNA was extracted from PC12 cells with the Invitrogen MicroFast Track 2.0 mRNA isolation system according to the manufacturer's protocol. Differentiated PC12 cell mRNA was subtractively hybridized to 12 h NGF-withdrawn PC12 cell mRNA with the CLONTECH PCR-Select cDNA Subtraction Kit according to the manufacturer's protocol. The PCR products generated from the subtraction were cloned in the vector pCR II (Invitrogen) and transformed into DH5α competent bacteria, and the resultant clones were picked for DNA sequence analysis. Forty clones identified by the subtractive hybridization were rescreened by Northern blot analysis to confirm induction during NGF withdrawal. Poly(A)+ mRNA (2–4 μg) was examined by Northern blot analysis with the use of probes random primer labeled with [α-32P]dATP. The probes were hybridized in 50% deionized formamide, 10% dextran sulfate, 1% SDS, 1 M NaCl, and 100 mg/ml of denatured, sonicated salmon sperm DNA overnight (12–16 h) at 42°C. The blots were washed twice in 2× SSC/0.1% SDS at 25°C, once in 0.2× SSC/0.1% SDS at 25°C, and once in 0.2× SSC/0.1% SDS at 42°C (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7).
Cell Death Determination.
Differentiated PC12 cells (6–7 days) were harvested, counted by trypan blue staining, replated (5 × 104 cells/ml), and differentiated for 5–7 additional days before NGF withdrawal. Cells were harvested by gentle pipetting, combined with the culture supernatant, centrifuged at 200 × g at 4°C, resuspended in 1 ml of 4°C PBS, and transferred to a prechilled microcentrifuge tube. Nucleosome fragmentation was measured with the Cell Death Detection ELISA assay (Roche Molecular Biochemicals) according to the manufacturer's protocol. Apoptosis was also examined by analysis of chromosome condensation by fluorescence microscopic examination of PC12 cells stained with 4,6-diamidino-2-phenylindole (26).
Measurement of MAO Activity.
MAO was assayed by the method of Wurtman and Axelrod with modifications (33). PC12 cells were harvested by gentle trituration, centrifuged at 200 × g (4°C) for 5 min, resuspended in 1 ml 0.01 M potassium phosphate buffer (pH 7.2), transferred to a microcentrifuge tube, and sheared by passage 10 times through a 20-gauge needle. The protein concentration was determined by the Bradford method, and equal amounts (20 mg) were added to 15 ml of 100 μM [14C]-5-hydroxytryptamine (5-HT) (100 μCi/ml) in 0.1 M potassium phosphate buffer (pH 7.4) (0.25 ml final volume). The reactions were terminated after 20 min at 37°C by the addition of 200 μl 2 M citric acid and mixed with 2 ml toluene:ethyl acetate (1:1). The organic phase containing the by-product of the MAO-catalyzed deamination of 5-HT was added to an equal volume of ScintiSafe Econo 1 scintillation fluid (Fisher Scientific), and radioactivity was quantitated with a liquid scintillation counter.
Results
Gene Expression Is Required for NGF Withdrawal-Induced Apoptosis.
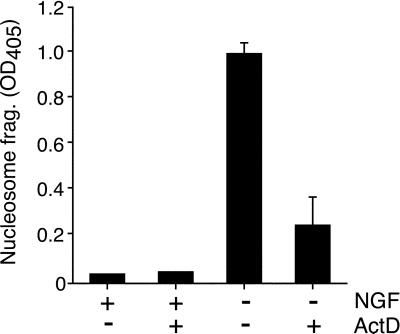
Neuronal PC12 cells undergo apoptosis when deprived of NGF (26). Previous studies have demonstrated that NGF withdrawal-induced death of PC12 cells (and primary neurons) can be blocked by inhibitors of transcription or translation (16, 34). We confirmed this observation by measuring nucleosome fragmentation of DNA with the use of the PC12 cells used in this study. Treatment with actinomycin D did not significantly affect NGF-treated PC12 cells, but this treatment dramatically reduced apoptosis caused by NGF deprivation (Fig. 1). These data suggest that gene expression is required to execute the apoptotic program induced by NGF withdrawal.
Figure 1.
Macromolecular synthesis dependence of NGF withdrawal-induced apoptosis of neuronal PC12 cells. Differentiated PC12 cells were incubated without and with NGF (75 ng/ml) in the presence or absence of 1 μg/ml of the transcriptional inhibitor actinomycin D (ActD). Cell lysates were prepared after 24 h and assayed for nucleosomal DNA fragmentation (mean ± SD; n = 3). Similar data were obtained in three independent experiments.
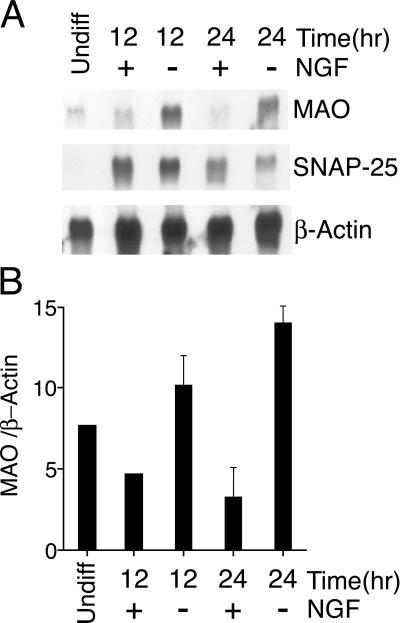
To identify genes induced during NGF withdrawal-induced apoptosis, we used subtractive hybridization of cDNA isolated from control neuronal PC12 cells and from PC12 cells 12 h after NGF withdrawal. cDNA clones identified in this assay were tested in a secondary screen by Northern blot analysis of polyA+ mRNA isolated from control and apoptotic PC12 cells. This strategy led to the identification of the A isoform of MAO (Fig. 2A). The MAO mRNA was induced 2.8- and 4.0-fold at 12 and 24 h, respectively, after NGF withdrawal (Fig. 2B). In contrast, the expression of SNAP-25 mRNA, which is selectively expressed in differentiated neuronal PC12 cells, was not induced by NGF withdrawal (Fig. 2B).
Figure 2.
MAO expression is induced after NGF withdrawal. Differentiated PC12 cells were incubated without or with NGF (12 or 24 h). Naive (undiff) PC12 cells were also examined. The expression of MAO, SNAP-25, and β-actin was examined by Northern blot analysis. The hybridization was detected by autoradiography (A) and was quantitated by PhosphorImager analysis (B). The ratio of MAO/β-actin is presented (arbitrary units) as a mean ± SE (n = 4).
MAPK Requirement for MAO Gene Expression.
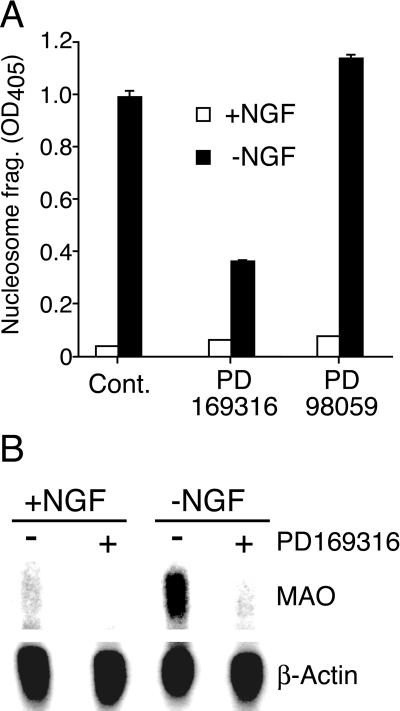
p38 MAPK [but not the extracellular signal-regulated kinase MAPK] is activated in response to trophic factor deprivation (26). To test the role of these MAPK signal transduction pathways in MAO gene expression after NGF withdrawal, we examined the effect of small molecules that are known to inhibit MAPK signal transduction. The extracellular signal-regulated kinase pathway inhibitor PD98059 (25 μM) blocked extracellular signal-regulated kinase activation (data not shown) but did not rescue PC12 cells from apoptosis (Fig. 3A) and did not prevent the expression of MAO mRNA in response to NGF withdrawal (data not shown). In contrast, the p38 MAPK inhibitor PD169316 (10 μM) blocked p38 activity (monitored by measurement of MAPKAPK-2 activity; data not shown), significantly reduced PC12 cell apoptosis (65% inhibition), and markedly reduced the basal expression of MAO (Fig. 3). Strikingly, the induction of MAO mRNA upon NGF deprivation was abolished by the p38 MAPK inhibitor PD169316 (Fig. 3B). These data suggest that p38 MAPK activity contributes to the expression of MAO in differentiated PC12 cells. Furthermore, these data indicate that p38 MAPK is required for the increased expression of MAO caused by NGF withdrawal.
Figure 3.
Role of MAPK in apoptosis and expression of MAO. Neuronal PC12 cells were incubated in the presence and absence of NGF (24 h). (A) Apoptosis was measured with a nucleosomal DNA fragmentation assay (mean ± SD; n = 3). The effect of treatment of the cells with inhibitors of p38 MAPK (PD169316) and the extracellular signal-regulated kinase pathway (PD98059) was examined. Similar data were obtained in three independent experiments. (B) MAO A and β-actin mRNA expression was examined by Northern blot analysis. The effect of treatment with the p38 inhibitor PD169316 is presented. Similar data were obtained in three independent experiments.
Effect of NGF Withdrawal on MAO Activity.
Two isoforms of MAO (A and B) are expressed in the brain (35, 36). MAO A is found predominantly in the substantia nigra (a region rich in dopaminergic neurons), the locus coeruleus, the striatum, and other areas containing catecholaminergic neurons (36, 37). MAO B is expressed at a lower level in the same brain regions as MAO A and is most abundant in the raphe nuclei and glial cells (35, 38). MAO A and B differ in their substrate preferences. MAO A has high activity toward the biogenic amines 5-HT, norepinephrine, and dopamine and is specifically inhibited by clorgyline (39). MAO B has high activity toward phenylethylamine and benzylamine as substrates and is inhibited by deprenyl, a drug with reported effects on Parkinson's disease (39).
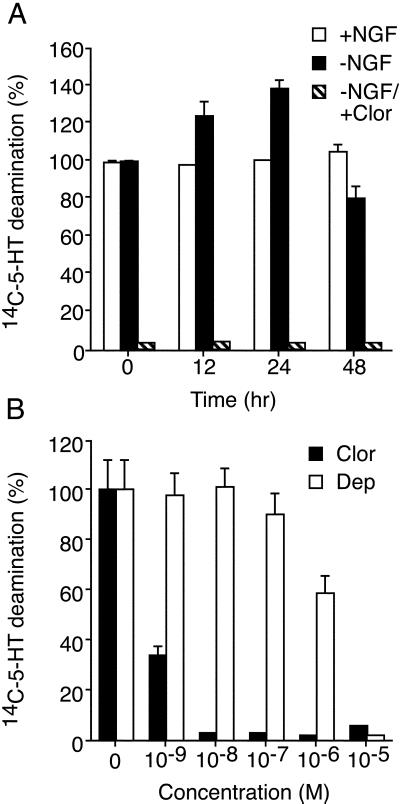
PC12 cells resemble catecholaminergic neurons by synthesizing, storing, and releasing dopamine (10, §). PC12 cells also contain predominantly the A isoform of MAO and undetectable levels of MAO B (41). To determine whether MAO activity is affected by the withdrawal of NGF, an enzyme assay was performed, with 5-HT as the substrate. A time course of NGF deprivation indicated that MAO substrate deamination (assayed in cell lysates) was increased after NGF withdrawal (Fig. 4A). The maximum 5-HT deamination was observed at 24 h after NGF withdrawal and was inhibited by the irreversible MAO A inhibitor clorgyline (Fig. 4A). Nanomolar concentrations of the MAO A inhibitor clorgyline effectively eliminated the metabolism of the 5-HT substrate, whereas the MAO B inhibitor deprenyl had no effect at the same concentration (Fig. 4B). As reported previously, micromolar and millimolar concentrations of deprenyl and clorgyline inhibit both MAO A and B isoforms (Fig. 4B). Collectively, these data demonstrate that withdrawal of NGF from differentiated PC12 cells causes an increase in MAO activity, and addition of the irreversible MAO inhibitor clorgyline strongly inhibits MAO activity in PC12 cells.
Figure 4.
MAO activity is increased in apoptotic PC12 cells. (A) Neuronal PC12 cells were incubated in the presence and absence of NGF (12, 24 or 48 h). The effect of addition of the irreversible MAO inhibitor clorgyline (10−7 M) was examined. Extracts were prepared from the cells, and MAO activity was measured. The data presented are the rates of 5-HT deamination (mean ± SD; n = 3) and are normalized to the rate of deamination measured in control neuronal PC12 cells. Similar data were obtained in three independent experiments. (B) Comparison of clorgyline (Clor) and deprenyl (Dep) on MAO activity in neuronal PC12 cell lysates. Clorgyline and deprenyl are selective inhibitors of the MAO isoforms A and B, respectively. The effect of different concentrations of the drugs on MAO activity was examined. The data presented are the rates of 5-HT deamination (mean ± SD; n = 3) and are normalized to the rate of deamination measured in control cell lysates. Similar data were obtained in three independent experiments.
MAO Inhibition Rescues NGF Withdrawal-Induced Apoptosis of PC12 Cells.
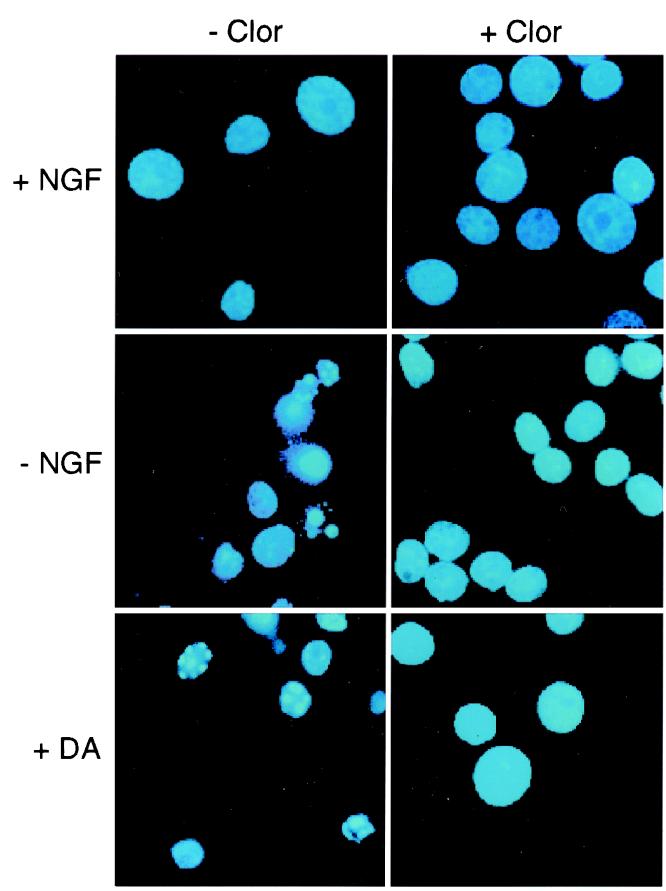
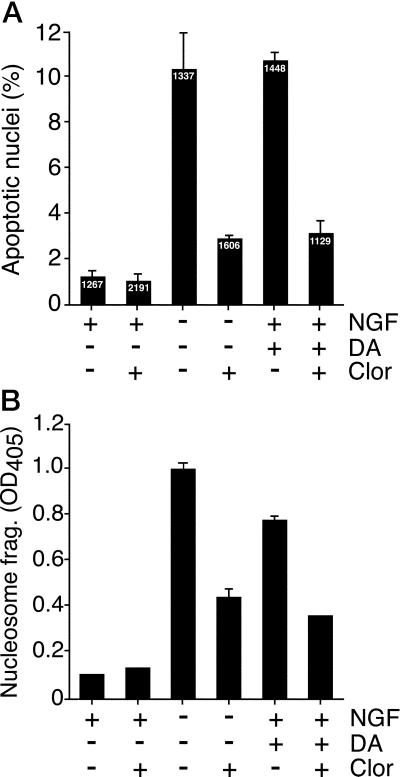
MAO expression and activity are increased after NGF withdrawal. To determine whether MAO activity was relevant to NGF withdrawal-induced apoptosis, we examined the effect of the MAO inhibitor clorgyline. Neuronal PC12 cells were deprived of NGF and treated with or without clorgyline for 24 h and then stained with 4,6-diamidino-2-phenylindole. Cells exhibiting chromatin condensation, visualized by fluorescence microscopy, were scored as apoptotic (Fig. 5). At least 1,000 cells were examined in each treatment group. NGF deprivation caused a 10-fold increase in apoptotic cells after 24 h (Fig. 6A). Addition of clorgyline to NGF-treated cells had no effect on apoptosis (Fig. 6A). In contrast, clorgyline treatment resulted in a 70% reduction of NGF withdrawal-induced apoptosis (Fig. 6A). These observations were confirmed by measurement of nucleosome fragmentation of DNA, a different parameter of cell death (Fig. 6B). Collectively, these data indicate that MAO activity potentiates the NGF withdrawal-induced death of PC12 cells. Furthermore, MAO inhibition causes reduced apoptosis.
Figure 5.
Effect of dopamine and NGF withdrawal on nuclear morphology. Neuronal PC12 cells were incubated (24 h) in the presence and absence of NGF or in the presence of NGF plus 300 μM dopamine (DA). The effect of the addition of 0.1 μM clorgyline (Clor) was examined. The cells were fixed, stained with 4,6-diamidino-2-phenylindole, and inspected by fluorescence microscopy. Representative images of PC12 cells are shown. Similar data were obtained in three independent experiments.
Figure 6.
Effect of MAO inhibition on dopamine and NGF withdrawal-induced apoptosis. Neuronal PC12 cells were incubated (24 h) in the presence and absence of NGF, or in the presence of NGF plus 300 μM dopamine (DA). The effect of the addition of 0.1 μM clorgyline (Clor) was examined. (A) The cells were fixed, stained with 4,6-diamidino-2-phenylindole, and inspected by fluorescence microscopy. Cells with condensed chromatin and/or fragmented nuclei were scored as apoptotic. The number of cells examined for each treatment group is shown (Inset). The percentage of apoptotic cells (mean ± SEM; n = 3) is presented. (B) Apoptosis was examined by measurement of nucleosomal DNA fragmentation (mean ± SD; n = 3). Similar data were obtained in three independent experiments.
MAO Inhibition Rescues Dopamine-Induced Apoptosis of PC12 Cells.
Substrates of MAO include the neurotransmitter dopamine. MAO deaminates dopamine, producing 3,4-dihydroxyphenyl-acetic acid and the by-product H2O2. Whereas the effects of 3,4-dihydroxyphenyl-acetic acid on cell survival have not been established, the potent and deleterious effects of H2O2 and related reactive oxygen species are established (42). Low concentrations (<1 μM) of exogenous dopamine have been shown to promote PC12 proliferation and survival, but higher concentrations (>100 μM) kill PC12 cells, sympathetic ganglia neurons, and cerebellar granule neurons (43). To investigate the effect of MAO inhibition on dopamine-induced apoptosis, 300 μM dopamine was added to NGF-maintained PC12 cells. The effect of addition of the MAO inhibitor clorgyline was examined. Exogenous dopamine caused a robust increase in PC12 cell apoptosis, measured by chromosome condensation (staining with 4,6-diamidino-2-phenylindole) and the nucleosomal DNA fragmentation assay (Figs. 5 and 6). Furthermore, dopamine-induced apoptosis was inhibited by the addition of the MAO inhibitor clorgyline (Fig. 6). Together, these data indicate that the dopamine-induced death of neuronal PC12 cells is reduced by the MAO inhibitor clorgyline and suggest that MAO-catalyzed deamination of dopamine contributes to PC12 cell apoptosis.
Discussion
The p38 MAPK Pathway Is Required for Pro-Apoptotic Expression of MAO.
JNK and p38 MAPK are activated by many stimuli that cause apoptosis (31). JNK phosphorylation and activation of the transcription factor c-Jun during cell death is well established (31). However, the roles of other substrates of JNK are not defined (31). Similarly, the targets of the p38 MAPK pathway in apoptosis are poorly understood. It is likely that these MAPK signal transduction pathways contribute to the control of gene expression in apoptotic cells. Gene expression is known to be required for neurotrophic factor withdrawal-induced apoptosis of PC12 cells (17). Candidate genes that may be induced by pro-apoptotic signal transduction by the JNK pathway include FasL (32). However, genes that are expressed by a p38 MAPK-dependent mechanism in apoptotic cells have not been described. Here we report that MAO expression is increased in neuronal PC12 cells after NGF withdrawal and that this expression of MAO requires the p38 MAPK signal transduction pathway.
The p38 MAPK inhibitor PD169316 prevents increased MAO expression by PC12 cells after NGF withdrawal and suppresses apoptosis. These data, together with the observation that NGF withdrawal causes p38 MAPK activation (26, 29), suggest that the p38 MAPK signaling pathway functions as a regulator of MAO gene expression in apoptotic cells. Further studies are required to identify the mechanism by which the p38 MAPK pathway increases the expression of MAO mRNA. It is possible that p38 MAPK may regulate MAO mRNA stability. Alternatively, p38 MAPK may regulate transcription of the MAO gene. The observation that p38 MAPK phosphorylates and activates a number of transcription factors (e.g., ATF2, Elk-1, Ets, and MEF2) and that p38-activated protein kinases can phosphorylate and stimulate the transcription factor cAMP response binding protein (CREB) suggests that the effects of p38 MAPK on MAO gene expression may be complex. Nevertheless, MAO expression depends on the p38 MAPK signal transduction pathway (Fig. 3B).
MAO Activity Contributes to PC12 Cell Apoptosis.
The observation that MAO expression and activity are increased in response to NGF withdrawal indicates that this oxidase may play a role in trophic factor deprivation-induced apoptosis of PC12 cells. Indeed, the by-products of MAO-catalyzed deamination of dopamine cause apoptosis of neuronal cells (42, 44). Whereas both autooxidation and MAO-catalyzed deamination of dopamine have been reported to cause apoptotic death (45), the results of the present study indicate that MAO activity does contribute to PC12 cell death (Figs. 4 and 5). Dopaminergic neurons and PC12 cells sequester millimolar concentrations of the neurotransmitter dopamine in releasable pools of synaptic vesicles.§ Apoptosis-induced defects in exocytosis, transporter molecules, or synaptic vesicle integrity may lead to increased cytosolic dopamine. Cytoplasmic dopamine may also be accumulated from exogenous sources (e.g., release from neighboring cells). MAO catalyzes substrate deamination in the cytoplasm, releasing hydrogen peroxide as a by-product. Therefore increased dopamine availability during the apoptotic process may lead to the generation of harmful reactive oxygen species by MAO. Other sources of reactive oxygen species include NADH oxidases (46). Inhibition of MAO may therefore protect cells from apoptosis by decreasing the formation of reactive oxygen species. This proposed mechanism implies that the role of MAO is to potentiate apoptosis rather than acting directly in the apoptotic death mechanism (e.g., caspase activation). The possibility that MAO may contribute to the death of dopaminergic neurons after the death of neighboring neurons (and subsequent release of dopamine) is intriguing because it suggests a potential mechanism for neurodegeneration.
MAO is a resident protein of the outer mitochondrial membrane (49). This localization of MAO is interesting because an important role for mitochondria in apoptosis has been established (50). This subcellular localization suggests that MAO may alter mitochondrial function. Indeed, it has been demonstrated that MAO activity can cause mitochondrial dysfunction (40). Further studies are required to demonstrate that MAO does cause mitochondrial dysfunction in apoptotic cells.
MAO A deficiency in mice and humans is associated with increased aggressive behavior (47, 48). Studies of MAO A knockout mice indicate elevated brain levels of dopamine, and this neurotransmitter mediates, in part, a stress response (48). A link between aggression and neuronal apoptosis has not been established. However, the results of our study suggest that apoptotic defects may contribute to the phenotype of MAO A-deficient mice. Nevertheless, the results of the present study establish that MAO is a target of pro-apoptotic signal transduction by the p38 MAPK signal transduction pathway.
Acknowledgments
We thank T. Barrett for DNA sequencing and K. Gemme for administrative assistance. This study was supported, in part, by a grant from the National Institutes of Health. R.J.D. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- 5-HT
5-hydroxytryptamine
- JNK
c-Jun NH2-terminal kinase
- MAPK
mitogen-activated protein kinase
- MAO
monoamine oxidase
- NGF
nerve growth factor
- PC12 cells
pheochromocytoma cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Lim, R. R., Bloomfield, M. R., Johnson, A. M. & Allen, J. M. (1995) Biochem. Soc. Trans. 23, 46.S (abstr.).
References
- 1.Oppenheim R W. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 2.Portera-Cailliau C, Hedreen J C, Price D L, Koliatsos V E. J Neurosci. 1995;15:3775–3787. doi: 10.1523/JNEUROSCI.15-05-03775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mutoh T, Tokuda A, Marini A M, Fujiki N. Brain Res. 1994;661:51–55. doi: 10.1016/0006-8993(94)91179-7. [DOI] [PubMed] [Google Scholar]
- 4.Tatton N A, Kish S J. Neuroscience. 1997;77:1037–1048. doi: 10.1016/s0306-4522(96)00545-3. [DOI] [PubMed] [Google Scholar]
- 5.Estus S, Tucker H M, van Rooyen C, Wright S, Brigham E F, Wogulis M, Rydel R E. J Neurosci. 1997;17:7736–7745. doi: 10.1523/JNEUROSCI.17-20-07736.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez I, Xu C J, Juo P, Kakizaka A, Blenis J, Yuan J. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 7.Tatton N A, Maclean-Fraser A, Tatton W G, Perl D P, Olanow C W. Ann Neurol. 1998;44:S142–S148. doi: 10.1002/ana.410440721. [DOI] [PubMed] [Google Scholar]
- 8.Ziv I, Barzilai A, Offen D, Nardi N, Melamed E. J Neural Transm Suppl. 1997;49:69–76. doi: 10.1007/978-3-7091-6844-8_7. [DOI] [PubMed] [Google Scholar]
- 9.Greene L A, Tischler A S. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene L A, Aletta J M, Rukenstein A, Green S H. Methods Enzymol. 1987;147:207–216. doi: 10.1016/0076-6879(87)47111-5. [DOI] [PubMed] [Google Scholar]
- 11.Offen D, Ziv I, Barzilai A, Gorodin S, Glater E, Hochman A, Melamed E. Neurochem Int. 1997;31:207–216. doi: 10.1016/s0197-0186(96)00150-7. [DOI] [PubMed] [Google Scholar]
- 12.Ivins K J, Ivins J K, Sharp J P, Cotman C W. J Biol Chem. 1999;274:2107–2112. doi: 10.1074/jbc.274.4.2107. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, Chrest F J, Horton W E, Jr, Sisodia S S, Kusiak J W. J Neurosci Res. 1997;47:253–263. [PubMed] [Google Scholar]
- 14.Park D S, Stefanis L, Yan C Y I, Farinelli S E, Greene L A. J Biol Chem. 1996;271:21898–21905. doi: 10.1074/jbc.271.36.21898. [DOI] [PubMed] [Google Scholar]
- 15.Park D S, Morris E J, Stefanis L, Troy C M, Shelanski M L, Geller H M, Greene L A. J Neurosci. 1998;18:830–840. doi: 10.1523/JNEUROSCI.18-03-00830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin D P, Schmidt R E, DiStefano P S, Lowry O H, Carter J G, Johnson E M., Jr J Cell Biol. 1988;106:829–844. doi: 10.1083/jcb.106.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pittman R N, Wang S, DiBenedetto A J, Mills J C. J Neurosci. 1993;13:3669–3680. doi: 10.1523/JNEUROSCI.13-09-03669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imaizumi K, Tsuda M, Imai Y, Wanaka A, Takagi T, Tohyama M. J Biol Chem. 1997;272:18842–18848. doi: 10.1074/jbc.272.30.18842. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Dibenedetto A J, Pittman R N. Dev Biol. 1997;188:322–336. doi: 10.1006/dbio.1997.8655. [DOI] [PubMed] [Google Scholar]
- 20.Freeman R S, Estus S, Johnson E M., Jr Neuron. 1994;12:343–355. doi: 10.1016/0896-6273(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 21.Estus S, Zaks W J, Freeman R S, Gruda M, Bravo R, Johnson E M., Jr J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrens A, Sibilia M, Wagner E F. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 23.Watson A, Eilers A, Lallemand D, Kyriakis J, Rubin L L, Ham J. J Neurosci. 1998;18:751–762. doi: 10.1523/JNEUROSCI.18-02-00751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ham J, Babij C, Whitfield J, Pfarr C M, Lallemand D, Yaniv M, Rubin L L. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 25.Eilers A, Whitfield J, Babij C, Rubin L L, Ham J. J Neurosci. 1998;18:1713–1724. doi: 10.1523/JNEUROSCI.18-05-01713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 27.Yang D D, Kuan C Y, Whitmarsh A J, Rincon M, Zheng T S, Davis R J, Rakic P, Flavell R A. Nature (London) 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki H, Morooka T, Shimohama S, Kimura J, Hirano T, Gotoh Y, Nishida E. J Biol Chem. 1997;272:18518–18521. doi: 10.1074/jbc.272.30.18518. [DOI] [PubMed] [Google Scholar]
- 29.Kummer J L, Rao P K, Heidenreich K A. J Biol Chem. 1997;272:20490–20494. doi: 10.1074/jbc.272.33.20490. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 31.Davis R J. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 32.Le-Niculescu H, Bonfoco E, Kasuya Y, Claret F X, Green D R, Karin M. Mol Cell Biol. 1999;19:751–763. doi: 10.1128/mcb.19.1.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wurtman R J, Axelrod J. Biochem Pharmacol. 1964;12:1439–1441. doi: 10.1016/0006-2952(63)90215-6. [DOI] [PubMed] [Google Scholar]
- 34.Mesner P W, Winters T R, Green S H. J Cell Biol. 1992;119:1669–1680. doi: 10.1083/jcb.119.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westlund K N, Denney R M, Rose R M, Abell C W. Neuroscience. 1988;25:439–456. doi: 10.1016/0306-4522(88)90250-3. [DOI] [PubMed] [Google Scholar]
- 36.Westlund K N, Krakower T J, Kwan S W, Abell C W. Brain Res. 1993;612:221–230. doi: 10.1016/0006-8993(93)91664-e. [DOI] [PubMed] [Google Scholar]
- 37.Di Monte D A, DeLanney L E, Irwin I, Royland J E, Chan P, Jakowec M W, Langston J W. Brain Res. 1996;738:53–59. doi: 10.1016/0006-8993(96)00761-5. [DOI] [PubMed] [Google Scholar]
- 38.Konradi C, Svoma E, Jellinger K, Riederer P, Denney R, Thibault J. Neuroscience. 1988;26:791–802. doi: 10.1016/0306-4522(88)90099-1. [DOI] [PubMed] [Google Scholar]
- 39.Shih J C, Chen K, Ridd M J. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen G, Farooqui R, Kesler N. Proc Natl Acad Sci USA. 1997;94:4890–4894. doi: 10.1073/pnas.94.10.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youdim M B. Eur J Pharmacol. 1991;192:201–202. doi: 10.1016/0014-2999(91)90094-7. [DOI] [PubMed] [Google Scholar]
- 42.Olanow C W. Ann Neurol. 1992;32:S2–S9. doi: 10.1002/ana.410320703. [DOI] [PubMed] [Google Scholar]
- 43.Zilkha-Falb R, Ziv I, Nardi N, Offen D, Melamed E, Barzilai A. Cell Mol Neurobiol. 1997;17:101–118. doi: 10.1023/A:1026333222008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawase M, Motohashi N, Kurihara T, Inagaki M, Satoh K, Sakagami H. Anticancer Res. 1998;18:1069–1074. [PubMed] [Google Scholar]
- 45.Graham D G. Neurotoxicology. 1984;5:83–95. [PubMed] [Google Scholar]
- 46.Tammariello S P, Quinn M T, Estus S. J Neurosci (Online) 2000;20:RC53. doi: 10.1523/JNEUROSCI.20-01-j0006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brunner H G, Nelen M, Breakefield X O, Ropers H H, van Oost B A. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 48.Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, Muller U, Aguet M, Babinet C, Shih J C, et al. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenawalt J W, Schnaitman C. J Cell Biol. 1970;46:173–179. doi: 10.1083/jcb.46.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desagher S, Martinou J C. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]