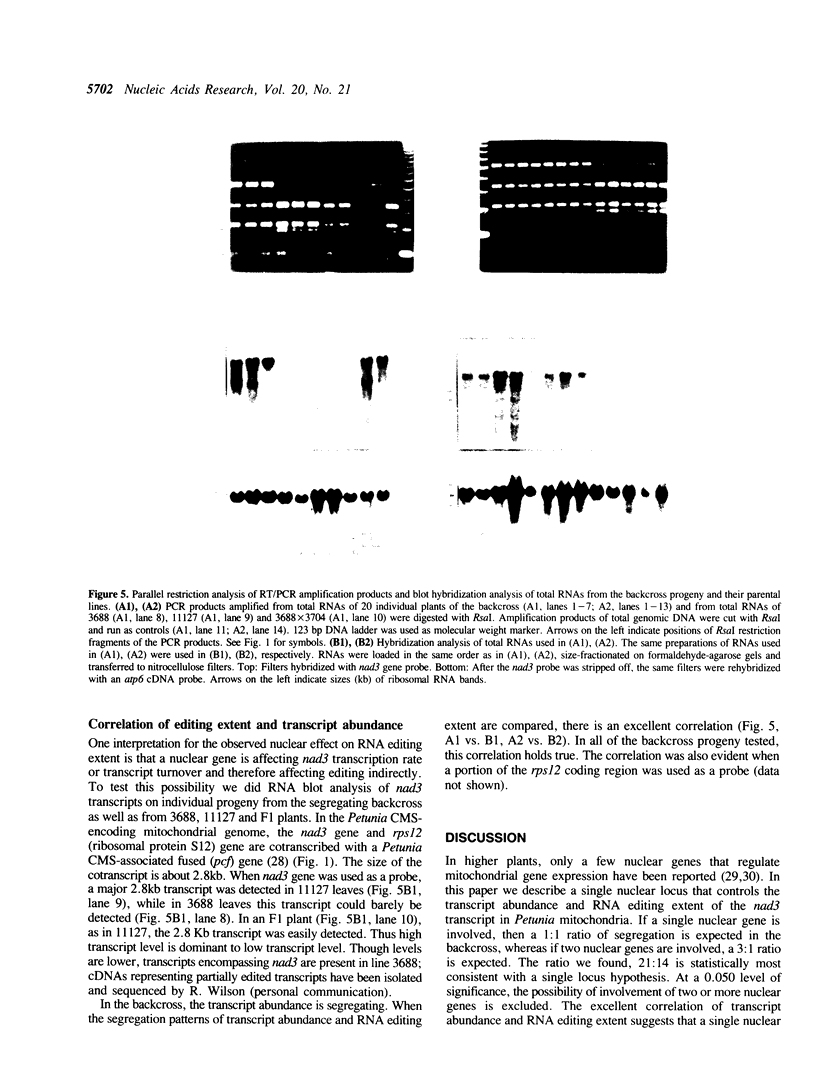
Abstract
A number of cytosines are altered to be recognized as uridines in transcripts of the NADH-dehydrogenase subunit 3 (nad3) gene in the mitochondria of the higher plant Petunia hybrida. Here we show that the extent of editing for three of the edit sites, all of which change the encoded amino acid, varies between different Petunia lines. Genetic analysis indicates that a single nuclear gene is responsible for this variation. Interestingly, according to RNA blot hybridization analysis, RNA editing extent and transcript abundance are correlated. This observation is consistent with the hypothesis that RNA editing is a post-transcriptional event.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beier H., Lee M. C., Sekiya T., Kuchino Y., Nishimura S. Two nucleotides next to the anticodon of cytoplasmic rat tRNA(Asp) are likely generated by RNA editing. Nucleic Acids Res. 1992 Jun 11;20(11):2679–2683. doi: 10.1093/nar/20.11.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Cattaneo R. Different types of messenger RNA editing. Annu Rev Genet. 1991;25:71–88. doi: 10.1146/annurev.ge.25.120191.000443. [DOI] [PubMed] [Google Scholar]
- Cattaneo R. RNA editing: in Chloroplast and brain. Trends Biochem Sci. 1992 Jan;17(1):4–5. doi: 10.1016/0968-0004(92)90415-6. [DOI] [PubMed] [Google Scholar]
- Cooper P., Butler E., Newton K. J. Identification of a maize nuclear gene which influences the size and number of cox2 transcripts in mitochondria of perennial ++teosintes. Genetics. 1990 Oct;126(2):461–467. doi: 10.1093/genetics/126.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P., Newton K. J. Maize nuclear background regulates the synthesis of a 22-kDa polypeptide in Zea luxurians mitochondria. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7423–7426. doi: 10.1073/pnas.86.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res. 1990 Sep 11;18(17):5189–5196. doi: 10.1093/nar/18.17.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Bonnard G., Lamattina L., Grienenberger J. M. Expression of the wheat mitochondrial nad3-rps12 transcription unit: correlation between editing and mRNA maturation. Plant Cell. 1991 Oct;3(10):1109–1120. doi: 10.1105/tpc.3.10.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Brennicke A. Cytochrome oxidase subunit II mRNAs in Oenothera mitochondria are edited at 24 sites. Curr Genet. 1990 Nov;18(4):371–375. doi: 10.1007/BF00318219. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991 Sep 12;353(6340):178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Kempken F., Mullen J. A., Pring D. R., Tang H. V. RNA editing of sorghum mitochondrial atp6 transcripts changes 15 amino acids and generates a carboxy-terminus identical to yeast. Curr Genet. 1991 Nov;20(5):417–422. doi: 10.1007/BF00317071. [DOI] [PubMed] [Google Scholar]
- Kudla J., Igloi G. L., Metzlaff M., Hagemann R., Kössel H. RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. EMBO J. 1992 Mar;11(3):1099–1103. doi: 10.1002/j.1460-2075.1992.tb05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendran R., Spottswood M. R., Miller D. L. RNA editing by cytidine insertion in mitochondria of Physarum polycephalum. Nature. 1991 Jan 31;349(6308):434–438. doi: 10.1038/349434a0. [DOI] [PubMed] [Google Scholar]
- Maier R. M., Hoch B., Zeltz P., Kössel H. Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell. 1992 May;4(5):609–616. doi: 10.1105/tpc.4.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rasmussen J., Hanson M. R. A NADH dehydrogenase subunit gene is co-transcribed with the abnormal Petunia mitochondrial gene associated with cytoplasmic male sterility. Mol Gen Genet. 1989 Jan;215(2):332–336. doi: 10.1007/BF00339738. [DOI] [PubMed] [Google Scholar]
- Salazar R. A., Pring D. R., Kempken F. Editing of mitochondrial atp9 transcripts from two sorghum lines. Curr Genet. 1991 Dec;20(6):483–486. doi: 10.1007/BF00334776. [DOI] [PubMed] [Google Scholar]
- Schuster W., Wissinger B., Unseld M., Brennicke A. Transcripts of the NADH-dehydrogenase subunit 3 gene are differentially edited in Oenothera mitochondria. EMBO J. 1990 Jan;9(1):263–269. doi: 10.1002/j.1460-2075.1990.tb08104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Sutton C. A., Conklin P. L., Pruitt K. D., Hanson M. R. Editing of pre-mRNAs can occur before cis- and trans-splicing in Petunia mitochondria. Mol Cell Biol. 1991 Aug;11(8):4274–4277. doi: 10.1128/mcb.11.8.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. RNA editing fixes problems in plant mitochondrial transcripts. Trends Genet. 1991 Feb;7(2):37–39. doi: 10.1016/0168-9525(91)90225-F. [DOI] [PubMed] [Google Scholar]
- Ward G. C., Levings C. S., 3rd The protein-encoding gene T-urf13 is not edited in maize mitochondria. Plant Mol Biol. 1991 Nov;17(5):1083–1088. doi: 10.1007/BF00037148. [DOI] [PubMed] [Google Scholar]
- Wintz H., Hanson M. R. A termination codon is created by RNA editing in the petunia atp9 transcript. Curr Genet. 1991 Jan;19(1):61–64. doi: 10.1007/BF00362089. [DOI] [PubMed] [Google Scholar]
- Yang A. J., Mulligan R. M. RNA editing intermediates of cox2 transcripts in maize mitochondria. Mol Cell Biol. 1991 Aug;11(8):4278–4281. doi: 10.1128/mcb.11.8.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]