Abstract
In the title compound, C60H50B2P4·1.5C4H8O, the diphosphadiborane molecule lies on an inversion centre, whereas the disordered tetrahydrofuran solvent molecule is in a general position with a partial occupancy of 0.75. The diphosphadiborane molecule consists of an ideal planar four-membered B2P2 ring with an additional phenyl and a –PPh2 group attached to each B atom.
Related literature
For the structure of a monomeric diphosphaborane molecule, see: Bartlett et al. (1988 ▶). For assumed monomeric PhB(PPh2)2, see: Coates & Livingstone (1961 ▶). For the structures of other dimeric boron-bridged bisphosphine compounds, see: Herdtweck et al. (1997 ▶); Kaufmann et al. (1997 ▶); Nöth (1987 ▶).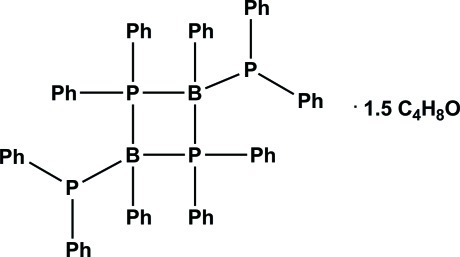
Experimental
Crystal data
C60H50B2P4·1.5C4H8O
M r = 1024.66
Orthorhombic,

a = 19.2421 (4) Å
b = 11.6938 (2) Å
c = 24.9769 (5) Å
V = 5620.13 (19) Å3
Z = 4
Mo Kα radiation
μ = 0.18 mm−1
T = 150 K
0.35 × 0.28 × 0.20 mm
Data collection
Stoe IPDS II diffractometer
Absorption correction: numerical (X-SHAPE and X-RED32; Stoe & Cie, 2005 ▶) T min = 0.927, T max = 0.986
91233 measured reflections
6705 independent reflections
4392 reflections with I > 2σ(I)
R int = 0.072
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.096
S = 0.87
6705 reflections
343 parameters
9 restraints
H-atom parameters constrained
Δρmax = 0.64 e Å−3
Δρmin = −0.29 e Å−3
Data collection: X-AREA (Stoe & Cie, 2005 ▶); cell refinement: X-AREA; data reduction: X-RED32 (Stoe & Cie, 2005 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812011361/yk2048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812011361/yk2048Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the Leibniz-Institut für Katalyse e.V. an der Universität Rostock.
supplementary crystallographic information
Comment
We became interested in such a class of compounds, because the boron-bridged bisphosphines could be potential ligands for the chromium catalyzed selective oligomerization of ethene, like PNP. Unfortunately the synthesis of PhB(PPh2)2, according to the literature (Coates & Livingstone, 1961), failed and always ended up with insoluble polymer. Therefore we changed the procedure using LiPPh2 instead of HPPh2. Examples of structurally characterized borone-bridged bisphosphines are known (Herdtweck et al., 1997; Kaufmann et al., 1997; Nöth, 1987). Only bulky substituents at the boron lead to a monomeric structure (Bartlett et al., 1988). In the present publication, we report on the formation of the dimeric C60H50B2P4. In the structure of the title compound, the diphosphadiborane molecule occupies the position at an inversion center, whereas the solvent molecule of tetrahydrofuran lies in general position with partial occupancy equal to 0.75. In the tetrahydrofuran molecule the O atom is disordered over two sites with occupancies of 0.341 (9): 0.409 (9). All P—B distances in the four-membered ring are essentially identical [B1—P2 = 2.030 (3) Å and B1—P2i = 2.036 (2) Å], and also the B1—P1 bond distance of 2.043 (2) Å is not significantly different. In the B2P2 ring, angles of nearly 90° were observed [P2—B1—P2i = 87.24 (9)° and B1—P2—B1i = 92.75°].
Experimental
PhBCl2 (0.817 ml, 6.3 mmol) was added to a solution of 25 mL Ph2PLi (0.5M in thf) in 20 ml of thf at -40°C and the resulting solution was stirred at room temperature for 48 h. Subsequently, the formed light brown solution was filtered, reduced to the half, over-layered with n-hexane and stored at 0°C. Crystals of the title compound appeared, which were suitable for crystal structure analysis. The white compound was fully characterized by standard analytical methods e.g.31P-NMR: (C6D6): -10.7(br), -42.2(tr) p.p.m..
Refinement
H atoms were placed in idealized positions with d(C—H) = 0.95 Å (CH), 0.99 Å (CH2) and refined using a riding model with Uiso(H) fixed at 1.2 Ueq(C).
Figures
Fig. 1.
The structure of the diphosphadiborane molecule showing the atom-labelling scheme. Hydrogen atoms are omitted for clarity. Displacement ellipsoids are drawn at the 30% probability level.
Crystal data
| C60H50B2P4·1.5C4H8O | Dx = 1.211 Mg m−3 |
| Mr = 1024.66 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 6569 reflections |
| a = 19.2421 (4) Å | θ = 1.6–28.0° |
| b = 11.6938 (2) Å | µ = 0.18 mm−1 |
| c = 24.9769 (5) Å | T = 150 K |
| V = 5620.13 (19) Å3 | Prism, colourless |
| Z = 4 | 0.35 × 0.28 × 0.20 mm |
| F(000) = 2160 |
Data collection
| Stoe IPDS II diffractometer | 6705 independent reflections |
| Radiation source: fine-focus sealed tube | 4392 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.072 |
| ω scans | θmax = 28.0°, θmin = 2.2° |
| Absorption correction: numerical (X-SHAPE and X-RED32; Stoe & Cie, 2005) | h = −25→25 |
| Tmin = 0.927, Tmax = 0.986 | k = −15→15 |
| 91233 measured reflections | l = −32→32 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.096 | H-atom parameters constrained |
| S = 0.87 | w = 1/[σ2(Fo2) + (0.0536P)2 + 0.0P] where P = (Fo2 + 2Fc2)/3 |
| 6705 reflections | (Δ/σ)max = 0.002 |
| 343 parameters | Δρmax = 0.64 e Å−3 |
| 9 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1A | 0.1872 (2) | 0.6123 (6) | 0.2276 (2) | 0.061 (2)* | 0.341 (9) |
| O1B | 0.1940 (2) | 0.5614 (5) | 0.24577 (17) | 0.0613 (19)* | 0.409 (9) |
| C31 | 0.2198 (2) | 0.5279 (4) | 0.1944 (2) | 0.0981 (17) | 0.75 |
| H31A | 0.1909 | 0.5086 | 0.1629 | 0.118* | 0.341 (9) |
| H31B | 0.2305 | 0.4572 | 0.2147 | 0.118* | 0.341 (9) |
| H31C | 0.1834 | 0.5419 | 0.1672 | 0.118* | 0.409 (9) |
| H31D | 0.2297 | 0.4448 | 0.1948 | 0.118* | 0.409 (9) |
| C32 | 0.2832 (2) | 0.5897 (4) | 0.17909 (16) | 0.0913 (16) | 0.75 |
| H32A | 0.2749 | 0.6378 | 0.1471 | 0.110* | 0.75 |
| H32B | 0.3214 | 0.5356 | 0.1711 | 0.110* | 0.75 |
| C33 | 0.3004 (2) | 0.6610 (4) | 0.22580 (13) | 0.0766 (13) | 0.75 |
| H33A | 0.3059 | 0.7420 | 0.2151 | 0.092* | 0.75 |
| H33B | 0.3441 | 0.6346 | 0.2427 | 0.092* | 0.75 |
| C34 | 0.24158 (17) | 0.6479 (4) | 0.26295 (13) | 0.0627 (10) | 0.75 |
| H34A | 0.2513 | 0.5891 | 0.2905 | 0.075* | 0.341 (9) |
| H34B | 0.2301 | 0.7211 | 0.2808 | 0.075* | 0.341 (9) |
| H34C | 0.2595 | 0.6279 | 0.2989 | 0.075* | 0.409 (9) |
| H34D | 0.2167 | 0.7217 | 0.2659 | 0.075* | 0.409 (9) |
| C1 | 1.06155 (9) | −0.01162 (16) | 0.34268 (7) | 0.0246 (4) | |
| C2 | 1.00019 (10) | 0.02724 (18) | 0.31988 (7) | 0.0307 (4) | |
| H2 | 0.9719 | 0.0795 | 0.3392 | 0.037* | |
| C3 | 0.97947 (11) | −0.00883 (19) | 0.26950 (8) | 0.0364 (5) | |
| H3 | 0.9371 | 0.0185 | 0.2547 | 0.044* | |
| C4 | 1.01996 (11) | −0.0841 (2) | 0.24080 (8) | 0.0369 (5) | |
| H4 | 1.0057 | −0.1089 | 0.2063 | 0.044* | |
| C5 | 1.08146 (11) | −0.12344 (19) | 0.26268 (8) | 0.0369 (5) | |
| H5 | 1.1099 | −0.1749 | 0.2430 | 0.044* | |
| C6 | 1.10164 (10) | −0.08786 (17) | 0.31321 (7) | 0.0308 (4) | |
| H6 | 1.1437 | −0.1161 | 0.3280 | 0.037* | |
| C7 | 1.13848 (10) | 0.16430 (16) | 0.39173 (7) | 0.0257 (4) | |
| C8 | 1.20183 (10) | 0.18941 (18) | 0.41622 (7) | 0.0299 (4) | |
| H8 | 1.2214 | 0.1365 | 0.4408 | 0.036* | |
| C9 | 1.23674 (11) | 0.29054 (19) | 0.40521 (9) | 0.0376 (5) | |
| H9 | 1.2794 | 0.3070 | 0.4228 | 0.045* | |
| C10 | 1.20972 (12) | 0.36709 (19) | 0.36884 (9) | 0.0414 (5) | |
| H10 | 1.2332 | 0.4369 | 0.3617 | 0.050* | |
| C11 | 1.14820 (11) | 0.34169 (18) | 0.34279 (9) | 0.0383 (5) | |
| H11 | 1.1302 | 0.3933 | 0.3169 | 0.046* | |
| C12 | 1.11277 (10) | 0.24195 (18) | 0.35416 (8) | 0.0319 (4) | |
| H12 | 1.0704 | 0.2259 | 0.3362 | 0.038* | |
| C13 | 0.98211 (9) | 0.19751 (16) | 0.43708 (7) | 0.0240 (4) | |
| C14 | 1.01833 (11) | 0.29722 (16) | 0.45140 (7) | 0.0304 (4) | |
| H14 | 1.0579 | 0.2909 | 0.4740 | 0.036* | |
| C15 | 0.99832 (12) | 0.40388 (18) | 0.43376 (9) | 0.0390 (5) | |
| H15 | 1.0236 | 0.4696 | 0.4448 | 0.047* | |
| C16 | 0.94210 (12) | 0.41587 (19) | 0.40029 (9) | 0.0427 (6) | |
| H16 | 0.9291 | 0.4892 | 0.3874 | 0.051* | |
| C17 | 0.90491 (12) | 0.32050 (19) | 0.38570 (8) | 0.0382 (5) | |
| H17 | 0.8659 | 0.3282 | 0.3627 | 0.046* | |
| C18 | 0.92386 (10) | 0.21293 (17) | 0.40430 (7) | 0.0280 (4) | |
| H18 | 0.8967 | 0.1484 | 0.3946 | 0.034* | |
| C19 | 0.86269 (9) | −0.06475 (16) | 0.46070 (6) | 0.0237 (4) | |
| C20 | 0.82663 (9) | 0.03413 (16) | 0.47541 (7) | 0.0263 (4) | |
| H20 | 0.8517 | 0.1014 | 0.4844 | 0.032* | |
| C21 | 0.75461 (10) | 0.03493 (18) | 0.47697 (8) | 0.0314 (4) | |
| H21 | 0.7306 | 0.1022 | 0.4876 | 0.038* | |
| C22 | 0.71758 (10) | −0.06199 (19) | 0.46314 (8) | 0.0344 (4) | |
| H22 | 0.6682 | −0.0612 | 0.4639 | 0.041* | |
| C23 | 0.75253 (10) | −0.15974 (18) | 0.44826 (8) | 0.0328 (4) | |
| H23 | 0.7271 | −0.2259 | 0.4381 | 0.039* | |
| C24 | 0.82462 (10) | −0.16246 (17) | 0.44795 (7) | 0.0280 (4) | |
| H24 | 0.8482 | −0.2313 | 0.4390 | 0.034* | |
| C25 | 0.98791 (10) | −0.18291 (16) | 0.42454 (7) | 0.0243 (4) | |
| C26 | 0.95519 (11) | −0.21012 (17) | 0.37607 (7) | 0.0295 (4) | |
| H26 | 0.9138 | −0.1714 | 0.3660 | 0.035* | |
| C27 | 0.98292 (11) | −0.29323 (18) | 0.34282 (8) | 0.0359 (5) | |
| H27 | 0.9600 | −0.3124 | 0.3104 | 0.043* | |
| C28 | 1.04372 (12) | −0.34830 (17) | 0.35668 (8) | 0.0385 (5) | |
| H28 | 1.0624 | −0.4054 | 0.3338 | 0.046* | |
| C29 | 1.07740 (11) | −0.32094 (17) | 0.40355 (8) | 0.0337 (5) | |
| H29 | 1.1197 | −0.3581 | 0.4126 | 0.040* | |
| C30 | 1.04967 (10) | −0.23923 (16) | 0.43754 (7) | 0.0268 (4) | |
| H30 | 1.0729 | −0.2213 | 0.4700 | 0.032* | |
| B1 | 1.01311 (10) | 0.07717 (18) | 0.45461 (7) | 0.0212 (4) | |
| P1 | 1.09614 (2) | 0.02764 (4) | 0.409099 (17) | 0.02250 (11) | |
| P2 | 0.95699 (2) | −0.06707 (4) | 0.467327 (17) | 0.02053 (10) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C31 | 0.079 (3) | 0.089 (4) | 0.126 (4) | −0.017 (3) | −0.022 (3) | −0.053 (3) |
| C32 | 0.098 (4) | 0.128 (4) | 0.048 (2) | −0.003 (3) | 0.015 (2) | −0.035 (3) |
| C33 | 0.081 (3) | 0.080 (3) | 0.069 (3) | −0.027 (2) | 0.015 (2) | −0.019 (2) |
| C34 | 0.048 (2) | 0.089 (3) | 0.0510 (19) | −0.013 (2) | 0.0032 (16) | −0.0253 (19) |
| C1 | 0.0280 (9) | 0.0259 (10) | 0.0200 (8) | −0.0036 (8) | 0.0021 (7) | 0.0005 (7) |
| C2 | 0.0311 (10) | 0.0355 (11) | 0.0256 (9) | 0.0051 (9) | −0.0015 (8) | −0.0013 (8) |
| C3 | 0.0369 (11) | 0.0464 (13) | 0.0260 (9) | 0.0042 (10) | −0.0062 (8) | 0.0006 (9) |
| C4 | 0.0420 (12) | 0.0480 (13) | 0.0207 (9) | −0.0042 (10) | −0.0003 (8) | −0.0058 (9) |
| C5 | 0.0399 (12) | 0.0422 (13) | 0.0285 (10) | 0.0029 (10) | 0.0061 (8) | −0.0082 (9) |
| C6 | 0.0287 (10) | 0.0343 (11) | 0.0294 (9) | 0.0007 (9) | 0.0010 (8) | −0.0022 (8) |
| C7 | 0.0261 (9) | 0.0268 (10) | 0.0243 (9) | −0.0006 (8) | 0.0043 (7) | −0.0015 (7) |
| C8 | 0.0293 (10) | 0.0337 (11) | 0.0267 (9) | −0.0032 (8) | 0.0019 (8) | 0.0003 (8) |
| C9 | 0.0321 (11) | 0.0420 (12) | 0.0387 (11) | −0.0092 (9) | 0.0044 (9) | −0.0064 (10) |
| C10 | 0.0395 (12) | 0.0311 (12) | 0.0535 (13) | −0.0086 (10) | 0.0155 (10) | 0.0006 (10) |
| C11 | 0.0370 (12) | 0.0322 (12) | 0.0458 (12) | 0.0028 (9) | 0.0100 (10) | 0.0113 (9) |
| C12 | 0.0271 (10) | 0.0330 (11) | 0.0355 (10) | −0.0004 (8) | 0.0028 (8) | 0.0068 (9) |
| C13 | 0.0284 (9) | 0.0237 (9) | 0.0199 (8) | 0.0006 (8) | 0.0053 (7) | 0.0009 (7) |
| C14 | 0.0389 (11) | 0.0246 (10) | 0.0276 (9) | −0.0031 (9) | 0.0080 (8) | 0.0011 (8) |
| C15 | 0.0532 (13) | 0.0237 (11) | 0.0401 (11) | −0.0005 (10) | 0.0192 (10) | 0.0018 (9) |
| C16 | 0.0525 (13) | 0.0302 (12) | 0.0455 (12) | 0.0149 (10) | 0.0232 (10) | 0.0159 (10) |
| C17 | 0.0359 (11) | 0.0462 (13) | 0.0324 (10) | 0.0167 (10) | 0.0111 (9) | 0.0150 (9) |
| C18 | 0.0290 (10) | 0.0321 (10) | 0.0229 (8) | 0.0056 (8) | 0.0054 (7) | 0.0043 (8) |
| C19 | 0.0248 (8) | 0.0265 (9) | 0.0198 (8) | −0.0023 (8) | −0.0019 (7) | 0.0022 (7) |
| C20 | 0.0278 (9) | 0.0252 (10) | 0.0257 (8) | −0.0013 (8) | −0.0036 (7) | 0.0019 (7) |
| C21 | 0.0294 (10) | 0.0317 (11) | 0.0331 (10) | 0.0033 (8) | −0.0045 (8) | 0.0013 (8) |
| C22 | 0.0230 (9) | 0.0424 (12) | 0.0379 (10) | −0.0015 (9) | −0.0057 (8) | 0.0018 (9) |
| C23 | 0.0290 (10) | 0.0330 (11) | 0.0363 (10) | −0.0093 (9) | −0.0073 (8) | 0.0008 (9) |
| C24 | 0.0289 (10) | 0.0267 (10) | 0.0285 (9) | −0.0021 (8) | −0.0035 (8) | 0.0012 (8) |
| C25 | 0.0300 (10) | 0.0207 (9) | 0.0221 (8) | −0.0051 (8) | 0.0038 (7) | −0.0002 (7) |
| C26 | 0.0335 (10) | 0.0294 (10) | 0.0255 (9) | −0.0058 (9) | 0.0008 (8) | −0.0008 (7) |
| C27 | 0.0471 (13) | 0.0333 (11) | 0.0271 (9) | −0.0123 (10) | 0.0023 (9) | −0.0075 (8) |
| C28 | 0.0524 (13) | 0.0265 (11) | 0.0367 (11) | −0.0033 (10) | 0.0130 (10) | −0.0102 (9) |
| C29 | 0.0394 (11) | 0.0244 (10) | 0.0375 (11) | 0.0005 (8) | 0.0080 (9) | 0.0003 (8) |
| C30 | 0.0306 (10) | 0.0236 (10) | 0.0263 (9) | −0.0034 (8) | 0.0037 (8) | 0.0015 (7) |
| B1 | 0.0235 (9) | 0.0220 (10) | 0.0182 (9) | −0.0021 (8) | −0.0006 (7) | 0.0005 (7) |
| P1 | 0.0239 (2) | 0.0231 (2) | 0.0206 (2) | 0.0002 (2) | −0.00034 (18) | 0.00079 (18) |
| P2 | 0.0229 (2) | 0.0195 (2) | 0.01925 (19) | −0.00212 (19) | −0.00134 (18) | −0.00060 (17) |
Geometric parameters (Å, º)
| O1A—C34 | 1.431 (3) | C12—H12 | 0.9500 |
| O1A—C31 | 1.433 (4) | C13—C18 | 1.400 (3) |
| O1B—C34 | 1.429 (3) | C13—C14 | 1.405 (3) |
| O1B—C31 | 1.430 (4) | C13—B1 | 1.590 (3) |
| C31—C32 | 1.470 (3) | C14—C15 | 1.378 (3) |
| C31—H31A | 0.9900 | C14—H14 | 0.9500 |
| C31—H31B | 0.9900 | C15—C16 | 1.374 (3) |
| C31—H31C | 0.9900 | C15—H15 | 0.9500 |
| C31—H31D | 0.9900 | C16—C17 | 1.374 (3) |
| C32—C33 | 1.471 (3) | C16—H16 | 0.9500 |
| C32—H32A | 0.9900 | C17—C18 | 1.390 (3) |
| C32—H32B | 0.9900 | C17—H17 | 0.9500 |
| C33—C34 | 1.472 (3) | C18—H18 | 0.9500 |
| C33—H33A | 0.9900 | C19—C24 | 1.394 (3) |
| C33—H33B | 0.9900 | C19—C20 | 1.398 (3) |
| C34—H34A | 0.9900 | C19—P2 | 1.8222 (18) |
| C34—H34B | 0.9900 | C20—C21 | 1.386 (3) |
| C34—H34C | 0.9900 | C20—H20 | 0.9500 |
| C34—H34D | 0.9900 | C21—C22 | 1.383 (3) |
| C1—C2 | 1.387 (3) | C21—H21 | 0.9500 |
| C1—C6 | 1.390 (3) | C22—C23 | 1.377 (3) |
| C1—P1 | 1.8455 (18) | C22—H22 | 0.9500 |
| C2—C3 | 1.386 (3) | C23—C24 | 1.388 (3) |
| C2—H2 | 0.9500 | C23—H23 | 0.9500 |
| C3—C4 | 1.377 (3) | C24—H24 | 0.9500 |
| C3—H3 | 0.9500 | C25—C30 | 1.397 (3) |
| C4—C5 | 1.382 (3) | C25—C26 | 1.401 (2) |
| C4—H4 | 0.9500 | C25—P2 | 1.8251 (19) |
| C5—C6 | 1.385 (3) | C26—C27 | 1.385 (3) |
| C5—H5 | 0.9500 | C26—H26 | 0.9500 |
| C6—H6 | 0.9500 | C27—C28 | 1.380 (3) |
| C7—C8 | 1.395 (3) | C27—H27 | 0.9500 |
| C7—C12 | 1.396 (3) | C28—C29 | 1.376 (3) |
| C7—P1 | 1.8454 (19) | C28—H28 | 0.9500 |
| C8—C9 | 1.388 (3) | C29—C30 | 1.385 (3) |
| C8—H8 | 0.9500 | C29—H29 | 0.9500 |
| C9—C10 | 1.377 (3) | C30—H30 | 0.9500 |
| C9—H9 | 0.9500 | B1—P2 | 2.028 (2) |
| C10—C11 | 1.383 (3) | B1—P2i | 2.0361 (18) |
| C10—H10 | 0.9500 | B1—P1 | 2.045 (2) |
| C11—C12 | 1.381 (3) | P2—B1i | 2.0363 (18) |
| C11—H11 | 0.9500 | ||
| C34—O1A—C31 | 103.7 (3) | C9—C10—H10 | 120.2 |
| C34—O1B—C31 | 104.0 (3) | C11—C10—H10 | 120.2 |
| O1B—C31—C32 | 112.7 (3) | C12—C11—C10 | 120.5 (2) |
| O1A—C31—C32 | 100.1 (4) | C12—C11—H11 | 119.8 |
| O1B—C31—H31A | 125.6 | C10—C11—H11 | 119.8 |
| O1A—C31—H31A | 111.8 | C11—C12—C7 | 120.84 (19) |
| C32—C31—H31A | 111.8 | C11—C12—H12 | 119.6 |
| O1B—C31—H31B | 81.0 | C7—C12—H12 | 119.6 |
| O1A—C31—H31B | 111.8 | C18—C13—C14 | 116.06 (17) |
| C32—C31—H31B | 111.8 | C18—C13—B1 | 125.14 (17) |
| H31A—C31—H31B | 109.5 | C14—C13—B1 | 118.58 (16) |
| O1B—C31—H31C | 109.0 | C15—C14—C13 | 122.1 (2) |
| O1A—C31—H31C | 88.5 | C15—C14—H14 | 119.0 |
| C32—C31—H31C | 109.0 | C13—C14—H14 | 119.0 |
| H31B—C31—H31C | 129.5 | C16—C15—C14 | 120.5 (2) |
| O1B—C31—H31D | 109.0 | C16—C15—H15 | 119.8 |
| O1A—C31—H31D | 138.9 | C14—C15—H15 | 119.8 |
| C32—C31—H31D | 109.0 | C15—C16—C17 | 119.23 (19) |
| H31A—C31—H31D | 83.9 | C15—C16—H16 | 120.4 |
| H31C—C31—H31D | 107.8 | C17—C16—H16 | 120.4 |
| C31—C32—C33 | 105.0 (3) | C16—C17—C18 | 120.6 (2) |
| C31—C32—H32A | 110.8 | C16—C17—H17 | 119.7 |
| C33—C32—H32A | 110.8 | C18—C17—H17 | 119.7 |
| C31—C32—H32B | 110.8 | C17—C18—C13 | 121.5 (2) |
| C33—C32—H32B | 110.8 | C17—C18—H18 | 119.3 |
| H32A—C32—H32B | 108.8 | C13—C18—H18 | 119.3 |
| C32—C33—C34 | 105.6 (3) | C24—C19—C20 | 118.51 (16) |
| C32—C33—H33A | 110.6 | C24—C19—P2 | 122.12 (14) |
| C34—C33—H33A | 110.6 | C20—C19—P2 | 118.87 (14) |
| C32—C33—H33B | 110.6 | C21—C20—C19 | 120.61 (18) |
| C34—C33—H33B | 110.6 | C21—C20—H20 | 119.7 |
| H33A—C33—H33B | 108.8 | C19—C20—H20 | 119.7 |
| O1B—C34—C33 | 112.1 (3) | C22—C21—C20 | 120.17 (19) |
| O1A—C34—C33 | 101.7 (3) | C22—C21—H21 | 119.9 |
| O1B—C34—H34A | 80.7 | C20—C21—H21 | 119.9 |
| O1A—C34—H34A | 111.4 | C23—C22—C21 | 119.75 (18) |
| C33—C34—H34A | 111.4 | C23—C22—H22 | 120.1 |
| O1B—C34—H34B | 127.1 | C21—C22—H22 | 120.1 |
| O1A—C34—H34B | 111.4 | C22—C23—C24 | 120.57 (18) |
| C33—C34—H34B | 111.4 | C22—C23—H23 | 119.7 |
| H34A—C34—H34B | 109.3 | C24—C23—H23 | 119.7 |
| O1B—C34—H34C | 109.2 | C23—C24—C19 | 120.35 (18) |
| O1A—C34—H34C | 138.2 | C23—C24—H24 | 119.8 |
| C33—C34—H34C | 109.2 | C19—C24—H24 | 119.8 |
| H34B—C34—H34C | 82.8 | C30—C25—C26 | 118.43 (17) |
| O1B—C34—H34D | 109.2 | C30—C25—P2 | 119.41 (13) |
| O1A—C34—H34D | 86.8 | C26—C25—P2 | 121.88 (15) |
| C33—C34—H34D | 109.2 | C27—C26—C25 | 120.28 (19) |
| H34A—C34—H34D | 130.1 | C27—C26—H26 | 119.9 |
| H34C—C34—H34D | 107.9 | C25—C26—H26 | 119.9 |
| C2—C1—C6 | 117.71 (17) | C28—C27—C26 | 120.24 (19) |
| C2—C1—P1 | 126.48 (14) | C28—C27—H27 | 119.9 |
| C6—C1—P1 | 115.81 (14) | C26—C27—H27 | 119.9 |
| C3—C2—C1 | 121.20 (19) | C29—C28—C27 | 120.29 (19) |
| C3—C2—H2 | 119.4 | C29—C28—H28 | 119.9 |
| C1—C2—H2 | 119.4 | C27—C28—H28 | 119.9 |
| C4—C3—C2 | 120.30 (19) | C28—C29—C30 | 120.0 (2) |
| C4—C3—H3 | 119.9 | C28—C29—H29 | 120.0 |
| C2—C3—H3 | 119.9 | C30—C29—H29 | 120.0 |
| C3—C4—C5 | 119.42 (18) | C29—C30—C25 | 120.69 (18) |
| C3—C4—H4 | 120.3 | C29—C30—H30 | 119.7 |
| C5—C4—H4 | 120.3 | C25—C30—H30 | 119.7 |
| C4—C5—C6 | 120.05 (19) | C13—B1—P2 | 125.41 (13) |
| C4—C5—H5 | 120.0 | C13—B1—P2i | 114.90 (12) |
| C6—C5—H5 | 120.0 | P2—B1—P2i | 87.27 (8) |
| C5—C6—C1 | 121.32 (19) | C13—B1—P1 | 113.00 (12) |
| C5—C6—H6 | 119.3 | P2—B1—P1 | 105.52 (9) |
| C1—C6—H6 | 119.3 | P2i—B1—P1 | 107.17 (9) |
| C8—C7—C12 | 117.86 (18) | C7—P1—C1 | 99.40 (8) |
| C8—C7—P1 | 117.71 (14) | C7—P1—B1 | 103.32 (8) |
| C12—C7—P1 | 124.38 (15) | C1—P1—B1 | 106.76 (8) |
| C9—C8—C7 | 121.03 (19) | C19—P2—C25 | 106.41 (9) |
| C9—C8—H8 | 119.5 | C19—P2—B1 | 120.25 (8) |
| C7—C8—H8 | 119.5 | C25—P2—B1 | 110.61 (8) |
| C10—C9—C8 | 120.13 (19) | C19—P2—B1i | 111.67 (8) |
| C10—C9—H9 | 119.9 | C25—P2—B1i | 115.19 (8) |
| C8—C9—H9 | 119.9 | B1—P2—B1i | 92.73 (8) |
| C9—C10—C11 | 119.6 (2) |
Symmetry code: (i) −x+2, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: YK2048).
References
- Bartlett, R. U., Dias, H. V. R. & Power, P. P. (1988). Inorg. Chem. 27, 3919–3922.
- Coates, G. E. & Livingstone, J. G. (1961). J. Chem. Soc. pp. 5053–5055.
- Herdtweck, E., Jäckle, F. & Wagner, M. (1997). Organometallics, 16, 4737–4745.
- Kaufmann, B., Jetzfellner, R., Leissring, E., Issleib, K., Nöth, H. & Schmidt, M. (1997). Chem. Ber. 130, 1677–1692.
- Nöth, H. (1987). Z. Anorg. Allg. Chem. 555, 79–84.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Stoe & Cie (2005). X-SHAPE, X-RED32 and X-AREA Stoe & Cie, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812011361/yk2048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812011361/yk2048Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



