Abstract
The family of p21-activated protein kinases (PAKs) is composed of serine–threonine kinases whose activity is regulated by the small guanosine triphosphatases (GTPases) Rac and Cdc42. In mammalian cells, PAKs have been implicated in the regulation of mitogen-activated protein cascades, cellular morphological and cytoskeletal changes, neurite outgrowth, and cell apoptosis. Although the ability of Cdc42 and Rac GTPases to activate PAK is well established, relatively little is known about the negative regulation of PAK or the identity of PAK cellular targets. Here, we describe the identification and characterization of a human PAK-interacting protein, hPIP1. hPIP1 contains G protein β-like WD repeats and shares sequence homology with the essential fission yeast PAK regulator, Skb15, as well as the essential budding yeast protein, MAK11. Interaction of hPIP1 with PAK1 inhibits the Cdc42/Rac-stimulated kinase activity through the N-terminal regulatory domains of PAK1. Cotransfection of hPIP1 in mammalian cells inhibits PAK-mediated c-Jun N-terminal kinase and nuclear factor κ B signaling pathways. Our results demonstrate that hPIP1 is a negative regulator of PAK and PAK signaling pathways.
The p21-activated kinases (PAKs) are members of a family of serine–threonine protein kinases that are regulated by the small GTPases, Cdc42 and Rac (1–4). There are four known PAK isoforms (PAK 1–4) that are differentially expressed in mammalian tissues (3, 4). Ste20, Cla4, and Shks are PAK homologs in the budding yeast and the fission yeast, respectively (5, 6). PAKs regulate diverse cellular functions, including gene expression, cytoskeletal actin assembly, mitogen-activated protein kinase (MAPK) pathways, neurite outgrowth, cell cycle control, and cell apoptosis (7–16). In mammalian cells, PAKs have been reported to activate the c-Jun N-terminal kinase (JNK) and nuclear factor κ B (NF-κB) pathways under different conditions (9, 17–20). Constitutively activated PAK mutants can activate the JNK–MAPK cascade, leading to transcriptional control (9). In HIV1-infected cells, PAKs bind the HIV Nef protein and play a critical role in cellular activation by Nef and in HIV pathogenesis (21, 22).
PAKs share a common structural organization consisting of an N-terminal half that includes autoregulatory and interaction segments and a C-terminal kinase domain. Both the crystal and solution structures of PAK show that the protein forms dimers (two autoregulatory fragments and two kinase domains) held together by the interaction of the autoregulatory fragments (23). Binding of GTPases (Cdc42/Rac) to the N-terminal regulatory domains triggers a series of conformational changes that lead to the disruption of the PAK dimer and the activation of the kinase. Regulation of PAK activity depends on both the autoregulatory domains of the molecule and the state of Thr-423 phosphorylation in the kinase-activation domain (24, 25). Mutations within the autoregulatory domains and at the autophosphorylation site (Ser-423 → Glu) produce constitutively active PAK (8, 24, 26).
The apparent multiplicity of PAK structure and PAK-mediated signaling pathways suggests that PAK activity must be tightly regulated. PAKs can be activated in vivo and in vitro by binding to GTP-bound Cdc42 and Rac. In response to tyrosine phosphorylation, adaptor protein Nck can recruit PAK1 to the membrane to increase its activity and specificity (27–29). In the budding yeast Saccharomyces cerevisiae, the PAK1 homologue Ste20 is involved in transmitting the mating-pheromone signal from the Gβγ-subunits of a heterotrimeric G protein to a downstream MAPK cascade (30). Binding of Gβ with the noncatalytic C-terminal region of Ste20 is essential for the activation of the MAPK cascade in signaling from the mating factor receptor (31). Recently, a family of PAK-interacting exchange factors (PIXs) for Rac and Cdc42 were identified as binding tightly to the fourth proline-rich domain in the N terminus of PAK (32, 33). PIX can regulate PAK activity both by catalyzing GTP exchange on Cdc42/Rac and by direct binding to PAK (34). However, we still know little about the negative modulation of PAK and PAK-mediated signaling pathways. In this report, we describe the identification and characterization of a human PAK1-interacting protein, hPIP1. Sequence analysis of hPIP1 indicates that the protein contains five G protein β-like WD repeats and shares sequence homology with the protein MAK11 in budding yeast (35) and with the PAK regulator Skb15 in fission yeast (37), both of which are essential for cell growth. hPIP1 interacts with PAK1 both in vivo and in vitro. Furthermore, transfection of hPIP1 in mammalian cells inhibits the Cdc42-stimulated PAK kinase activity, PAK-mediated JNK, and NF-κB signaling pathways presumably through binding to the N-terminal regulatory domains of PAK1. These results suggest that hPIP1 is a negative regulator of PAK and PAK signaling pathways.
Materials and Methods
Cloning of hPIP1 and Construction of Expression Plasmids.
A yeast two-hybrid system was used to clone hPIP1. Phospholipase C (PLC)-β3 was subcloned into the Gal4 DNA-binding domain in the vector pGBKT7. The bait vector was transformed into the yeast strain AH109. A pretransformed human heart MatchMaker cDNA library (CLONTECH) was screened. Interacting proteins were selected on synthetic plates lacking four different nutrients (tryptophan, leucine, histidine, and adenine). The interaction was further tested by the activation of a second reporter gene, lacZ, by filter assays. For expression of hPIP1 in bacteria, hPIP1 was expressed as glutathione S-transferase (GST)-fusion protein by subcloning into pGEX-2T (Amersham Pharmacia). For mammalian expression, hPIP1 was subcloned into a FLAG-tagged vector, pCMV-Tag2 (Stratagene).
Protein Kinase Assays.
Cdc42V12 (1 μg) was cotransfected into COS-7 cells with PAK1 (2 μg) or the mutant PAK1T423E (2 μg) and hPIP1 (2 μg) expression vectors, where appropriate. Hemagglutinin (HA)-tagged PAK1 or HA-JNK was immunoprecipitated by using anti-HA monoclonal antibody 12CA5 (Roche Molecular Biochemicals) or rabbit anti-PAK1 antibody. The immunoprecipitates were washed three times in Nonidet P-40 lysis buffer, once in 0.5 M LiCl, and then once in protein kinase buffer (50 mM Hepes, pH 7.5/10 mM MgCl2/1 mM DTT/0.05% Triton X-100). The immunoprecipitates were then incubated at 30°C for 30 min in 50 μl of protein kinase buffer. For in vitro PAK1 activation, 4 μg of Cdc42 bound to the γ-thio analog of GTP (GTPγS-Cdc42) was added to the reaction with or without 4 μg of hPIP1. Samples were resolved on SDS/12% PAGE and processed for autoradiography.
In Vitro Translation, Binding Assays, and Immunoprecipitation.
In vitro transcription and translation of proteins were performed with a kit essentially according to the manufacturer's instructions (Promega). Briefly, plasmids (1–2 μg per reaction) were added to a reaction mixture that included 25 μl of rabbit reticulocyte lysate, 2 μl of reaction buffer, 1 μl of T3 RNA polymerase, 1 μl of amino acid mixture minus methionine (1 mM), 4 μl of [35S]methionine (10 mCi/ml; 1 Ci = 37 GBq), ribonuclease inhibitor (40 units/μl), and nuclease-free H2O to a final volume of 50 μl. The reactions were incubated at 30°C for 90 min. The in vitro translated proteins were then used for in vitro binding assays. The interactions of in vitro translated proteins with GST-PAK1 fusion proteins were directly examined by autoradiography.
Binding of PAK1 with hPIP1 in mammalian cells was examined by immunoprecipitation and by Western blot analysis. Briefly, cells were lysed with a cell extraction buffer (1% Nonidet P-40/20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM EDTA/1 mM Na3VO4/2 mM PMSF/20 μg/ml aprotinin/3 μg/ml leupeptin/3 μg/ml pepstatin) and immunoprecipitated with the indicated antibodies. Anti-PAK1 and anti-FLAG immunocomplexes were recovered by using protein A and G beads (Sigma). All immunoprecipitates were washed four times with lysis buffer and were separated by SDS/PAGE and then transferred to poly(vinylidene difluoride) (PVDF) membranes (Millipore). After incubation in TBST (20 mM Tris⋅HCl, pH 7.5/500 mM NaCl/0.02% Tween-20) containing 2% BSA or 10% dry milk powder, the membranes were probed with the indicated antibodies and visualized with the SuperSignal West Pico detection system (Pierce).
Transient Transfection and Reporter Assays.
COS-7 or mouse embryonic fibroblast cells were seeded in DMEM containing 10% (vol/vol) FBS 24 h before transfection. Cells were transfected with Lipofectamine (BRL) in serum-free Opti-MEM (BRL). A cytomegalovirus vector pCIS encoding β-galactosidase was used to maintain a constant amount of cDNA and equalize the amount of a particular cDNA in each set of experiments. For transcriptional reporter assays, the PathDetect AP-1 cis-Reporting System (pAP1-Luc, 7× AP-1 enhancer elements) and the PathDetect NF-κB cis-Reporting System (pNF-kB-Luc, 5× NF-κB enhancer elements) from Stratagene were used in the assays. Luciferase assays were performed with a kit as recommended by the manufacturer's instructions (Promega). The data presented are the mean of three individually transfected wells. The experiments were performed at least three times.
Immunocytochemistry.
Cells were fixed with 4% (vol/vol) paraformaldehyde for 20 min, blocked with 10% (wt/vol) BSA, and incubated with monoclonal antibody against the FLAG epitope (M2 monoclonal, Sigma). Actin filaments were labeled by rhodamine-conjugated phalloidin. Fluorescent images of cells were captured on a Sony charge-coupled device camera mounted on a Nikon E600 microscope using photoshop imaging software.
Results
Cloning and Expression of hPIP1.
To understand the regulation of PAK signaling pathways, we examined the interaction of PAK with a human protein, hPIP1 (human PAK/PLC-interacting protein 1). hPIP1 was initially identified from a yeast two-hybrid screen by using PLC-β3 as a bait. Sequence analysis revealed that hPIP1 contains five putative WD repeat domains like the G protein β-subunits (Fig. 1 A and B). On the other hand, hPIP1 also shares more than 25% sequence identity and a highly positively charged C-terminal domain with the protein MAK11 in the budding yeast Saccharomyces cerevisiae (35) and Skb15 (the yeast PAK-binding protein) in the fission yeast Schizosaccharomyces pombe (ref. 37; Fig. 1C). Sequence homology with the fission yeast PAK (Shk1)-binding protein Skb15 suggests that hPIP1 may interact with human PAK and modulate PAK-mediated signaling pathways.
Figure 1.
Amino acid sequence and domain structure of hPIP1. (A) Deduced amino acid sequence of hPIP1. (B) Comparison of WD repeats of hPIP1 with G protein β1-subunit. :, identical residues; +, conservative changes. (C) Domain structures of hPIP1 and two yeast homologs, Skb15 in Schizosaccharomyces pombe and MAK11 in Saccharomyces cerevisiae. These three proteins contain five or six WD repeats and a C-terminal positively charged domain.
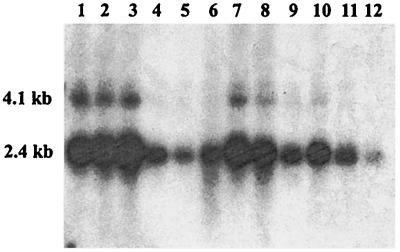
To analyze the tissue distribution of hPIP1, we examined the expression of the protein in different human tissues by using Northern blot analysis. As shown in Fig. 2, hPIP1 is expressed in most human tissues, with a higher level of expression in brain, heart, and muscle.
Figure 2.
Northern blot analysis of hPIP1 expression in different human tissues. Multiple Tissue Northern (MTN) blots were prepared from the purified poly(A)+ RNA and blotted onto a positively charged nylon membrane (CLONTECH). The amount of RNA on an MTN blot membrane is adjusted so that the β-actin hybridization signal is of comparable intensity in every lane (CLONTECH). RNA was isolated from following human tissues: 1, brain; 2, heart; 3, muscle; 4, colon; 5, thymus; 6, spleen; 7, kidney; 8, liver; 9, small intestine; 10, placenta; 11, lung; 12, peripheral blood leukocytes. The size of hPIP1 cDNA was estimated to be ≈2.4 kb with minor bands at 4.1 kb.
hPIP1 Interacts with PAK1 in Vitro and in Vivo.
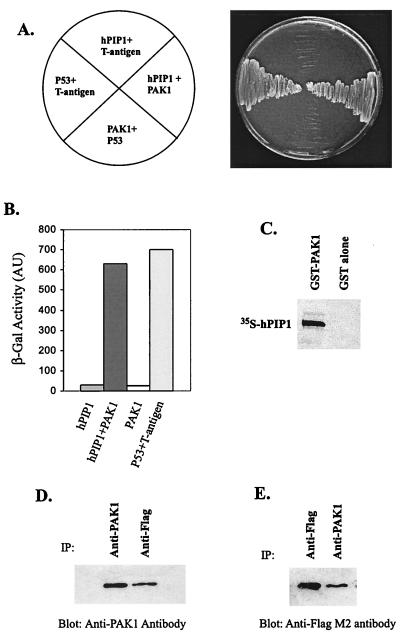
To determine whether hPIP1 interacts with PAK, we cotransformed a yeast two-hybrid test strain with vectors expressing Gal4 DNA-binding domain hPIP1 fusion protein (pGBD-hPIP1) and Gal4 activation domain PAK1 fusion protein (pGAD-PAK1). GBD-hPIP1 interacts with GAD-PAK1 in yeast cells as shown by activation of the nutritional reporter gene HIS3 (growth of yeast cells on selective medium plates deficient in tryptophan, leucine, and histidine) (Fig. 3A) and by activation of the lacZ reporter gene, which encodes β-galactosidase (Fig. 3B). To test whether hPIP1 can bind directly to PAK1 in vitro, PAK1 was expressed as GST fusion protein in bacteria and tested for interaction with in vitro transcribed and translated [35S]methionine-labeled hPIP1. GST alone failed to bind to hPIP1, whereas GST-PAK1 fusion protein bound labeled hPIP1 (Fig. 3C), suggesting that hPIP1 is able to interact with PAK1 in vitro. To further demonstrate that hPIP1 interacts with PAK1 in mammalian cells, we transiently coexpressed FLAG epitope-tagged hPIP1 and PAK1 in COS-7 cells and assayed for complex formation by using immunoprecipitation and Western blot analysis. As shown in Fig. 3 D and E, association of hPIP1 with PAK1 in COS-7 cells was demonstrated by coimmunoprecipitation using either anti-PAK1 antibody or anti-FLAG M2 monoclonal antibody, suggesting that hPIP1 interacts with PAK1 in mammalian cells.
Figure 3.
hPIP1 specifically binds to PAK1 in vitro and in vivo. (A) hPIP1 cotransformed with PAK1 or control plasmids, pVA3–1 and pTD1–1, respectively. pVA3–1 and pTD1–1 are positive control plasmids that encode the murine p53 protein and the simian virus 40 large T antigen. Only yeast cells cotransformed with hPIP1 and PAK1 grew on synthetic medium plates deficient in tryptophan, leucine, and histidine. (B) Activation of lacZ reporter gene in yeast cells cotransformed with hPIP1 and PAK1. p53 and large T antigen were used as a positive control in the assays. AU, arbitrary units. (C) Binding of GST-PAK1 with hPIP1 in vitro. (D and E) Coimmunoprecipitation of hPIP1 with PAK1 in mammalian cells. cDNAs encoding PAK1 and hPIP1 were cotransfected into COS-7 cells. The protein complex was immunoprecipitated (IP) by specific antibodies and blotted with anti-PAK1 antibody (D) or anti-FLAG M2 monoclonal antibody (E). Preimmune serum or unspecific immuneserum did not precipitate the two proteins in the assays.
Inhibition of PAK1 Activity by hPIP1.
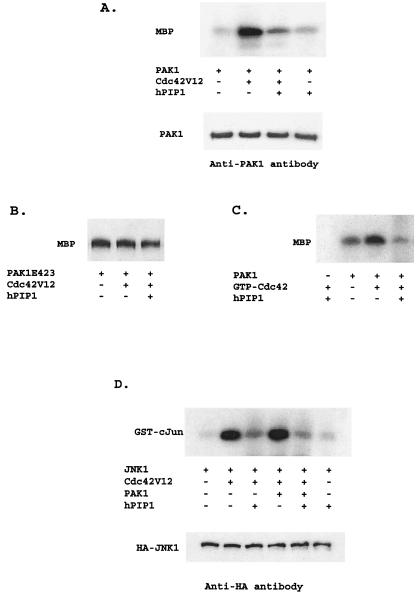
Because PAKs are directly activated by Cdc42/Rac, we performed a series of biochemical experiments to assess the effect of hPIP1 on Cdc42-mediated PAK activity (Fig. 4A). COS-7 cells were transfected with Myc-tagged PAK1 in the presence or absence of dominant active Cdc42 and hPIP1, respectively. Cells were lysed and assayed for PAK kinase activity by an immune complex kinase assay in which myelin basic protein was used as a substrate (24, 25). Cotransfection of PAK1 with Cdc42 dominant active mutant Cdc42V12 resulted in a 10-fold increase in PAK1 kinase activity (Fig. 4A). However, the presence of hPIP1 almost completely blocked the Cdc42-induced PAK1 activation (Fig. 4A). Titration of hPIP1 revealed progressive inhibition of the activity of PAK1 kinase (data not shown), whereas hPIP1 had little effect on a constitutively active PAK1 kinase mutant (PAK1T423E; Fig. 4B). These results suggest that hPIP1 inhibits the activation of PAK1 by activated Cdc42 protein in mammalian cells. To confirm the inhibitory effect of hPIP1 on PAK1 activation, we further tested whether PAK1 kinase activity could be inhibited in vitro by the hPIP1. Purified PAK1 was mixed with recombinant GST-Cdc42 and hPIP1. Fig. 4C demonstrates that the stimulation of PAK1 kinase by GTPγS-Cdc42 was reduced by the addition of hPIP1. Together, these data indicate that hPIP1 inhibits the specific activation of PAK1 by the Cdc42 GTPases.
Figure 4.
hPIP1 inhibits Cdc42-stimulated PAK1 activity. (A) Inhibition of Cdc42-stimulated PAK1 activity by hPIP1 in mammalian cells. Cell lysates from COS-7 cells expressing proteins as shown were immunoprecipitated with specific anti-PAK1 antibody to isolate PAK1, and were then subjected to protein kinase assays using myelin basic protein (MBP) as substrate. Bottom band shows that the amount of PAK1 used in the reaction was constant, as assayed by Western blot analysis. (B) hPIP1 had little effect on the kinase activity of PAK1 mutant (PAK1T423E). (C) hPIP1 inhibited GTPγS-Cdc42-mediated PAK1 activity in vitro. A fixed amount of PAK1 was activated by 2 μg of GTPγS-loaded Cdc42 in the presence or absence of hPIP1 (4 μg per reaction) with MBP as a substrate (4 μg per reaction). (D) Inhibition of Cdc42- and PAK1-mediated JNK activation by hPIP1. Cotransfection of hPIP1 greatly inhibited the activation of HA-JNK by Cdc42 (V12) or Cdc42 together with PAK1. JNK kinase was immunoprecipitated by anti-HA monoclonal antibody, and GST-cJun (2 μg, amino acids 1–79) was used as a phosphorylation substrate. Equal amounts of JNK were used in the assays, as shown in the bottom band by Western blot analysis.
hPIP1 Regulates PAK1-Induced JNK.
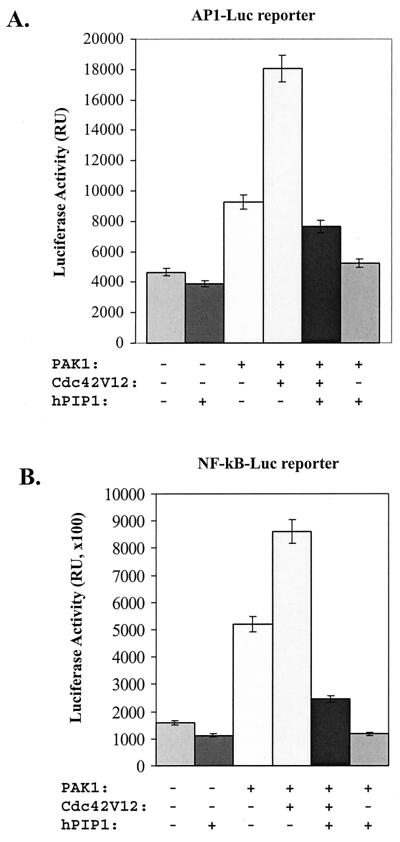
PAKs have been shown to activate JNK in the MAPK pathways, as well as the NF-κB signaling pathway. To define the role of hPIP1 on PAK-mediated JNK-signal transduction, we tested whether hPIP1 affects the activation of JNK by PAK. As shown in Fig. 4D, overexpression of activated Cdc42 and PAK1 significantly increased the kinase activity of JNK by ≈10-fold as assayed by the phosphorylation of GST-cJun. Activation of JNK by dominant activated Cdc42 mutant Cdc42V12 alone occurred presumably through the activation of endogenous PAK. However, coexpression of hPIP1 dramatically blocked the activation of JNK either by activated Cdc42 (Fig. 4D, lane 3) or by Cdc42 and PAK (Fig. 4D, lane 5), suggesting that hPIP1 inhibits both the endogenous and the transiently expressed Cdc42/PAK-induced JNK signaling. To further confirm the effect of hPIP1 on JNK signaling, we used a transcriptional reporter (pAP 1-Luc) to assay for activation of the JNK signaling pathway. Expression of PAK1 with the AP 1-Luc reporter increased the luciferase activity by approximately 2-fold, whereas cotransfection of PAK1 with a dominant active mutant of Cdc42 V12 into the cells enhanced the PAK-induced AP-1 activation by ≈4- to 5-fold (Fig. 5A). However, in the presence of hPIP1, the Cdc42-PAK-mediated AP-1 luciferase activity was completely inhibited (Fig. 5A), indicating that hPIP1 acts as a negative regulator of the PAK signaling pathway. A similar inhibitory effect on AP-1 activation by PAK was observed by using a dominant negative Rac mutant (RacN17), suggesting that activation of the AP-1 transcriptional reporter is PAK-specific in the assay. Together, these data demonstrate that hPIP1 can block Cdc42-mediated JNK activation by interacting with PAK1.
Figure 5.
Regulation of PAK1-induced AP-1 and NF-κB signaling pathways. (A) Overexpression of hPIP1 blocks Cdc42-PAK1-stimulated AP-1 transcriptional activity. COS-7 cells were cotransfected with either empty vector (lane 1) or cDNAs encoding the listed proteins, together with the reporter pAP1-Luc. RU, relative light units. (B) hPIP1 inhibited the activation of NF-κB transcriptional activity by PAK1 or PAK1 plus Cdc42 in the reporter assays. In the control experiment (lane 1), the cells were transfected with pCMV-lacZ plasmid. The results are expressed as the mean of values determined from at least three independent transfection assays.
hPIP1 Inhibited PAK-Induced NF-κB Activity.
PAKs are also known to be involved in the activation of NF-κB by interacting with the NF-κB-inducing kinase (NIK) (19, 20). Transient transfection of PAK1 activated the expression of an NF-κB-dependent reporter gene in COS-7 cells (Fig. 5B). Dominant activators of PAK1, Cdc42V12 or RacV12, further activated the NF-κB-dependent transcriptional activation (Fig. 5B). However, cotransfection of hPIP1 with PAK1 in the cell blocked the PAK1-induced NF-κB transcriptional activation as assayed by the luciferase activity (Fig. 5B), suggesting that hPIP1 inhibits the PAK1-coupled NF-κB signaling pathway.
hPIP1 Interacts with the N-Terminal Regulatory Domain of PAK1 to Modulate Its Function.
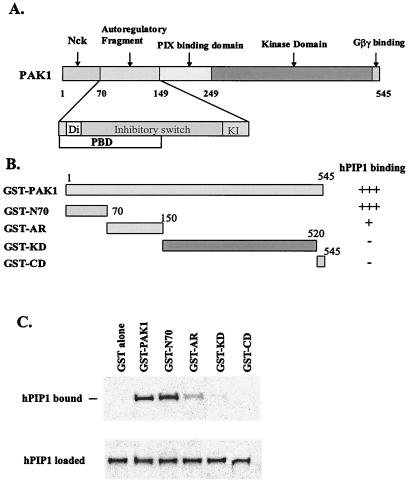
Recent studies demonstrate that PAK forms an asymmetric dimer in the autoinhibited conformation, held together by the interaction of the autoregulatory fragments (23). The domain structure and organization of PAK proteins suggest multiregulatory mechanisms for the activation of the kinases (Fig. 6A). Binding of Cdc42 or Rac to PAK disrupts the “inhibitory switch” (IS) domain, thereby relieving the kinase inhibition by breaking the dimer contact (23). A similar mechanism of activation by Cdc42/Rac was demonstrated for Wiskott–Aldrich syndrome protein (36). To identify which domain(s) of PAK1 interacts with hPIP1, we generated a number of GST fusion proteins corresponding to different domains of PAK1 (Fig. 6B). The purified GST-PAK fusion proteins were used in GST pull-down assays with in vitro transcribed and translated hPIP1. As shown in Fig. 6C, hPIP1 binds strongly with full-length PAK1 and the PAK1 N-terminal domain (residues 1–70), and to a lesser degree with the autoregulatory domain (residues 70–150). Little or no interaction was detected for the PIX-binding domain, the kinase domain, and the putative Gβγ-binding motif of PAK1, suggesting that hPIP1 interacts with PAK1 through the N-terminal regulatory region of the PAK1 protein. The N-terminal region of PAK1 (residues 1–149) contains the Nck binding motif, the dimerization domain (Di), the inhibitory switch (IS) domain, and the kinase inhibitor segment (KI). Intact PAK1 forms an inactive dimer held together by the autoregulatory fragments with the IS domain inhibiting the kinase with one surface and anchoring the dimer contact with another (23). Other studies have suggested a role for the N-terminal regions of PAKs in negatively regulating kinase activity (24). Therefore, interaction of hPIP1 with the N-terminal regulatory domain of PAK1 could stabilize the protein in the inactive dimeric conformation.
Figure 6.
Identification of PAK1 domain(s) responsible for the interaction with hPIP1. (A) Domain structure of the PAK1 polypeptide chain. The N terminus (residues 1–69) was identified as interacting with Nck. The autoregulatory regions (residues 70–149) contain the dimerization segment (Di), the p21 binding domain (PBD), the inhibitory switch domain (IS), and the kinase inhibitory segment (KI). The C terminus contains the kinase domain and the Gβγ-binding motif. (B) GST-PAK1 fusion proteins. Numerals indicate residue numbers at the boundaries of various subdivisions. (C) Interaction of different GST-PAK1 domains with in vitro translated hPIP1. HPIP1 interacts with the full length PAK1, the N-terminal domain (residues 1–70), and the autoregulatory domain (residues 70–149). The PIX-binding domain and the C-terminal region, including the kinase domain and the Gβγ-binding motif, are not involved in the hPIP1 association. Bottom band shows that the same amount of 35S-labeled hPIP1 was used in the binding assays.
Overexpression of hPIP1 Affects Cell Shape and Cytoskeletal Organization.
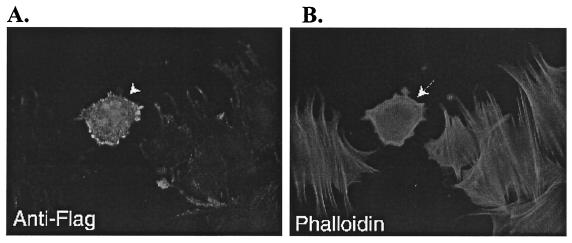
To study the potential role of hPIP1 in regulating cytoskeletal organization, we examined cellular morphology and cytoskeletal organization in mouse embryonic fibroblasts expressing FLAG-tagged hPIP1. As shown in Fig. 7, the cell expressing the FLAG-tagged hPIP1 has an abnormal morphology as compared with cells not transfected by hPIP1 (Fig. 7A). We further examined the organization of the actin filaments in the cells expressing FLAG-hPIP1 by using rhodamine-conjugated phalloidin (Fig. 7B). Cells expressing hPIP1 showed much more rounded cell shape and reorganized actin stress fibers, suggesting a potential role of this protein in actin cytoskeletal organization.
Figure 7.
Effects of hPIP1 overexpression in mouse embryo fibroblasts. (A) Cells transfected with FLAG-tagged hPIP1 and stained with anti-FLAG monoclonal antibody (M2, Sigma). Arrow indicates the cell overexpressing FLAG-hPIP1. (B) Immunofluorescence image of actin filaments with rhodamine-phalloidin staining. Arrow indicates the fibroblast cell overexpressing hPIP1 protein.
Discussion
Protein kinases are often subject to multiple levels of regulation. The involvement of PAKs in gene expression, cytoskeletal architecture, neuronal cell growth, and cellular apoptosis requires that their activities be tightly controlled. We have described the identification and characterization of a human PAK1-interacting protein, hPIP1, that contains five Gβ-like WD repeats and a C-terminal positively charged domain. The interaction of hPIP1 with PAK1 and the negative regulation of PAK1 activity have important implications for the understanding of PAK and PAK-mediated signaling pathways. Activation of PAK and PAK signaling pathways has been reported for a number of different proteins (Cdc42/Rac, adaptor protein Nck, exchanger factors PIX and Cool, and Gβγ in yeast cells; refs. 1, 27, and 31–33). However, relatively little is known regarding the negative regulation of PAK. Our data demonstrate that hPIP1 interacts with the N-terminal regulatory domain of PAK1 and negatively modulates PAK and PAK-mediated signaling pathways in mammalian cells. In fission yeast, a null mutation of the hPIP1 homolog, the Skb15 gene, is lethal and results in severe deregulation of both actin and microtubule cytoskeletons (37). hPIP1 was identified initially as interacting with PLC-β3 in the yeast two-hybrid assays. The exact role of this protein in PLC-Ca2+ signaling is not clear.
It has been demonstrated that PAK1 exists as a dimer in an autoinhibitory conformation. Mutation at a specific Thr site of PAK1 (T423E) could disrupt the dimeric structure and change the autoinhibitory conformation of the PAK protein, leading to the autophosphorylation of PAK1 at multiple sites, and consequently, the creation of a constitutively active protein kinase. If hPIP1 binds to the inactive conformation of PAK1 and is involved in the activation process by Cdc42/Rac, then we expect that hPIP1 has little effect on the constitutively active mutant of PAK1 (PAK1T423E), as shown in our experimental results. The ability of the hPIP1 protein to interact with PAK1 through the N-terminal regulatory domain provides a mechanism for the inhibition of the kinase at the molecular level. The fact that hPIP1 negatively modulates PAK-mediated JNK and NF-κB signaling pathways suggests a new mode of regulation for PAK-mediated signal transduction in mammalian cells. Furthermore, overexpression of hPIP1 into fibroblastic cells results in reorganization of actin cytoskeleton, suggesting that hPIP1 may be an important regulator for PAK-mediated cytoskeletal organization. Therefore, understanding of how cells regulate their cytoskeleton dynamics and morphogenesis through PAK and other signaling pathways is of great importance for providing basic molecular mechanisms of cellular functions.
Acknowledgments
We thank Dr. Jonathan Chernoff at the Fox Chase Cancer Center, Philadelphia, PA, for human PAK1 constructs. This work was supported in part by a Scientist Development grant from the American Heart Association National and by the Basil O'Connor Starter Scholar Research Award from the March of Dimes Foundation (to M.L.), an American Heart Association Southeastern Affiliate Grant-In-Aid (to W.C.X.), and National Institutes of Health Grant ROI GM53239 (to S. M.).
Abbreviations
- PAK
p21-activated kinase
- MAPK
mitogen-activated protein kinase
- JNK
c-Jun N-terminal kinase
- NF-κB
nuclear factor κ B
- WD
tryptophan–aspartic acid
- PIX
PAK-interacting exchange factor
- PLC
phospholipase C
- GST
glutathione S-transferase
- HA
hemagglutinin
- GTPγS
γ-thio-GTP
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF283303).
References
- 1.Manser E, Leung T, Salihuddin H, Zhao Z S, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 2.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 3.Daniels R H, Bokoch G M. Trends Biochem Sci. 1999;24:350–355. doi: 10.1016/s0968-0004(99)01442-5. [DOI] [PubMed] [Google Scholar]
- 4.Bagrodia S, Cerione R A. Trends Cell Biol. 1999;9:350–355. doi: 10.1016/s0962-8924(99)01618-9. [DOI] [PubMed] [Google Scholar]
- 5.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottilie S, Miller P J, Johnson D I, Creasy C L, Sells M A, Bagrodia S, Forsburg S L, Chernoff J. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders L C, Matsumura F, Bokoch G M, de Lanerolle P. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 8.Sells M A, Boyd J T, Chernoff J. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagrodia S, Derijard B, Davis R J, Cerione R A. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 11.Faure S, Vigneron S, Doree M, Morin N. EMBO J. 1997;16:5550–5561. doi: 10.1093/emboj/16.18.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manser E, Huang H Y, Loo T H, Chen X Q, Dong J M, Leung T, Lim L. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels R H, Hall P S, Bokoch G M. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudel T, Bokoch G M. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 15.Howe A K, Juliano R L. Nat Cell Biol. 2000;2:593–600. doi: 10.1038/35023536. [DOI] [PubMed] [Google Scholar]
- 16.Schurmann A, Mooney A F, Sanders L C, Sells M A, Wang H G, Reed J C, Bokoch G M. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minden A, Lin A, Claret F X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 18.Frost J A, Xu S, Hutchison M R, Marcus S, Cobb M H. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost J A, Swantek J L, Stippec S, Yin M J, Gaynor R, Cobb M H. J Biol Chem. 2000;275:19693–19699. doi: 10.1074/jbc.M909860199. [DOI] [PubMed] [Google Scholar]
- 20.Foryst-Ludwig A, Naumann M. J Biol Chem. 2000;275:39779–39785. doi: 10.1074/jbc.M007617200. [DOI] [PubMed] [Google Scholar]
- 21.Brown A, Wang X, Sawai E, Cheng-Mayer C. J Virol. 1999;73:9899–9907. doi: 10.1128/jvi.73.12.9899-9907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fackler O T, Lu X, Frost J A, Geyer M, Jiang B, Luo W, Abo A, Alberts A S, Peterlin B M. Mol Cell Biol. 2000;20:2619–2627. doi: 10.1128/mcb.20.7.2619-2627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei M, Lu W, Meng W, Parrini M C, Eck M J, Mayer B J, Harrison S C. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Z S, Manser E, Chen X Q, Chong C, Leung T, Lim L. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zenke F T, King C C, Bohl B P, Bokoch G M. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- 26.Tu H, Wigler M. Mol Cell Biol. 1999;19:602–611. doi: 10.1128/mcb.19.1.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 28.Galisteo M L, Chernoff J, Su Y C, Skolnik E Y, Schlessinger J. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 29.Lu W, Mayer B J. Oncogene. 1999;18:797–806. doi: 10.1038/sj.onc.1202361. [DOI] [PubMed] [Google Scholar]
- 30.Leberer E, Dignard D, Thomas D Y, Leeuw T. Biol Chem Hoppe-Seyler. 2000;381:427–431. doi: 10.1515/BC.2000.055. [DOI] [PubMed] [Google Scholar]
- 31.Leeuw T, Wu C, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Nature (London) 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 32.Manser E, Loo T H, Koh C G, Zhao Z S, Chen X Q, Tan L, Tan I, Leung T, Lim L. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 33.Bagrodia S, Taylor S J, Jordon K A, Van Aelst L, Cerione R A. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- 34.Daniels R H, Zenke F T, Bokoch G M. J Biol Chem. 1999;274:6047–6050. doi: 10.1074/jbc.274.10.6047. [DOI] [PubMed] [Google Scholar]
- 35.Icho T, Wickner R B. J Biol Chem. 1988;263:1467–1475. [PubMed] [Google Scholar]
- 36.Abdul-Manan N, Aghazadeh B, Liu G A, Majumdar A, Ouerfelli O, Siminovitch K A, Rosen M K. Nature (London) 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 37.Kim, H. W., Yang, P., Qyang, Y., Lai, H., Du, H., Henkel, J. S., Kumar, K., Bao, S., Liu, M. & Marcus, S. (2001) Mol. Cell, in press. [DOI] [PubMed]