Abstract
Streptomyces lincolnensis is a producer of lincomycin, which is a lincosamide antibiotic for the treatment of infective diseases caused by Gram-positive bacteria. S. lincolnensis is refractory to introducing plasmid DNA into cells because of resistance of foreign DNAs and poor sporulation. In this study, a simple and efficient method of transferring plasmids into S. lincolnensis through the intergeneric Escherichia coli-mycelia conjugation was established and optimized for the first time. The recipient mycelia of S. lincolnensis were prepared in liquid SM medium containing 10.3% sucrose for three days. The dispersed mycelia were conjugated with competent E. coli donor cells. The exconjugants were regenerated efficiently on solid mannitol soya flour (MS) medium containing 20 mM MgCl2. The average conjugation frequency was observed at 1.1 × 10−4 per input donor cell and validated functionally by transferring two types of vectors containing lincomycin resistance genes lmrA, lmrB and lmrC into S. lincolnensis mycelia. The data of fermentation in shaking flasks showed the lincomycin yield of the exconjugants increased by 52.9% for the multiple copy vector and 38.3% for the integrative one, compared with the parental strain. The efficient and convenient method of intergeneric E. coli-mycelia conjugation in this study provides a promising procedure to introduce plasmid DNA into other refractory streptomycetes.
Keywords: Streptomyces lincolnensis, lincomycin, intergeneric conjugation, E. coli-mycelia conjugation, lmrABC
1. Introduction
Lincomycin and its 7-chloro-7-deoxy derivative clindamycin are lincosamide antibiotics composed of propylproline and methylthiolincosamide [1]. Lincomycin, produced by Streptomyces lincolnensis, is used as a major antibiotic for the treatment of diseases caused by Gram-positive bacteria [1]. Clindamycin is usually used to treat infections caused by anaerobic bacteria and some protozoal diseases, such as malaria, infections of the respiratory tract, skin and soft tissue infections, and peritonitis [2]. It is a common topical treatment for acne [3] and can be useful against some methicillin-resistant Staphylococcus aureus infections [2]. Lincomycin biosynthetic gene cluster had been cloned and characterized [4,5]. The importance of fermentative production of lincosamide antibiotics [6] and the lack of efficient techniques to introduce plasmid DNA into S. lincolnensis, have encouraged the development of an efficient DNA manipulation technology to improve productivity and to analyze functionality of the secondary metabolite genes of interest in S. lincolnensis.
Two genetic transformation systems have been developed based on the model strain Streptomyces coelicolor A3(2) [7]. The protoplast transformation is widely used as the most common method for transferring foreign plasmid to streptomycetes. However, the efficiency of the protoplast transformation system varies significantly from one species to another, so it is necessary to improve the experimental procedures for a new species with a strong restriction to foreign DNAs. Few studies have reported methods of transferring DNA into S. lincolnensis mainly because of the resistance and modification of foreign DNAs [8]. The intergeneric conjugation between E. coli and streptomycetes was first reported by Mazodier [9] and spore conjugation methods have successfully been applied in various Streptomyces species [10–13]. For streptomycetes with poor sporulation, spore conjugation was not always successful or had extremely low conjugation frequency. Mycelia are the vegetative and propagation forms of actinobacteria, and have potency in the intergeneric conjugation. Some research results concerning plasmid transferring methods using mycelia have been reported for several Streptomyces [14–16], Streptosporangiaceae [13] and Saccharopolyspora spinosa [17] from E. coli. In our early studies, we tried to transfer foreign plasmid into S. lincolnensis through the two available methods. However, a few (usually less than five) transformants came up in each plate through the protoplast transformation, and no exconjugants were obtained when S. lincolnensis spores were used in conjugation with E. coli cells. In this study, by using the mycelia rather than the spores as recipients, with E. coli as the donor, we established and optimized an effective method of transferring DNA into S. lincolnensis through intergeneric conjugation for the first time.
2. Materials and Methods
2.1. Strains and Plasmids
E. coli DH5α was used as general host for cloning. The methylation-deficient E. coli ET12567 (dam-13::Tn9, dcm-6, hsdM, hsdS), containing pUZ8002 which is a RK2 derivative, was used as the donor in intergeneric conjugation [18]. Plasmid pIJ773 was used to obtain the apramycin resistance marker aac(3)IV and the oriT fragment [19]. Plasmid pUWL201 (provided by Professor Huizhan Zhang, East China University of Science and Technology, Shanghai, China), a high-copy-number Streptomyces plasmid, was used as the expression vector in streptomycetes [20]. Plasmid pIB139 (provided by Professor Hongyu Ou, Shanghai Jiaotong University, China), carriying ϕC31 int/att system functions, was used as the integrative expression vector in streptomycetes. S. lincolnensis ATCC25466 was reserved in our laboratory.
2.2. Media and Culture Conditions
E. coli stains were cultured in Luria Bertani (LB) medium [21] at 37 °C, with shaking at 220 rpm, supplemented with appropriate antibiotics as required. The spores of the S. lincolnensis, which grew at 30 °C on modified Gauze’s Medium No.1 (MGM, containing 2% soluble starch, 0.5% soybean, 0.1% KNO3, 0.05% NaCl, 0.05% MgSO4, 0.05% K2HPO4, 0.001% FeSO4, 2% agar, pH 7.0) for 7 days, were scraped and stored in 20% glycerol at −80 °C. Liquid SM medium (containing 1% glucose, 0.4% yeast extract, 0.4% peptone, 0.4% K2HPO4, 0.2% KH2PO4, 0.05% MgSO4, pH 7.0) [22] was used for S. lincolnensis culturing to collect mycelia. 2× YT medium [7] and mannitol soya flour (MS) medium [7] containing 20 mM of MgCl2 were used for the intergeneric conjugation in this study. For shake flask fermentation, we used 25 mL of seed medium (containing 2% soluble starch, 1% glucose, 1% soybean, 3% cream corn, 0.15% (NH4)2SO4, 0.4% CaCO3, with/without the apramycin) in 250 mL flask at 30 °C with shaking at 220 rpm for 2 days. And 2 mL of the seed cultures was transferred into 25 mL of fermentation medium (10% glucose, 2% soybean, 0.15% cream corn, 0.8% NaNO3, 0.5% NaCl, 0.6% (NH4)2SO4, 0.03% K2HPO4, 0.8% CaCO3, with/without the apramycin) and incubated under the same conditions of seed culture but for 7 days.
2.3. Conjugation Method
E. coli ET12567/pUZ8002/pUWL201apr (or other plasmid containing the oriT for conjugation) cells were prepared as previously described [7] with the minor modification. 50 μL of overnight culture was inoculated to 5 mL of LB (containing 25 mg/L kanamycin, 25 mg/L chloramphenicol, 50 mg/L apramycin, and 10 mM MgCl2), and incubated at 37 °C with shaking at 220 rpm for 3–4 h to OD600 0.4–0.6. To remove the antibiotics, the cells were collected at 4000 rpm and washed twice with an equal volume of LB (containing 10 mM MgCl2) without antibiotics. E. coli cells were counted by microscopy, resuspended in 0.5 mL of LB (containing 10 mM MgCl2), and used as the donor cells. S. lincolnensis mycelia were prepared from 5 mL on day 3 of liquid cultures. The mycelia were collected at 4000 rpm and washed with the equal volume of 10% of glycerol once and 2× YT twice (vortex strongly each time). Finally the mycelia were resuspended in 0.5 mL of 2× YT and used as the recipient.
The prepared E. coli cells were transferred into the resuspended mycelia to achieve homogeneity with vortex briefly. The mixture was collected by centrifugation at 4000 rpm, and 400–600 μL of supernatant was removed. The residue was spread on the MS plate (containing 0–50 mM MgCl2). The plate was dried in the hoodbench with sterile air, and then incubated overnight in 30 °C with the upside-down. One millilitre of mixed antibiotics (0.5 mg nalidixic acid and 1 mg apramycin) was evenly overlaid on the plate and the surface was dried. The plate was incubated in 30 °C until the exconjugants showed up. The single clones were spread on the new plates containing 25 mg/L of nalidixic acid and 50 mg/L of apramycin to confirm the exconjugants.
2.4. PCR Cloning of Lincomycin Resistance Genes and Construction of Over-Expression Vectors
The genomic DNA of S. lincolnensis was isolated by Kirby mix procedure [7]. E. coli plasmid preparation, restriction digestion, agarose gel electrophoresis and ligation reaction procedures were conducted according to the standard protocols [21]. Streptomycetes plasmids were isolated conducted according to the neutral lysis procedure [7]. The three lincomycin resistance genes were amplified from the genomic DNA by polymerase chain reaction (PCR). Six primers were designed, based on the sequence of S. lincolnensis ATCC25466 [5]: rA-F, 5′-GCCCAAGCTTGGAGTCATCTCTCCATGTCT-3′ (with a HindIII site, underlined) and rA-R, 5′-GCCGGAATTCCTGGAAGTTATCCGGGAGT-3′ (with a EcoRI site, underlined) for amplifying lmrA; rB-F, 5′-CACCGCTTGAATTCGTTCCATCTGGAGTG-3′ (with a EcoRI site, underlined) and rB-R, 5′-GGAAGATCTCCCGGATGTGCTTCTACGA-3′ (with a BglII site, underlined) for amplifying lmrB; rC-F, 5′-GCCGGAATTCCGGAAGATCTCCGACTCCTCCCTGGGAAT-3′ (with a EcoRI and a bglII site, underlined) and rC-R, 5′-GCTAGACTAGTCGGCCGATTAACACGCAAGACGCCCTCC-3′ (with a SpeI site, underlined) for amplifying lmrC. Sequence similarities in databases were searched with the Blast program through the website at EMBL. Clustal W was used for the multiple alignment of amino acid and DNA sequences.
The PCR reaction (50 μL) contained 2.5 U TransStart FastPfu DNA Polymerase (TRANSGEN), 2 mM of MgCl2, 10 μL of 5× TransStart FastPfu buffer, 0.5 mM of dNTPs, 100 pM of primer each, 200 ng of template DNA, and 5 μL of DMSO. The PCR onditions were as follows: pre-denaturation for 5 min at 97 °C; denaturation for 30 s at 95 °C, annealing for 30 s at the suitable temperature according to the primers and extension at 72 °C for suitable time according to the length of lmrA, lmrB, and lmrC, repeated for 30 cycles; followed by a final extension for 5 min at 72 °C. The PCR products were purified. DNA sequencing, using the dideoxynucleotide chain termination method with an automated ABI 377A sequencer (Applied Biosystems, USA), was carried out to confirm the correctness of cloned gene.
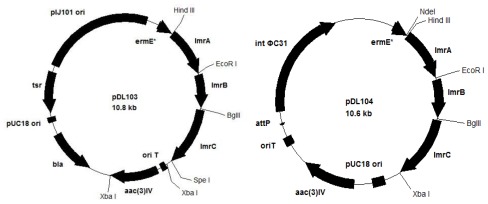
The aac(3)IV and oriT fragment was obtained from pIJ773 after digested with XbaI and ligated to pUWL201, yielding pUWL201apr. Genes of lmrA, lmrB, and lmrC, amplified from PCR, were ligated into pUWL201apr one after another by restriction enzyme digestion and ligation reaction procedure, yielding vector pDL103. Using the similar procedure, lmrA, lmrB, and lmrC were ligated into pIB139, resulting vector pDL104 (Figure 1).
Figure 1.
The map of expression vectors pDL103 and pDL104.
2.5. Lincomycin Extraction and HPLC Analysis
The fermentation broth was centrifuged at 8000 rpm for 5 min after cultivation, and the supernatant was extracted with 1.5 volumes of methanol. After centrifuged, the extract was freeze-dried for 24 h until no flowed liquid. And the residue was dissolved in methanol to be used.
The concentration of lincomycin was determined by the high pressure liquid chromatography (HPLC) with 18 alkylsilane bonded silica as the stationary phase. 0.05 mol/L borax solution (adjusted pH with 85% phosphoric acid to 6.0): methanol (with the ratio 4:6) was used as the mobile phase. The liquid chromatography was equipped with a 214 nm detector and maintained at a temperature of 30 °C. The flow rate was 0.8 mL per minute. The injection volume was 20 μL.
3. Results and Discussion
3.1. Optimization of Liquid Media for Recipient Mycelia Growth of S. lincolnensis
E. coli ET12567/pUZ8002 (containing pUWL201apr) was used as the donor cells in the following studies to determine the optimal condition for intergeneric conjugation.
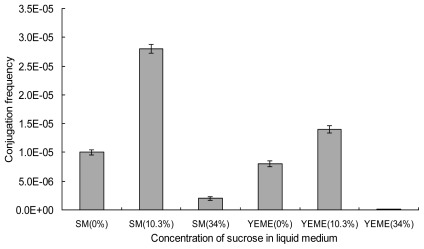
To select the liquid medium for S. lincolnensis mycelia growth, four representative media, SM, YEME, TSB and DNB [7] were tested for their effects on the conjugation efficiency. Exconjugants only appeared when mycelia were collected from liquid SM or YEME. As in the protoplast transformation in other Streptomyces [7,23,24], the concentration of sucrose added in the liquid medium was an effective factor for mycelia competence of S. lincolnensis. The liquid media added with 10.3% of sucrose led to a significant increase in conjugation frequency (Figure 2). 10.3% of sucrose was favorable for obtaining dispersed S. lincolnensis mycelia by loosening cell wall formation, which could have contact with donor cells and absorb plasmids adequately. But high concentration (34%) of sucrose added in both SM and YEME showed adverse results. Mycelia growth was heavily restrained by the high osmotic pressure caused by the high concentration of sucrose. SM medium with 10.3% of sucrose added was optimal for culturing recipient mycelia, compared with YEME, and the conjugation frequency reached 2.8 × 10−5, which is almost one fold higher.
Figure 2.
Effects of mycelia growth media on intergeneric conjugation. The competent mycelia were resuspended in 5 mL of 2× YT after being washed with an equal volume of 10% glycerol once and 2× YT twice. 0.5 mL of mycelia suspension and 0.5 mL of approximately 107 of the donor cells were thoroughly mixed and used for the intergeneric conjugation. The conjugation frequency was calculated as the number of exconjugants per input donor cell. Results were the average of three independent experiments. The error bars represent standard deviation.
3.2. Optimization of Recipient Mycelia Growth Conditions
Mycelia age played an important role in competence and transformation. Mycelia incubated for 34 h was suitable for the intergeneric conjugation in S. peucetius [16], whereas mycelia during the exponential phase of Nonomuraea were optimal [24]. S. lincolnensis mycelia of various growing times (2–6 days) were conjugated with E. coli cells. According to our study, mycelia of the earlier (1–2 days) or later (5–6 days) stages were not suitable for intergeneric conjugation, with a frequency rate lower than 1.0 × 10−6. S. lincolnensis mycelia grown for three days at the exponential stage were well dispersed in culture, and this competent stage was favorable for conjugation. The conjugation frequency (7.6 × 10−5) was one fold higher than the four-day-old mycelia.
3.3. Optimization of Solid Media for Exconjugants Regeneration
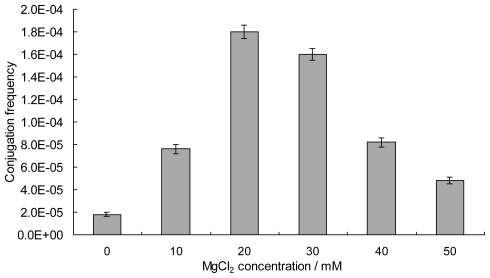
The conjugation medium has a significant influence on the conjugation frequency in streptomycetes [10,18,25–27]. Five applicable solid media, MGM, MS, R2YE [7], ISP Medium 2 (Difco), ISP Medium 4 (Difco), were tested to determine the appropriate medium. Exconjugants were obtained on MS and MGM media, while no exconjugants appeared when using R2YE, ISP2 or ISP4 as the conjugation medium. MS medium, on which the conjugation frequency (7.6 × 10−5) increased 3.2-fold compared to the MGM medium (1.8 × 10−5), proved to be the most appropriate medium for conjugate regeneration. The conjugation MS medium was further optimized with a supplement of 10, 20, 30, 40, 50 mM of MgCl2, respectively. When 20 mM of MgCl2 was added to the MS medium, the conjugation frequency of S. lincolnensis mycelia increased 9-fold compared without MgCl2 (Figure 3). However, the conjugation frequency decreased significantly as the concentration of MgCl2 increased, considering the spore formation and mycelia growth were severely inhibited at concentrations of 40 mM or higher for S. pristinaespiralis [28]. Thus, MS medium supplemented with 20 mM of MgCl2 proved to be optimal for the conjugation of E. coli-S. lincolnensis mycelia.
Figure 3.
Effects of MgCl2 concentration of MS on intergeneric conjugation. Exconjugants were counted after incubation on MS medium (containing 0–50 mM of MgCl2) at 30 °C for 7 days. The conjugation frequency was calculated as the number of exconjugants per input donor cell. Results were the average of three independent experiments. The error bars represent standard deviation.
3.4. Ratio of Donor Cell Numbers to Recipient Mycelia
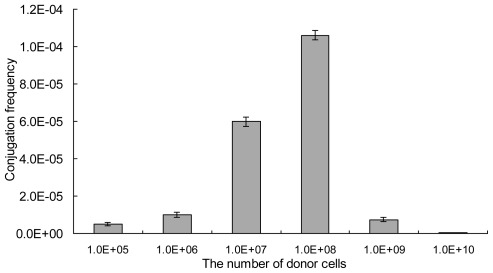
The ratio of donor cell numbers and recipient mycelia was a crucial parameter in the intergeneric conjugation of streptomycetes [29]. A stationary volume of 0.5 mL of mycelia suspension on third day and a range of E. coli donor cells (105–1010) were tested. As shown in Figure 4, few exconjugants were obtained when the number of donor cells was less than 106. The conjugation frequency significantly increased with increasing number of donor cells. When the number of the donor cells was up to 108, the conjugation frequency reached the highest frequency of 1.1 × 10−4. And this indicated that 108 of donor cells were optimal for the intergeneric conjugation from E. coli to S. lincolnensis mycelia. Similar results had been observed in the study of actinomycete Kitasatosporasetae, where a certain number of donor cells was required to achieve the highest frequency [25].
Figure 4.
Effects of the number of donor cells on intergeneric conjugation. 0.5 mL of mycelia suspension on the 3rd day and 105–1010 of E. coli donor cells were used. The conjugation frequency was calculated as the number of exconjugants per input donor cell. Results were the average of three independent experiments. The error bars represent standard deviation.
3.5. Over-Expression of lmrABC in S. lincolnensis
Further studies were carried out in order to practice the reliability of this intergeneric conjugation. The multiple copy vector pDL103 and the integrative vector pDL104 expressing lmrABC driven by the promoter ermE* were constructed. These two types of expression vectors were subsequently transformed into S. lincolnensis through the intergeneric conjugation as described above. In the three separate conjugation experiments, the average conjugation frequencies were 1.2 × 10−4 for pDL103 and 9.6 × 10−5 pDL104, respectively. The multiple copy and integrative vectors affected the intergeneric E. coli-mycelia conjugation least.
The resulting exconjugate strain, over-expression of lmrABC in S. lincolnensis, named D201 (containing pDL103) and D202 (integrated with pDL104), were easily screened and selected with apramycin and nalidixic acid after conjugation. For growth status, exconjugants of S. lincolnensis carrying pDL103 or pDL104 did not differ from the parent strain in the growth rate and colony size. The culture of the flask fermentation was extracted and the lincomycin titer was measured by HPLC. The production of lincomycin in fermentation broth of strains D201 and D202 were 94.8 ± 3.6 mg/L and 85.7 ± 3.4 mg/L, with a ratio of 52.9% and 38.3% higher than the parent strain (62.0 ± 2.9 mg/L), respectively.
4. Conclusions
In conclusion, a simple and efficient intergeneric conjugation method of transferring pasmid DNA from E. coli to S. lincolnensis using mycelia was developed and optimized for the first time. Recently, a few intergeneric conjugation methods using mycelia for plasmid transferring have been reported in poorly sporulating S. fradiae [14] and S. peucetius [16], and in reluctant PEG-mediated protoplast transformation of S. rimosus [15]. Without preparation of protoplasts or spores, the use of mycelia streamlines conjugation with Escherichia coli. The E. coli-mycelia conjugation method from our results combined with those reports [13–17], could be an attractive biotechnology for introducing foreign DNA to mycelia organisms, especially for refractory streptomycetes, on account of its timesaving, convenience, and high transformation frequency advantages.
Acknowledgment
This study was supported by the National Basic Research Program of China (2012CB721105, 2011CBA00802, 2007CB714301), and Research Found for the Doctoral Program of Higher Education (20090032110015).
References
- 1.Spížek J., Řezanka T. Lincomycin, clindamycin and their applications. Appl. Microbiol. Biotechnol. 2004;64:455–464. doi: 10.1007/s00253-003-1545-7. [DOI] [PubMed] [Google Scholar]
- 2.Daum R.S. Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N. Engl. J. Med. 2007;357:380–390. doi: 10.1056/NEJMcp070747. [DOI] [PubMed] [Google Scholar]
- 3.Feldman S., Careccia R.E., Barham K.L., Hancox J. Diagnosis and treatment of acne. Am. Fam. Physician. 2004;69:2123–2130. [PubMed] [Google Scholar]
- 4.Peschke U., Schmidt H., Zhang H.Z., Piepersberg W. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 1995;16:1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 5.Koberska M., Kopecky J., Olsovska J., Jelinkova M., Ulanova D., Man P., Flieger M., Janata J. Sequence analysis and heterologous expression of the lincomycin biosynthetic cluster of the type strain Streptomyces lincolnensis ATCC 25466. Folia Microbiol. 2008;53:395–401. doi: 10.1007/s12223-008-0060-8. [DOI] [PubMed] [Google Scholar]
- 6.Li X.B., Zhao G.R., Zheng H., Yuan Y.J. Improved industrial fermentation of lincomycin by phosphorus feeding. Process Biochem. 2007;42:662–668. [Google Scholar]
- 7.Kieser T., Bibb M.J., Buttner M.J., Chater K., Hopwood D.A. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 8.Chung S.T., Manis J.J., McWethy S.J., Patt T.E., Witz D.F., Wolf H.J., Wovcha M.G. Fermentation, Biosynthesis, and Molecular Genetics of Lincomycin. In: Strohl W.R., editor. Biotechnology of Industrial Antibiotics. 2nd ed. Dekker; New York, NY, USA: 1997. pp. 165–186. [Google Scholar]
- 9.Mazodier P., Petter R., Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flett F., Mersinias V., Smith C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 11.Luzhetskii A.N., Ostash B.E., Fedorenko V.A. Intergeneric conjugation Escherichia coli-Streptomyces globisporus 1912 using integrative plasmid pSET152 and its derivatives. Russ. J. Genet. 2001;37:1123–1129. [PubMed] [Google Scholar]
- 12.Enriquez L.L., Mendes M.V., Anton N., Tunca S., Guerra S.M., Martin J.F., Aparicio J.F. An efficient gene transfer system for the pimaricin producer Streptomyces natalensis. FEMS Microbiol. Lett. 2006;257:312–318. doi: 10.1111/j.1574-6968.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 13.Marcone G.L., Foulston L., Binda E., Marinelli F., Bibb M., Beltrametti F. Methods for the genetic manipulation of Nonomuraea sp ATCC 39727. J. Ind. Microbiol. Biot. 2010;37:1097–1103. doi: 10.1007/s10295-010-0807-5. [DOI] [PubMed] [Google Scholar]
- 14.Bierman M., Logan R., Obrien K., Seno E.T., Rao R.N., Schoner B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 15.Phornphisutthimas S., Sudtachat N., Bunyoo C., Chotewutmontri P., Panijpan B., Thamchaipenet A. Development of an intergeneric conjugal transfer system for rimocidin-producing Streptomyces rimosus. Lett. Appl. Microbiol. 2010;50:530–536. doi: 10.1111/j.1472-765X.2010.02835.x. [DOI] [PubMed] [Google Scholar]
- 16.Paranthaman S., Dharmalingam K. Intergeneric conjugation in Streptomyces peucetius and Streptomyces sp. strain C5: Chromosomal integration and expression of recombinant plasmids carrying the chiC gene. Appl. Environ. Microbiol. 2003;69:84–91. doi: 10.1128/AEM.69.1.84-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushima P., Broughton M.C., Turner J.R., Baltz R.H. Conjugal transfer of cosmid DNA from Escherichia coli to Saccharopolyspora spinosa: Effects of chromosomal insertions on macrolide A83543 production. Gene. 1994;146:39–45. doi: 10.1016/0378-1119(94)90831-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim M.K., Ha H.S., Choi S.U. Conjugal transfer using the bacteriophage phi C31 att/int system and properties of the attB site in Streptomyces ambofaciens. Biotechnol. Lett. 2008;30:695–699. doi: 10.1007/s10529-007-9586-0. [DOI] [PubMed] [Google Scholar]
- 19.Gust B., Kieser T., Chater K. PCR Targeting System in Streptomyces coelicolor. 2. A3. Norwich Research Park; Norwich, UK: 2002. [Google Scholar]
- 20.Doumith M., Weingarten P., Wehmeier U.F., Salah-Bey K., Benhamou B., Capdevila C., Michel J.M., Piepersberg W., Raynal M.C. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Mol. Genet. Genomics. 2000;264:477–485. doi: 10.1007/s004380000329. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Russell D.W., Sambrook J. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 22.Zhang H.Z., Schmidt H., Piepersberg W. Molecular cloning and characterization of two lincomycin-resistance genes, lmrA and lmrB, from Streptomyces lincolnensls 78-11. Mol. Microbiol. 1992;6:2147–2157. doi: 10.1111/j.1365-2958.1992.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 23.Marcone G.L., Carrano L., Marinelli F., Beltrametti F. Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. 2010;63:83–88. doi: 10.1038/ja.2009.127. [DOI] [PubMed] [Google Scholar]
- 24.Luo J.M., Li J.S., Wang Y.T., Luo S.H., Wang M. Protoplast Formation and Regeneration Conditions of Streptomyces gilvosporeus. Proceedings of 2009 3rd International Conference on Bioinformatics and Biomedical Engineering; Beijing, China. 11–13 June 2009; Piscataway, NJ, USA: IEEE; 2009. pp. 1647–1650. [Google Scholar]
- 25.Choi S.U., Lee C.K., Hwang Y.I., Kinoshita H., Nihira T. Intergeneric conjugal transfer of plasmid DNA from Escherichia coli to Kitasatospora setae, a bafilomycin B1 producer. Arch. Microbiol. 2004;181:294–298. doi: 10.1007/s00203-004-0654-8. [DOI] [PubMed] [Google Scholar]
- 26.Kitani S., Bibb M.J., Nihira T. Conjugal transfer of plasmid DNA from Escherichia coli to Streptomyces lavendulae FRI-5. J. Microbiol. Biotechnol. 2000;10:535–538. [Google Scholar]
- 27.Guan D., Pettis G.S. Intergeneric conjugal gene transfer from Escherichia coli to the sweet potato pathogen Streptomyces ipomoeae. Lett. Appl. Microbiol. 2009;49:67–72. doi: 10.1111/j.1472-765X.2009.02619.x. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z.H., Jin X., Jin Q.C. Conjugal transferring of resistance gene ptr for improvement of pristinamycin-producing Streptomyces pristinaespiralis. Appl. Biochem. Biotechnol. 2010;160:1853–1864. doi: 10.1007/s12010-009-8691-z. [DOI] [PubMed] [Google Scholar]
- 29.Nikodinovic J., Barrow K.D., Chuck J.A. High frequency transformation of the amphotericin-producing bacterium Streptomyces nodosus. J. Microbiol. Methods. 2003;55:273–277. doi: 10.1016/s0167-7012(03)00160-x. [DOI] [PubMed] [Google Scholar]