Abstract
In the title compound, [NdCl3(C12H10N2O)3], the central NdIII ion is nine-coordinated by six N atoms from three bidentate chelate N-[phenyl(pyridin-2-yl)methylidene]hydroxylamine ligands and three Cl− ions, and displays a distorted tricapped trigonal prismatic geometry. The complex molecules are stabilized by intramolecular O—H⋯Cl hydrogen bonds.
Related literature
For complexes of oximes, see: Kukushkin & Pombeiro (1999 ▶); Milios et al. (2007 ▶); Fritsky et al. (2004 ▶); Xu et al. (2007 ▶); Papatriantafyllopoulou et al. (2009 ▶). For 3d-metal complexes of N-[phenyl(pyridine-2-yl)methylidene]hydroxylamine, see: Milios et al. (2003 ▶); Milios et al. (2004 ▶). For an Sm complex with this ligand, see: Lei et al. (2012 ▶).
Experimental
Crystal data
[NdCl3(C12H10N2O)3]
M r = 845.25
Triclinic,
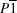
a = 8.6367 (17) Å
b = 10.460 (2) Å
c = 19.847 (4) Å
α = 91.87 (3)°
β = 94.38 (3)°
γ = 92.80 (3)°
V = 1784.4 (6) Å3
Z = 2
Mo Kα radiation
μ = 1.72 mm−1
T = 293 K
0.31 × 0.18 × 0.13 mm
Data collection
Bruker SMART CCD-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.617, T max = 0.807
30524 measured reflections
7775 independent reflections
7155 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.022
wR(F 2) = 0.059
S = 1.04
7775 reflections
445 parameters
H-atom parameters constrained
Δρmax = 0.50 e Å−3
Δρmin = −0.44 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812014055/zs2190sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812014055/zs2190Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Nd1—N2 | 2.604 (2) |
| Nd1—N1 | 2.661 (2) |
| Nd1—N5 | 2.680 (2) |
| Nd1—N4 | 2.6953 (19) |
| Nd1—N6 | 2.7018 (19) |
| Nd1—N3 | 2.742 (2) |
| Nd1—Cl3 | 2.7686 (8) |
| Nd1—Cl2 | 2.7903 (9) |
| Nd1—Cl1 | 2.8296 (10) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯Cl3 | 0.82 | 2.22 | 2.966 (2) | 152 |
| O2—H2A⋯Cl1 | 0.82 | 2.19 | 2.9290 (19) | 151 |
| O3—H3A⋯Cl2 | 0.82 | 2.19 | 2.930 (2) | 150 |
Acknowledgments
The author appreciates financial support from Yanan University (grant No. YD2011–20) and the Science and Technology Bureau of Yanan City (grant No. kn2009–16).
supplementary crystallographic information
Comment
The coordination chemistry of oximes (Kukushkin & Pombeiro, 1999; Milios et al., 2007) continues to attract considerable attention, with the efforts of several research groups driven by a number of considerations. These include the use of metal oxime complexes in supramolecular chemistry (Fritsky et al., 2004) and the employment of oximate ligands in the synthesis of complexes with interesting magnetic properties (Xu et al., 2007; Papatriantafyllopoulou et al., 2009; Milios et al., 2007). N-[phenyl(pyridine-2-yl)methylidene]hydroxylamine [(py)C(ph)NOH], is one of the oximes that is currently a popular ligand for synthesis of the 3d-metal complexes (Milios et al., 2003; Milios et al., 2004). However, the structures of rare earth metal complexes with this ligand are uncommon in the crystallographic literature. Here we report the structure of the neodymium complex with [(py)C(ph)NOH], the title compound [NdCl3(C12H10N2O)3], which was synthesized by the reaction of NdCl3 . 6H2O with the ligand under autogenous pressure. The title compound is isomorphous with the SmIII analogue (Lei et al., 2012).
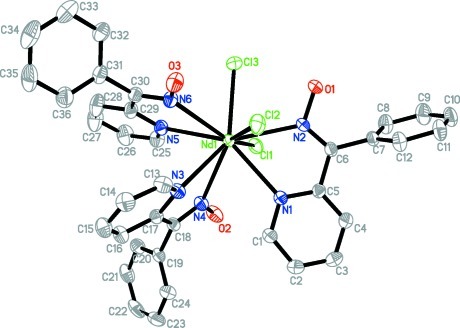
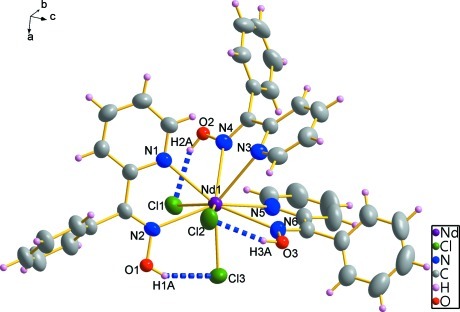
In the title complex, the central NdIII ion is nine-coordinated by six nitrogen atoms from three bidentate chelate ligands and three Cl- ions [Nd—N range, 2.604 (2)–2.742 (2) Å; Nd—Cl, 2.7686 (8)–2.8296 (10) Å (Table 1)] and displays a distorted tricapped trigonal prismatic geometry (Fig. 1). The discrete complex molecules are stabilized by intramolecular O—H···Cl hydrogen bonds (Table 2, Fig. 2).
Experimental
A mixture of phenyl-2-pyridyl ketone oxime (0.0198 g, 0.10 mmol), NdCl3 . 6H2O (0.0179 g, 0.05 mmol), and ethanol (2 mL) was sealed in a 6 mL Pyrex tube. The tube was heated at 80 °C for 4 days under autogenous pressure. Cooling of the resultant solution to room temperature gave colourless crystals of the product. The crystals were collected by filtration, washed with ethanol (2 mL) and dried in air. Yield: 54%. Anal. Calcd. for C36H30Cl3N6NdO3: C, 51.15; H, 3.58; N, 9.94%. Found: C, 50.93; H, 3.43; N, 9.76%.
Refinement
H atoms were placed in calculated positions and included in the refinement using a riding-model approximation, with C—H = 0.93 Å and O—H = 0.82 Å, and with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(O).
Figures
Fig. 1.
The molecular structure of the title complex, showing atom labels and 30% probability displacement ellipsoids.
Fig. 2.
Intramolecular hydrogen-bonding interactions in the title complex, with hydrogen bonds shown as dashed lines.
Crystal data
| [NdCl3(C12H10N2O)3] | Z = 2 |
| Mr = 845.25 | F(000) = 846 |
| Triclinic, P1 | char |
| Hall symbol: -P 1 | Dx = 1.573 Mg m−3 |
| a = 8.6367 (17) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.460 (2) Å | Cell parameters from 7737 reflections |
| c = 19.847 (4) Å | θ = 2.2–27.0° |
| α = 91.87 (3)° | µ = 1.72 mm−1 |
| β = 94.38 (3)° | T = 293 K |
| γ = 92.80 (3)° | Block, colourless |
| V = 1784.4 (6) Å3 | 0.31 × 0.18 × 0.13 mm |
Data collection
| Bruker SMART CCD-detector diffractometer | 7775 independent reflections |
| Radiation source: fine-focus sealed tube | 7155 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| φ and ω scans | θmax = 27.0°, θmin = 1.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | h = −11→11 |
| Tmin = 0.617, Tmax = 0.807 | k = −13→13 |
| 30524 measured reflections | l = −25→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.022 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.059 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0316P)2 + 0.3895P] where P = (Fo2 + 2Fc2)/3 |
| 7775 reflections | (Δ/σ)max = 0.002 |
| 445 parameters | Δρmax = 0.50 e Å−3 |
| 0 restraints | Δρmin = −0.44 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Nd1 | 0.170030 (12) | 0.594157 (10) | 0.248609 (5) | 0.03370 (4) | |
| Cl2 | 0.10720 (8) | 0.35456 (5) | 0.30113 (3) | 0.05379 (15) | |
| Cl1 | 0.27975 (7) | 0.70288 (6) | 0.13140 (3) | 0.05048 (14) | |
| Cl3 | 0.48680 (7) | 0.57109 (6) | 0.27455 (3) | 0.05349 (15) | |
| N2 | 0.2436 (2) | 0.41331 (18) | 0.16542 (10) | 0.0429 (4) | |
| N6 | 0.2375 (2) | 0.61037 (18) | 0.38390 (9) | 0.0437 (4) | |
| N1 | −0.0455 (2) | 0.48814 (17) | 0.15808 (9) | 0.0397 (4) | |
| N3 | −0.0819 (2) | 0.65372 (18) | 0.31740 (9) | 0.0415 (4) | |
| N5 | 0.2564 (2) | 0.81945 (17) | 0.31196 (10) | 0.0434 (4) | |
| C27 | 0.3334 (5) | 1.0579 (3) | 0.37346 (19) | 0.0884 (11) | |
| H27 | 0.3572 | 1.1381 | 0.3943 | 0.106* | |
| N4 | −0.0185 (2) | 0.77901 (18) | 0.20749 (9) | 0.0418 (4) | |
| C7 | 0.2007 (3) | 0.2515 (2) | 0.07372 (11) | 0.0416 (5) | |
| C18 | −0.1209 (2) | 0.8276 (2) | 0.24228 (11) | 0.0378 (4) | |
| O1 | 0.3882 (2) | 0.36271 (19) | 0.17316 (10) | 0.0627 (5) | |
| H1A | 0.4419 | 0.4034 | 0.2031 | 0.094* | |
| O2 | −0.0065 (2) | 0.83670 (19) | 0.14624 (9) | 0.0592 (5) | |
| H2A | 0.0687 | 0.8104 | 0.1282 | 0.089* | |
| C19 | −0.2129 (3) | 0.9365 (2) | 0.21929 (11) | 0.0397 (5) | |
| C5 | −0.0047 (3) | 0.4056 (2) | 0.10935 (11) | 0.0394 (5) | |
| O3 | 0.2290 (2) | 0.50316 (16) | 0.42307 (8) | 0.0576 (4) | |
| H3A | 0.2028 | 0.4398 | 0.3987 | 0.086* | |
| C17 | −0.1480 (2) | 0.7670 (2) | 0.30694 (11) | 0.0384 (5) | |
| C13 | −0.1152 (3) | 0.5941 (2) | 0.37345 (13) | 0.0502 (6) | |
| H13 | −0.0759 | 0.5139 | 0.3803 | 0.060* | |
| C31 | 0.2881 (3) | 0.7204 (3) | 0.49380 (12) | 0.0508 (6) | |
| C6 | 0.1547 (3) | 0.3581 (2) | 0.11766 (11) | 0.0399 (5) | |
| C21 | −0.2296 (4) | 1.1591 (3) | 0.19732 (14) | 0.0624 (7) | |
| H21 | −0.1853 | 1.2421 | 0.1996 | 0.075* | |
| C1 | −0.1928 (3) | 0.5235 (2) | 0.15376 (12) | 0.0459 (5) | |
| H1 | −0.2241 | 0.5772 | 0.1879 | 0.055* | |
| C4 | −0.1058 (3) | 0.3652 (3) | 0.05469 (12) | 0.0521 (6) | |
| H4 | −0.0734 | 0.3112 | 0.0210 | 0.063* | |
| C30 | 0.2678 (3) | 0.7144 (2) | 0.41872 (11) | 0.0440 (5) | |
| C24 | −0.3642 (3) | 0.9139 (3) | 0.19283 (13) | 0.0512 (6) | |
| H24 | −0.4110 | 0.8319 | 0.1923 | 0.061* | |
| C14 | −0.2040 (3) | 0.6440 (3) | 0.42155 (13) | 0.0569 (6) | |
| H14 | −0.2222 | 0.5994 | 0.4602 | 0.068* | |
| C25 | 0.2728 (3) | 0.9250 (2) | 0.27624 (14) | 0.0537 (6) | |
| H25 | 0.2582 | 0.9167 | 0.2294 | 0.064* | |
| C20 | −0.1452 (3) | 1.0601 (2) | 0.22234 (13) | 0.0516 (6) | |
| H20 | −0.0437 | 1.0759 | 0.2411 | 0.062* | |
| C29 | 0.2820 (3) | 0.8323 (2) | 0.37946 (12) | 0.0475 (5) | |
| C23 | −0.4461 (3) | 1.0143 (3) | 0.16705 (14) | 0.0630 (7) | |
| H23 | −0.5476 | 0.9994 | 0.1482 | 0.076* | |
| C32 | 0.4105 (3) | 0.6620 (3) | 0.52790 (13) | 0.0559 (6) | |
| H32 | 0.4837 | 0.6224 | 0.5036 | 0.067* | |
| C16 | −0.2384 (3) | 0.8229 (2) | 0.35265 (13) | 0.0516 (6) | |
| H16 | −0.2810 | 0.9013 | 0.3441 | 0.062* | |
| C33 | 0.4240 (4) | 0.6624 (3) | 0.59736 (15) | 0.0744 (9) | |
| H33 | 0.5055 | 0.6222 | 0.6199 | 0.089* | |
| C11 | 0.1861 (4) | 0.0299 (3) | 0.03858 (14) | 0.0635 (7) | |
| H11 | 0.1448 | −0.0530 | 0.0427 | 0.076* | |
| C12 | 0.1387 (3) | 0.1293 (2) | 0.07905 (13) | 0.0549 (6) | |
| H12 | 0.0648 | 0.1129 | 0.1098 | 0.066* | |
| C2 | −0.3004 (3) | 0.4845 (3) | 0.10144 (14) | 0.0554 (6) | |
| H2 | −0.4018 | 0.5110 | 0.1006 | 0.067* | |
| C22 | −0.3780 (4) | 1.1357 (3) | 0.16916 (14) | 0.0676 (8) | |
| H22 | −0.4332 | 1.2026 | 0.1513 | 0.081* | |
| C8 | 0.3121 (3) | 0.2733 (3) | 0.02820 (14) | 0.0605 (7) | |
| H8 | 0.3573 | 0.3552 | 0.0250 | 0.073* | |
| C9 | 0.3564 (4) | 0.1743 (3) | −0.01235 (15) | 0.0738 (9) | |
| H9 | 0.4298 | 0.1897 | −0.0434 | 0.089* | |
| C3 | −0.2561 (3) | 0.4062 (3) | 0.05076 (14) | 0.0612 (7) | |
| H3 | −0.3258 | 0.3808 | 0.0142 | 0.073* | |
| C15 | −0.2652 (3) | 0.7611 (3) | 0.41137 (14) | 0.0597 (7) | |
| H15 | −0.3238 | 0.7982 | 0.4435 | 0.072* | |
| C35 | 0.1970 (5) | 0.7814 (4) | 0.60097 (17) | 0.0867 (11) | |
| H35 | 0.1257 | 0.8220 | 0.6260 | 0.104* | |
| C26 | 0.3100 (4) | 1.0446 (3) | 0.30484 (18) | 0.0729 (8) | |
| H26 | 0.3191 | 1.1153 | 0.2780 | 0.087* | |
| C10 | 0.2927 (4) | 0.0537 (3) | −0.00695 (14) | 0.0663 (8) | |
| H10 | 0.3223 | −0.0128 | −0.0346 | 0.080* | |
| C36 | 0.1811 (4) | 0.7812 (3) | 0.53085 (15) | 0.0708 (8) | |
| H36 | 0.0992 | 0.8216 | 0.5088 | 0.085* | |
| C34 | 0.3174 (5) | 0.7219 (4) | 0.63336 (16) | 0.0888 (12) | |
| H34 | 0.3270 | 0.7220 | 0.6803 | 0.107* | |
| C28 | 0.3211 (4) | 0.9497 (3) | 0.41152 (16) | 0.0735 (9) | |
| H28 | 0.3391 | 0.9562 | 0.4583 | 0.088* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Nd1 | 0.03692 (7) | 0.02906 (7) | 0.03506 (7) | 0.00424 (4) | 0.00134 (4) | −0.00019 (4) |
| Cl2 | 0.0757 (4) | 0.0322 (3) | 0.0531 (3) | −0.0011 (3) | 0.0043 (3) | 0.0023 (2) |
| Cl1 | 0.0546 (3) | 0.0522 (3) | 0.0471 (3) | 0.0083 (3) | 0.0152 (3) | 0.0057 (3) |
| Cl3 | 0.0391 (3) | 0.0638 (4) | 0.0559 (3) | 0.0057 (3) | −0.0057 (2) | −0.0067 (3) |
| N2 | 0.0381 (9) | 0.0402 (10) | 0.0505 (11) | 0.0122 (8) | 0.0013 (8) | −0.0055 (8) |
| N6 | 0.0537 (11) | 0.0375 (10) | 0.0399 (10) | 0.0020 (8) | 0.0022 (8) | 0.0021 (8) |
| N1 | 0.0399 (9) | 0.0357 (10) | 0.0434 (10) | 0.0052 (8) | 0.0020 (8) | −0.0007 (8) |
| N3 | 0.0434 (10) | 0.0372 (10) | 0.0449 (10) | 0.0043 (8) | 0.0065 (8) | 0.0070 (8) |
| N5 | 0.0473 (10) | 0.0341 (10) | 0.0486 (11) | −0.0012 (8) | 0.0056 (8) | −0.0010 (8) |
| C27 | 0.132 (3) | 0.0428 (16) | 0.087 (2) | −0.0277 (18) | 0.021 (2) | −0.0182 (16) |
| N4 | 0.0471 (10) | 0.0395 (10) | 0.0406 (10) | 0.0087 (8) | 0.0080 (8) | 0.0085 (8) |
| C7 | 0.0463 (12) | 0.0391 (12) | 0.0395 (11) | 0.0083 (10) | 0.0026 (9) | −0.0033 (9) |
| C18 | 0.0390 (11) | 0.0327 (11) | 0.0421 (11) | 0.0045 (9) | 0.0043 (9) | 0.0014 (9) |
| O1 | 0.0439 (9) | 0.0694 (12) | 0.0732 (13) | 0.0254 (9) | −0.0082 (8) | −0.0251 (10) |
| O2 | 0.0709 (12) | 0.0669 (12) | 0.0464 (9) | 0.0307 (10) | 0.0216 (8) | 0.0219 (9) |
| C19 | 0.0465 (12) | 0.0367 (11) | 0.0378 (11) | 0.0112 (9) | 0.0092 (9) | 0.0038 (9) |
| C5 | 0.0432 (11) | 0.0347 (11) | 0.0400 (11) | 0.0033 (9) | 0.0012 (9) | 0.0004 (9) |
| O3 | 0.0852 (13) | 0.0419 (9) | 0.0445 (9) | 0.0005 (9) | −0.0032 (9) | 0.0069 (7) |
| C17 | 0.0398 (11) | 0.0352 (11) | 0.0402 (11) | 0.0009 (9) | 0.0047 (9) | 0.0012 (9) |
| C13 | 0.0487 (13) | 0.0478 (14) | 0.0557 (14) | 0.0033 (11) | 0.0073 (11) | 0.0153 (11) |
| C31 | 0.0542 (14) | 0.0535 (15) | 0.0435 (13) | −0.0110 (12) | 0.0077 (11) | −0.0074 (11) |
| C6 | 0.0438 (12) | 0.0349 (11) | 0.0412 (11) | 0.0060 (9) | 0.0044 (9) | −0.0029 (9) |
| C21 | 0.087 (2) | 0.0373 (13) | 0.0674 (17) | 0.0171 (13) | 0.0259 (16) | 0.0089 (12) |
| C1 | 0.0426 (12) | 0.0432 (13) | 0.0523 (13) | 0.0081 (10) | 0.0033 (10) | −0.0012 (10) |
| C4 | 0.0551 (14) | 0.0561 (15) | 0.0441 (13) | 0.0093 (12) | −0.0024 (11) | −0.0088 (11) |
| C30 | 0.0434 (12) | 0.0454 (13) | 0.0424 (12) | −0.0003 (10) | 0.0028 (9) | −0.0039 (10) |
| C24 | 0.0435 (13) | 0.0515 (14) | 0.0597 (15) | 0.0071 (11) | 0.0062 (11) | 0.0058 (12) |
| C14 | 0.0586 (15) | 0.0644 (17) | 0.0491 (14) | −0.0020 (13) | 0.0124 (12) | 0.0132 (12) |
| C25 | 0.0626 (15) | 0.0399 (13) | 0.0585 (15) | −0.0035 (11) | 0.0066 (12) | 0.0045 (11) |
| C20 | 0.0555 (14) | 0.0422 (13) | 0.0585 (15) | 0.0070 (11) | 0.0094 (12) | 0.0036 (11) |
| C29 | 0.0498 (13) | 0.0422 (13) | 0.0497 (13) | −0.0042 (10) | 0.0073 (11) | −0.0068 (10) |
| C23 | 0.0500 (14) | 0.084 (2) | 0.0590 (16) | 0.0265 (14) | 0.0082 (12) | 0.0115 (15) |
| C32 | 0.0616 (16) | 0.0540 (15) | 0.0503 (14) | −0.0100 (12) | 0.0015 (12) | −0.0005 (12) |
| C16 | 0.0565 (14) | 0.0481 (14) | 0.0521 (14) | 0.0096 (11) | 0.0131 (11) | 0.0009 (11) |
| C33 | 0.086 (2) | 0.077 (2) | 0.0556 (17) | −0.0239 (18) | −0.0106 (16) | 0.0101 (15) |
| C11 | 0.089 (2) | 0.0368 (13) | 0.0653 (17) | 0.0058 (13) | 0.0113 (15) | −0.0059 (12) |
| C12 | 0.0658 (16) | 0.0435 (14) | 0.0570 (15) | 0.0007 (12) | 0.0186 (12) | −0.0038 (11) |
| C2 | 0.0427 (13) | 0.0603 (16) | 0.0625 (16) | 0.0095 (12) | −0.0041 (11) | 0.0003 (13) |
| C22 | 0.085 (2) | 0.0652 (19) | 0.0611 (17) | 0.0458 (17) | 0.0277 (15) | 0.0205 (14) |
| C8 | 0.0739 (18) | 0.0513 (15) | 0.0576 (16) | −0.0027 (13) | 0.0204 (14) | −0.0051 (12) |
| C9 | 0.084 (2) | 0.078 (2) | 0.0618 (17) | 0.0028 (17) | 0.0327 (16) | −0.0137 (15) |
| C3 | 0.0541 (15) | 0.0714 (18) | 0.0550 (15) | 0.0049 (13) | −0.0141 (12) | −0.0037 (13) |
| C15 | 0.0648 (16) | 0.0647 (17) | 0.0521 (15) | 0.0055 (14) | 0.0203 (13) | 0.0008 (13) |
| C35 | 0.100 (3) | 0.098 (3) | 0.063 (2) | −0.018 (2) | 0.0377 (19) | −0.0240 (18) |
| C26 | 0.094 (2) | 0.0370 (14) | 0.088 (2) | −0.0098 (14) | 0.0204 (18) | 0.0034 (14) |
| C10 | 0.080 (2) | 0.0618 (18) | 0.0586 (16) | 0.0186 (15) | 0.0136 (15) | −0.0186 (14) |
| C36 | 0.0677 (18) | 0.082 (2) | 0.0618 (17) | −0.0014 (16) | 0.0121 (14) | −0.0123 (15) |
| C34 | 0.115 (3) | 0.103 (3) | 0.0442 (16) | −0.040 (2) | 0.0093 (19) | −0.0032 (17) |
| C28 | 0.102 (2) | 0.0538 (17) | 0.0621 (17) | −0.0224 (16) | 0.0136 (16) | −0.0154 (14) |
Geometric parameters (Å, º)
| Nd1—N2 | 2.604 (2) | C21—H21 | 0.9300 |
| Nd1—N1 | 2.661 (2) | C1—C2 | 1.377 (3) |
| Nd1—N5 | 2.680 (2) | C1—H1 | 0.9300 |
| Nd1—N4 | 2.6953 (19) | C4—C3 | 1.384 (4) |
| Nd1—N6 | 2.7018 (19) | C4—H4 | 0.9300 |
| Nd1—N3 | 2.742 (2) | C30—C29 | 1.485 (3) |
| Nd1—Cl3 | 2.7686 (8) | C24—C23 | 1.384 (4) |
| Nd1—Cl2 | 2.7903 (9) | C24—H24 | 0.9300 |
| Nd1—Cl1 | 2.8296 (10) | C14—C15 | 1.372 (4) |
| N2—C6 | 1.277 (3) | C14—H14 | 0.9300 |
| N2—O1 | 1.380 (2) | C25—C26 | 1.370 (4) |
| N6—C30 | 1.275 (3) | C25—H25 | 0.9300 |
| N6—O3 | 1.387 (2) | C20—H20 | 0.9300 |
| N1—C1 | 1.339 (3) | C29—C28 | 1.380 (4) |
| N1—C5 | 1.354 (3) | C23—C22 | 1.371 (4) |
| N3—C13 | 1.336 (3) | C23—H23 | 0.9300 |
| N3—C17 | 1.355 (3) | C32—C33 | 1.374 (4) |
| N5—C25 | 1.339 (3) | C32—H32 | 0.9300 |
| N5—C29 | 1.342 (3) | C16—C15 | 1.380 (4) |
| C27—C26 | 1.363 (5) | C16—H16 | 0.9300 |
| C27—C28 | 1.385 (4) | C33—C34 | 1.369 (5) |
| C27—H27 | 0.9300 | C33—H33 | 0.9300 |
| N4—C18 | 1.278 (3) | C11—C10 | 1.358 (4) |
| N4—O2 | 1.383 (2) | C11—C12 | 1.390 (4) |
| C7—C12 | 1.372 (3) | C11—H11 | 0.9300 |
| C7—C8 | 1.386 (3) | C12—H12 | 0.9300 |
| C7—C6 | 1.481 (3) | C2—C3 | 1.365 (4) |
| C18—C17 | 1.479 (3) | C2—H2 | 0.9300 |
| C18—C19 | 1.485 (3) | C22—H22 | 0.9300 |
| O1—H1A | 0.8200 | C8—C9 | 1.378 (4) |
| O2—H2A | 0.8200 | C8—H8 | 0.9300 |
| C19—C24 | 1.377 (3) | C9—C10 | 1.362 (4) |
| C19—C20 | 1.390 (3) | C9—H9 | 0.9300 |
| C5—C4 | 1.381 (3) | C3—H3 | 0.9300 |
| C5—C6 | 1.485 (3) | C15—H15 | 0.9300 |
| O3—H3A | 0.8200 | C35—C34 | 1.367 (5) |
| C17—C16 | 1.376 (3) | C35—C36 | 1.388 (4) |
| C13—C14 | 1.375 (4) | C35—H35 | 0.9300 |
| C13—H13 | 0.9300 | C26—H26 | 0.9300 |
| C31—C32 | 1.389 (4) | C10—H10 | 0.9300 |
| C31—C36 | 1.389 (4) | C36—H36 | 0.9300 |
| C31—C30 | 1.486 (3) | C34—H34 | 0.9300 |
| C21—C22 | 1.366 (4) | C28—H28 | 0.9300 |
| C21—C20 | 1.379 (4) | ||
| N2—Nd1—N1 | 60.38 (6) | C7—C6—C5 | 120.62 (19) |
| N2—Nd1—N5 | 146.60 (6) | C22—C21—C20 | 120.3 (3) |
| N1—Nd1—N5 | 140.98 (6) | C22—C21—H21 | 119.9 |
| N2—Nd1—N4 | 121.50 (6) | C20—C21—H21 | 119.9 |
| N1—Nd1—N4 | 72.25 (6) | N1—C1—C2 | 123.5 (2) |
| N5—Nd1—N4 | 68.74 (6) | N1—C1—H1 | 118.3 |
| N2—Nd1—N6 | 126.82 (6) | C2—C1—H1 | 118.3 |
| N1—Nd1—N6 | 139.68 (6) | C5—C4—C3 | 119.1 (2) |
| N5—Nd1—N6 | 59.14 (6) | C5—C4—H4 | 120.4 |
| N4—Nd1—N6 | 111.33 (6) | C3—C4—H4 | 120.4 |
| N2—Nd1—N3 | 137.44 (6) | N6—C30—C29 | 115.7 (2) |
| N1—Nd1—N3 | 83.49 (6) | N6—C30—C31 | 123.3 (2) |
| N5—Nd1—N3 | 75.89 (6) | C29—C30—C31 | 121.0 (2) |
| N4—Nd1—N3 | 58.57 (6) | C19—C24—C23 | 119.5 (3) |
| N6—Nd1—N3 | 67.56 (6) | C19—C24—H24 | 120.3 |
| N2—Nd1—Cl3 | 74.36 (5) | C23—C24—H24 | 120.3 |
| N1—Nd1—Cl3 | 134.58 (4) | C15—C14—C13 | 118.6 (2) |
| N5—Nd1—Cl3 | 78.41 (5) | C15—C14—H14 | 120.7 |
| N4—Nd1—Cl3 | 136.62 (5) | C13—C14—H14 | 120.7 |
| N6—Nd1—Cl3 | 71.65 (5) | N5—C25—C26 | 123.7 (3) |
| N3—Nd1—Cl3 | 138.76 (4) | N5—C25—H25 | 118.1 |
| N2—Nd1—Cl2 | 69.77 (5) | C26—C25—H25 | 118.1 |
| N1—Nd1—Cl2 | 77.37 (5) | C21—C20—C19 | 119.4 (3) |
| N5—Nd1—Cl2 | 130.24 (4) | C21—C20—H20 | 120.3 |
| N4—Nd1—Cl2 | 131.64 (4) | C19—C20—H20 | 120.3 |
| N6—Nd1—Cl2 | 71.34 (5) | N5—C29—C28 | 121.7 (2) |
| N3—Nd1—Cl2 | 81.64 (5) | N5—C29—C30 | 117.4 (2) |
| Cl3—Nd1—Cl2 | 91.19 (4) | C28—C29—C30 | 120.9 (2) |
| N2—Nd1—Cl1 | 70.22 (5) | C22—C23—C24 | 120.2 (3) |
| N1—Nd1—Cl1 | 81.67 (5) | C22—C23—H23 | 119.9 |
| N5—Nd1—Cl1 | 86.33 (5) | C24—C23—H23 | 119.9 |
| N4—Nd1—Cl1 | 70.85 (5) | C33—C32—C31 | 120.5 (3) |
| N6—Nd1—Cl1 | 138.42 (5) | C33—C32—H32 | 119.8 |
| N3—Nd1—Cl1 | 129.42 (4) | C31—C32—H32 | 119.8 |
| Cl3—Nd1—Cl1 | 79.77 (3) | C17—C16—C15 | 119.1 (2) |
| Cl2—Nd1—Cl1 | 139.95 (3) | C17—C16—H16 | 120.5 |
| C6—N2—O1 | 113.33 (18) | C15—C16—H16 | 120.5 |
| C6—N2—Nd1 | 126.43 (14) | C34—C33—C32 | 119.9 (3) |
| O1—N2—Nd1 | 120.13 (13) | C34—C33—H33 | 120.0 |
| C30—N6—O3 | 113.23 (18) | C32—C33—H33 | 120.0 |
| C30—N6—Nd1 | 125.00 (15) | C10—C11—C12 | 120.1 (3) |
| O3—N6—Nd1 | 121.56 (13) | C10—C11—H11 | 120.0 |
| C1—N1—C5 | 117.13 (19) | C12—C11—H11 | 120.0 |
| C1—N1—Nd1 | 122.39 (15) | C7—C12—C11 | 120.2 (2) |
| C5—N1—Nd1 | 120.07 (14) | C7—C12—H12 | 119.9 |
| C13—N3—C17 | 116.6 (2) | C11—C12—H12 | 119.9 |
| C13—N3—Nd1 | 121.58 (15) | C3—C2—C1 | 119.0 (2) |
| C17—N3—Nd1 | 119.27 (14) | C3—C2—H2 | 120.5 |
| C25—N5—C29 | 117.5 (2) | C1—C2—H2 | 120.5 |
| C25—N5—Nd1 | 120.04 (16) | C21—C22—C23 | 120.4 (2) |
| C29—N5—Nd1 | 122.42 (15) | C21—C22—H22 | 119.8 |
| C26—C27—C28 | 118.7 (3) | C23—C22—H22 | 119.8 |
| C26—C27—H27 | 120.6 | C9—C8—C7 | 120.3 (3) |
| C28—C27—H27 | 120.6 | C9—C8—H8 | 119.8 |
| C18—N4—O2 | 112.71 (17) | C7—C8—H8 | 119.8 |
| C18—N4—Nd1 | 125.20 (14) | C10—C9—C8 | 120.1 (3) |
| O2—N4—Nd1 | 122.00 (12) | C10—C9—H9 | 120.0 |
| C12—C7—C8 | 118.9 (2) | C8—C9—H9 | 120.0 |
| C12—C7—C6 | 120.9 (2) | C2—C3—C4 | 119.0 (2) |
| C8—C7—C6 | 120.1 (2) | C2—C3—H3 | 120.5 |
| N4—C18—C17 | 116.40 (18) | C4—C3—H3 | 120.5 |
| N4—C18—C19 | 122.86 (19) | C14—C15—C16 | 118.9 (2) |
| C17—C18—C19 | 120.71 (18) | C14—C15—H15 | 120.6 |
| N2—O1—H1A | 109.5 | C16—C15—H15 | 120.6 |
| N4—O2—H2A | 109.5 | C34—C35—C36 | 120.1 (3) |
| C24—C19—C20 | 120.1 (2) | C34—C35—H35 | 120.0 |
| C24—C19—C18 | 119.6 (2) | C36—C35—H35 | 120.0 |
| C20—C19—C18 | 120.2 (2) | C27—C26—C25 | 118.7 (3) |
| N1—C5—C4 | 122.2 (2) | C27—C26—H26 | 120.6 |
| N1—C5—C6 | 116.83 (19) | C25—C26—H26 | 120.6 |
| C4—C5—C6 | 120.9 (2) | C11—C10—C9 | 120.5 (3) |
| N6—O3—H3A | 109.5 | C11—C10—H10 | 119.8 |
| N3—C17—C16 | 122.7 (2) | C9—C10—H10 | 119.8 |
| N3—C17—C18 | 116.38 (19) | C35—C36—C31 | 119.7 (3) |
| C16—C17—C18 | 120.9 (2) | C35—C36—H36 | 120.2 |
| N3—C13—C14 | 124.0 (2) | C31—C36—H36 | 120.2 |
| N3—C13—H13 | 118.0 | C35—C34—C33 | 120.7 (3) |
| C14—C13—H13 | 118.0 | C35—C34—H34 | 119.7 |
| C32—C31—C36 | 119.1 (2) | C33—C34—H34 | 119.7 |
| C32—C31—C30 | 120.8 (2) | C29—C28—C27 | 119.5 (3) |
| C36—C31—C30 | 120.1 (3) | C29—C28—H28 | 120.2 |
| N2—C6—C7 | 124.1 (2) | C27—C28—H28 | 120.2 |
| N2—C6—C5 | 115.26 (19) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···Cl3 | 0.82 | 2.22 | 2.966 (2) | 152 |
| O2—H2A···Cl1 | 0.82 | 2.19 | 2.9290 (19) | 151 |
| O3—H3A···Cl2 | 0.82 | 2.19 | 2.930 (2) | 150 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZS2190).
References
- Bruker (2000). SADABS, SAINT and SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Fritsky, O., Swiatek-Kozlowska, J., Dobosz, A., Sliva, T. Y. & Dudarenko, N. M. (2004). Inorg. Chim. Acta, 357, 3746–3752.
- Kukushkin, Y. & Pombeiro, A. J. L. (1999). Coord. Chem. Rev. 181, 147–175.
- Lei, T., Chen, W., Chen, Y., Hu, B. & Li, Y. (2012). Acta Cryst. E68, m344–m345. [DOI] [PMC free article] [PubMed]
- Milios, C. J., Inglis, R., Vinslava, A., Bagai, R., Wernsdorfer, W., Parsons, S., Perlepes, S. P., Christou, G. & Brechin, E. K. (2007). J. Am. Chem. Soc. 129, 12505–12511. [DOI] [PubMed]
- Milios, C. J., Kefalloniti, E., Raptopoulou, C. P., Terzis, A., Vicente, R., Lalioti, N., Escuer, A. & Perlepes, S. P. (2003). Chem. Commun. pp. 819–821. [DOI] [PubMed]
- Milios, C. J., Stamatatos, T. C., Kyritsis, P., Terzis, A., Raptopoulou, C. P., Vicente, R., Escuer, A. & Perlepes, S. P. (2004). Eur. J. Inorg. Chem. pp. 2885–2901. [DOI] [PubMed]
- Papatriantafyllopoulou, C., Estrader, M., Efthymiou, C. G., Dermitzaki, D., Gkotsis, K., Terzis, A., Diaz, C. & Perlepes, S. P. (2009). Polyhedron, 28, 1652–1655.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xu, H. B., Wang, B. W., Pan, F., Wang, Z. M. & Gao, S. (2007). Angew. Chem. Int. Ed. 46, 7388–7392. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812014055/zs2190sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812014055/zs2190Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report